Integrated Chemical Characterization, Network Pharmacology and Transcriptomics to Explore the Mechanism of Sesquiterpenoids Isolated from Gynura divaricata (L.) DC. against Chronic Myelogenous Leukemia
Abstract
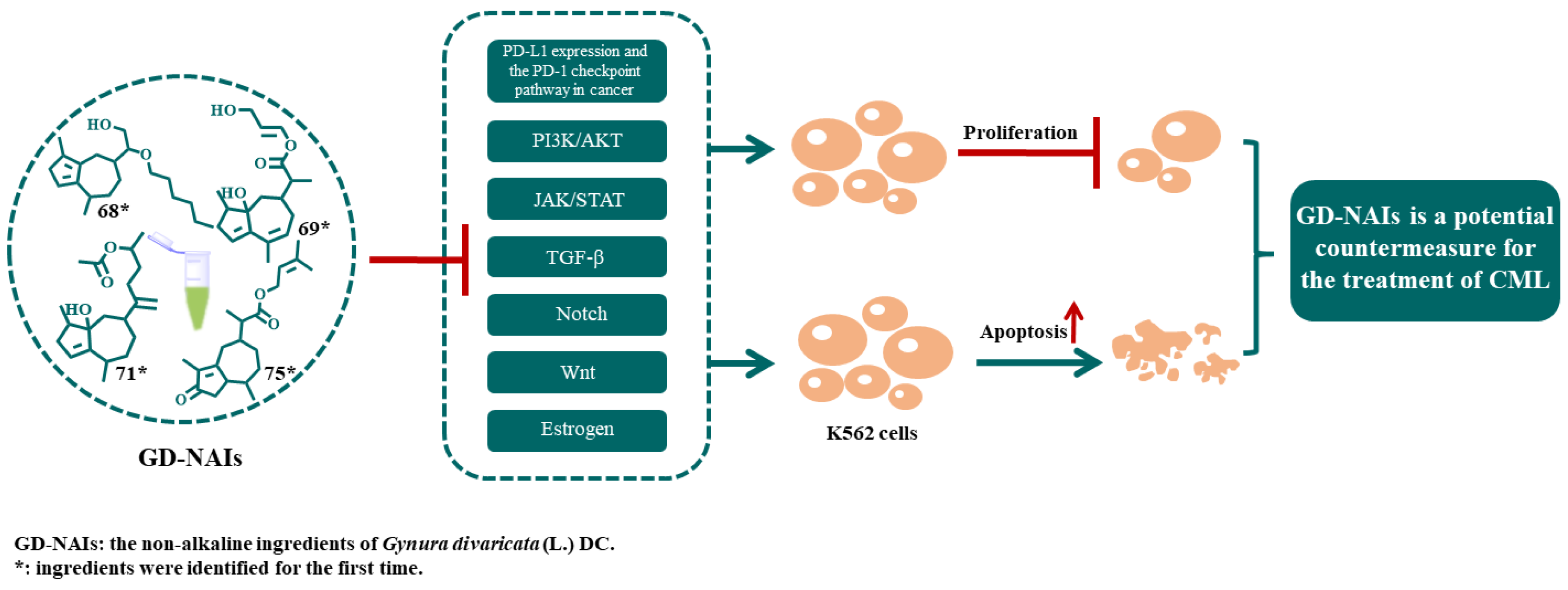
1. Introduction
2. Results
2.1. Characterization of GD-NAIs by UHPLC-HRMS-MN
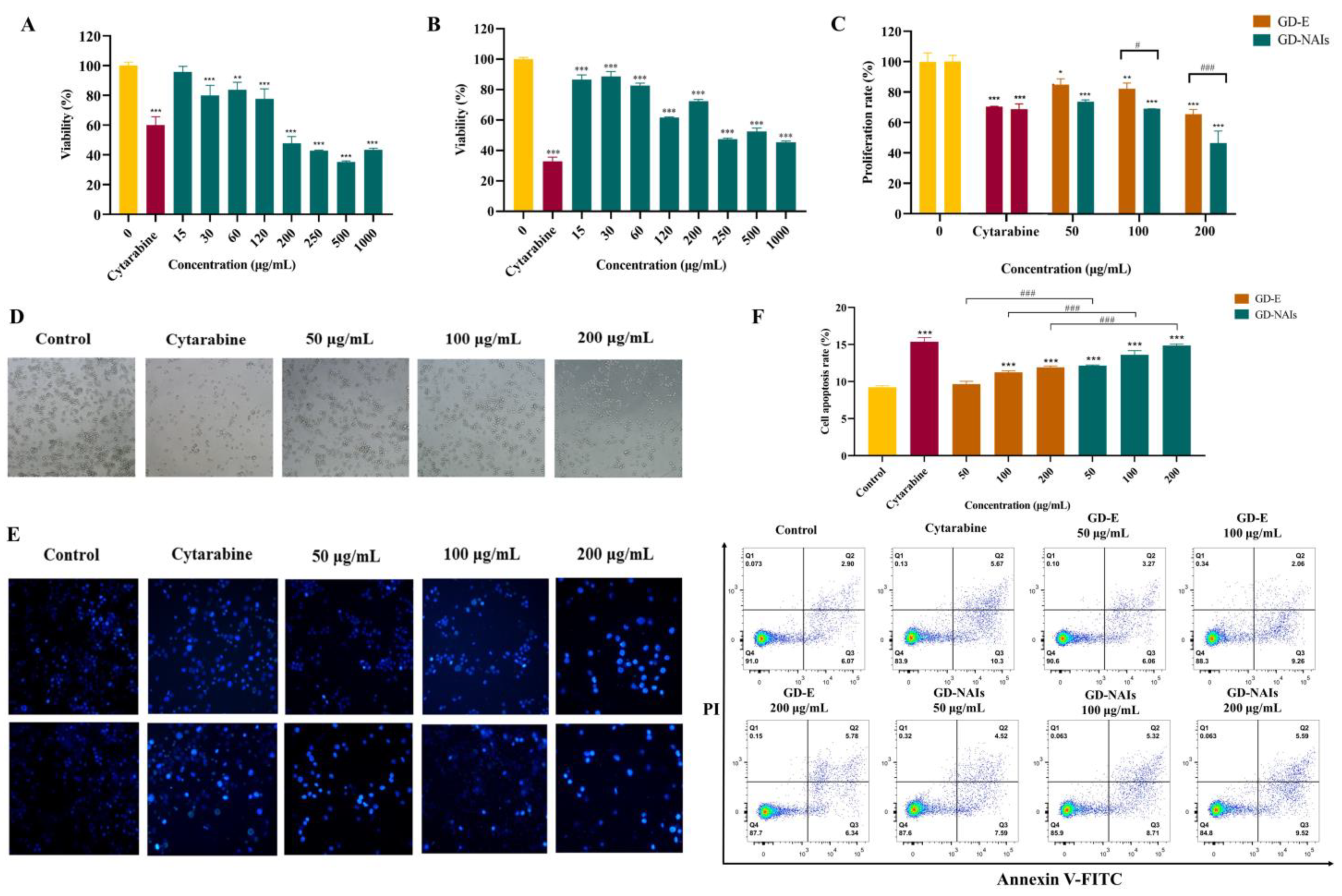
2.2. The Effect of GD-NAIs on Suppressing CML Cell Proliferation
2.3. The Effect of GD-NAIs on Inducing Cell Apoptosis
2.4. Network Pharmacology Analysis of GD-NAIs against CML
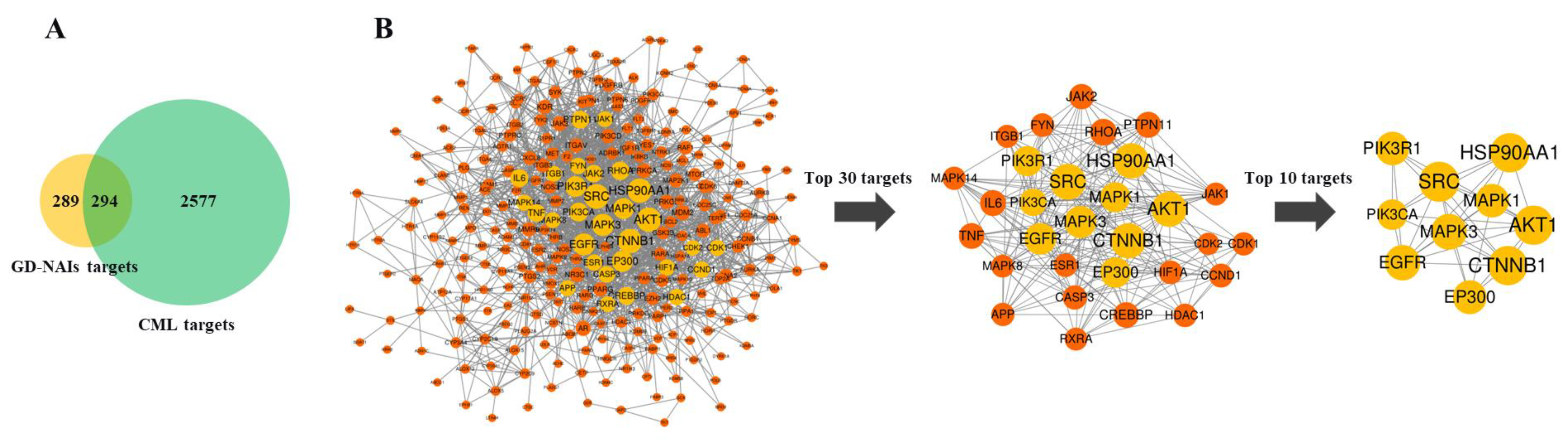
2.4.1. Target Prediction and Screening of GD-NAIs against CML
2.4.2. PPI Network of Targets of GD-NAIs against CML
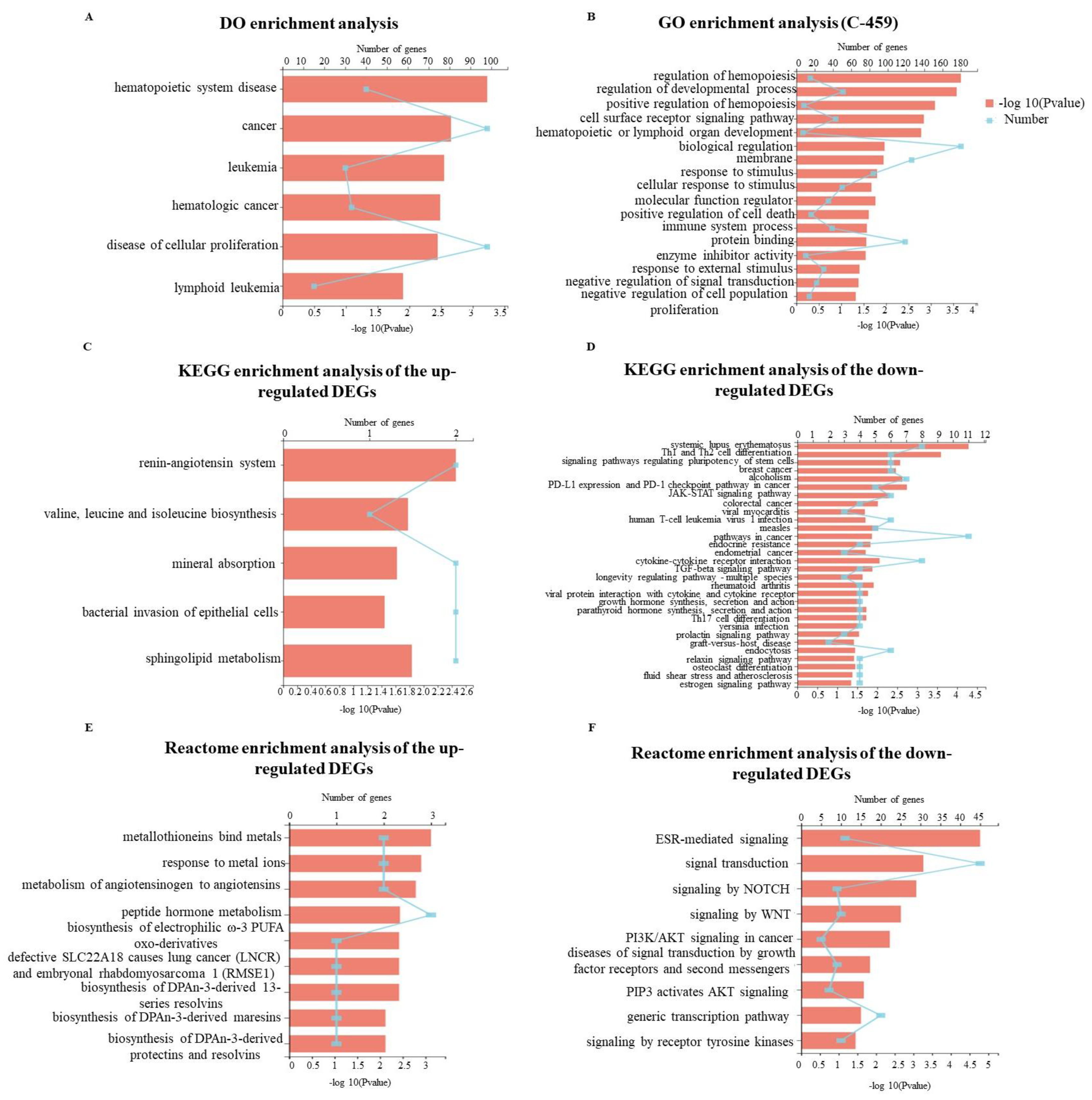
2.4.3. KEGG Pathway Enrichment Analysis of Targets of GD-NAIs against CML
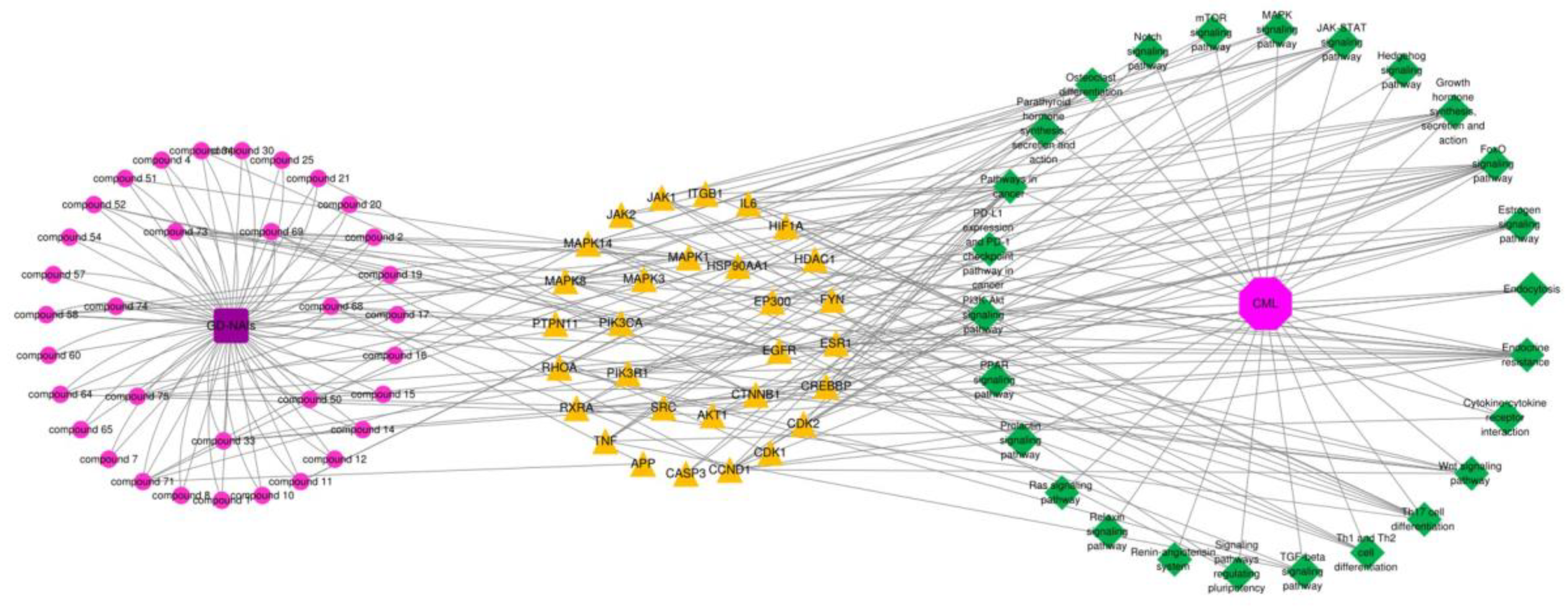
2.4.4. Compound-Target-Pathway Network and Analysis
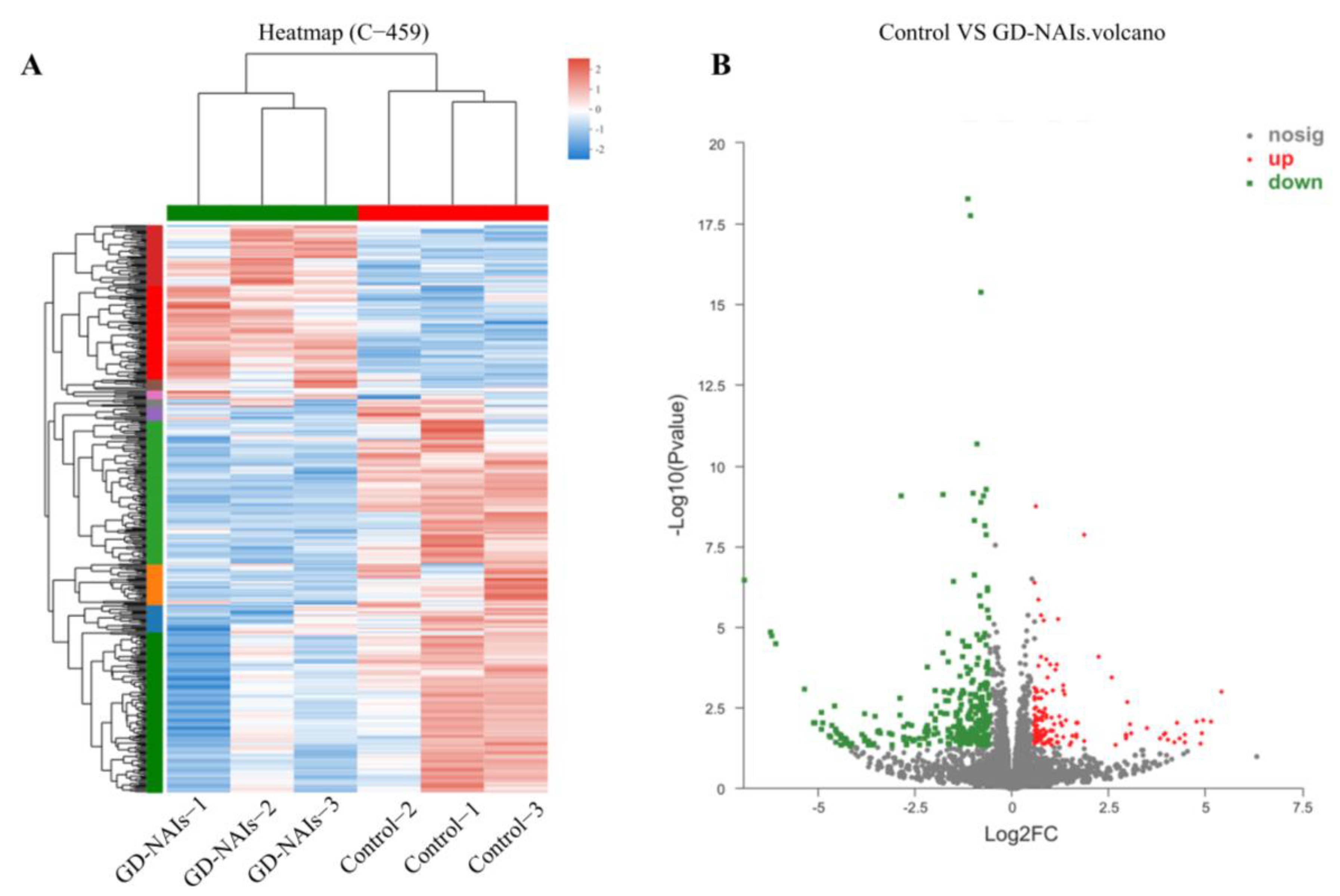
2.5. Gene Expression Profile Regulated by GD-NAIs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Chemicals and Materials
4.3. Preparation of GD-NAIs
4.4. UHPLC-HRMS Method, Data Acquisition and Preprocessing
4.5. Cell Viability Assay
4.6. Cell Proliferation Assay
4.7. Cell Apoptosis Assay
4.8. Network Pharmacology Analysis
4.8.1. Prediction of Targets of GD-NAIs
4.8.2. Collection of Predicted Targets for CML
4.8.3. Construction of Protein–Protein Interaction (PPI) Network
4.8.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
4.8.5. Construction of Compound–Target–Pathway Network
4.9. RNA-Seq and Data Analysis
4.10. Differential Expression Analysis and Functional Enrichment
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Abdulmawjood, B.; Costa, B.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic biomarkers in chronic myeloid leukemia: What have we learned so far? Int. J. Mol. Sci. 2021, 22, 15216. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, P.S.; Lemos, L.G.; de Moraes, G.N.; Maia, R.C. Unraveling survivin expression in chronic myeloid leukemia: Molecular interactions and clinical implications. Blood Rev. 2020, 43, 100671. [Google Scholar] [CrossRef] [PubMed]
- Ozgur Yurttas, N.; Eskazan, A.E. Novel therapeutic approaches in chronic myeloid leukemia. Leuk. Res. 2020, 91, 106337. [Google Scholar] [CrossRef]
- Al Baghdadi, T.; Abonour, R.; Boswell, H.S. Novel combination treatments targeting chronic myeloid leukemia stem cells. Clin. Lymph Myelom Leuk. 2012, 12, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Helgason, G.V.; Mukhopadhyay, A.; Karvela, M.; Salomoni, P.; Calabretta, B.; Holyoake, T.L. Autophagy in chronic myeloid leukaemia: Stem cell survival and implication in therapy. Curr. Cancer Drug Tar. 2013, 13, 724–734. [Google Scholar] [CrossRef]
- Mojtahedi, H.; Yazdanpanah, N.; Rezaei, N. Chronic myeloid leukemia stem cells: Targeting therapeutic implications. Stem Cell Res. Ther. 2021, 12, 603. [Google Scholar] [CrossRef]
- Soverini, S.; De Santis, S.; Monaldi, C.; Bruno, S.; Mancini, M. Targeting leukemic stem cells in chronic myeloid leukemia: Is it worth the effort? Int. J. Mol. Sci. 2021, 22, 7093. [Google Scholar] [CrossRef]
- More, M.P.; Pardeshi, S.R.; Pardeshi, C.V.; Sonawane, G.A.; Shinde, M.N.; Deshmukh, P.K.; Naik, J.B.; Kulkarni, A.D. Recent advances in phytochemical-based nano-formulation for drug-resistant cancer. Med. Drug Discov. 2021, 10, 100082. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, M.; Zhao, R.; Wang, D.; Ma, Y.; Li, A. Plant natural products: Promising resources for cancer chemoprevention. Molecules 2021, 26, 933. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, F.L.; Feng, T. Sesquiterpenoids specially produced by Fungi: Structures, biological activities, chemical and biosynthesis (2015–2020). J. Fungi 2021, 7, 1026. [Google Scholar] [CrossRef]
- Zhao, M.R.; Yang, C.Y.; Chai, S.Y.; Yuan, Y.J.; Zhang, J.; Cao, P.F.; Wang, Y.; Xiao, X.; Wu, K.; Yan, H.; et al. Curcumol and FTY720 synergistically induce apoptosis and differentiation in chronic myelomonocytic leukemia via multiple signaling pathways. Phytother. Res. 2021, 35, 2157–2170. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chong, L.; Li, X.C.; Khan, I.A.; Walker, L.A.; Khan, S.I. Selective inhibition of human leukemia cell growth and induction of cell cycle arrest and apoptosis by pseudolaric acid B. J. Cancer Res. Clin. 2010, 136, 1333–1340. [Google Scholar] [CrossRef]
- Lee, J.; Shen, P.Q.; Zhang, G.B.; Wu, X.H.; Zhang, X.G. Dihydroartemisinin inhibits the Bcr/Abl oncogene at the mRNA level in chronic myeloid leukemia sensitive or resistant to imatinib. Biomed. Pharmacother. 2013, 67, 157–163. [Google Scholar] [CrossRef] [PubMed]
- De Novais, L.M.; Ferreira, L.F.; de Sousa, P.T., Jr.; Ribeiro, T.A.; Jacinto, M.J.; Dos Santos, C.H.; de Carvalho, M.G.; Torquato, H.F.; Paredes-Gamero, E.J.; Silva, V.C. Eglerisine, a novel sesquiterpenoid tropolone from Dulacia egleri with antiproliferative Effect against an acute myeloi leukemia lineage. Planta Med. 2019, 86, 55–60. [Google Scholar] [CrossRef]
- Roeder, E.; Eckert, A.; Wiedenfeld, H. Pyrrolizidine alkaloids from Gynura divaricata. Planta Med. 1996, 62, 386. [Google Scholar] [CrossRef] [PubMed]
- Nadechanok, J.; Boonsom, L.; Saisunee, L.; John, K.; Stephen, G. The chemical constituents and biological activities of the essential oil and the extracts from Leaves of Gynura divaricata (L.) DC. growing in Thailand. J. Essent. Oil Bear. Plant 2015, 18, 543–555. [Google Scholar] [CrossRef]
- Wan, C.; Yu, Y.; Zhou, S.; Liu, W.; Tian, S.; Cao, S. Antioxidant activity and free radical-scavenging capacity of Gynura divaricata leaf extracts at different temperatures. Pharmacogn. Mag. 2011, 7, 40–45. [Google Scholar] [CrossRef]
- Chen, J.; Mangelinckx, S.; Ma, L.; Wang, Z.; Li, W.; De Kimpe, N. Caffeoylquinic acid derivatives isolated from the aerial parts of Gynura divaricata and their yeast alpha-glucosidase and PTP1B inhibitory activity. Fitoterapia 2014, 99, 1–6. [Google Scholar] [CrossRef]
- Dong, X.; Zhao, S.X.; Xu, B.Q.; Zhang, Y.Q. Gynura divaricata ameliorates hepatic insulin resistance by modulating insulin signalling, maintaining glycolipid homeostasis and reducing inflammation in type 2 diabetic mice. Toxicol. Res.-UK 2019, 8, 928–938. [Google Scholar] [CrossRef]
- Xu, B.Q.; Yang, P.; Zhang, Y.Q. Hypoglycemic activities of lyophilized powder of Gynura divaricata by improving antioxidant potential and insulin signaling in type 2 diabetic mice. Food Nutr. Res. 2015, 59, 29652. [Google Scholar] [CrossRef][Green Version]
- Xu, W.; Lu, Z.; Wang, X.; Cheung, M.H.; Lin, M.; Li, C.; Dong, Y.; Liang, C.; Chen, Y. Gynura divaricata exerts hypoglycemic effects by regulating the PI3K/AKT signaling pathway and fatty acid metabolism signaling pathway. Food Nutr. Res. 2020, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.L.; Xu, B.Q.; Zhang, Y.Q. Gynura divaricata rich in 3, 5-/4, 5-dicaffeoylquinic acid and chlorogenic acid reduces islet cell apoptosis and improves pancreatic function in type 2 diabetic mice. Nutr. Metab. 2018, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.H.; Jin, X.J.; Yoon, J.J.; Lee, Y.J.; Oh, H.C.; Lee, H.S.; Kim, H.Y.; Kang, D.G. Antihypertensive effects of Gynura divaricata (L.) DC in rats with renovascular hypertension. Nutrients 2020, 12, 3321. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Xiong, L.; Fu, Q.; Wang, B.; Wang, Y.; Zhang, K.; Yang, J.; Kantawong, F.; Kumsaiyai, W.; Zhou, J.; et al. Chemical characterization and DPP-IV inhibitory activity evaluation of tripeptides from Gynura divaricata (L.) DC. J. Ethnopharmacol. 2022, 292, 115203. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Wang, J.; Li, D.; Ni, W.; Qin, X.J.; Liu, H.; Yu, L.L.; Qiao, X.; Ji, Y.H.; He, L.; Nian, S.H.; et al. Molecular networking uncovers steroidal saponins of Paris tengchongensis. Fitoterapia 2020, 145, 104629. [Google Scholar] [CrossRef]
- Gu, D.Y.; Yang, Y.; Hang, B.; Lv, Q.Y.; Aisa, H.A. Characterization and identification of the chemical compositions in a traditional uighur medicine prescription yizhihao granule by LC-ESI-QTOF-MS. J. Liq. Chromatogr. Relat. Technol. 2014, 38, 229–242. [Google Scholar] [CrossRef]
- Ingert, N.; Bombarda, I.; Herbette, G.; Faure, R.; Moretti, C.; Raharivelomanana, P. Oleodaphnoic acid and coriaceol, two new natural products from the stem bark of Wikstroemia coriacea. Molecules 2013, 18, 2988–2996. [Google Scholar] [CrossRef]
- Banchelin, T.S.L.; Carret, S.; Giannini, A.; Deprés, J.P. Short and stereoselective total Synthesis of Δ-11,13-didehydroguaianes and-guaianolides: Synthesis of (±)-achalensolide and (±)-pechueloic acid; revision of the structure of (+)-rupestonic acid. Eur. J. Org. Chem. 2009, 2009, 3678–3682. [Google Scholar] [CrossRef]
- Coquerel, Y.; Greene, A.E.; Depre´s, J.P. New approach to bicyclo[5.3.0]decanes: Stereoselective guaiane synthesis. Org. Lett. 2003, 5, 4453–4455. [Google Scholar] [CrossRef]
- Cao, Y.; Zang, Y.; Huang, X.; Cheng, Z. Chemical constituents from Artemisia rupestris and their neuraminidase inhibitory activity. Nat. Prod. Res. 2021, 35, 1775–1782. [Google Scholar] [CrossRef]
- Perold, G.W. The structure of geigerin. J. Chem. Soc. 1957, 9, 47–51. [Google Scholar] [CrossRef]
- Barrero, A.F.; Sánchez, J.F.; Altarejos, J.; Zafra, M.J. Homoditerpenes from the essential oil of Tanacetum annuum. Phytochemistry 1992, 31, 1727–1730. [Google Scholar] [CrossRef]
- Caine, D.; Ingwalson, P.F. The influence of electron-withdrawing substituents on the photochemical behavior of bicyclic 6/6-fused cross-conjugated cyclohexadienones. J. Org. Chem. 1973, 38, 3751–3752. [Google Scholar] [CrossRef]
- Broka, C.A. Photorearrangements of carbomethoxy-substituted cyclohexadienones in neutral media. J. Org. Chem. 1988, 53, 575–583. [Google Scholar] [CrossRef]
- Yong, J.P.; Aisa, H.A. Chemical modification of rupestonic acid and preliminarily In vitro antiviral activity against influenza A3and B viruses. Bull. Korean Chem. Soc. 2011, 32, 1293–1297. [Google Scholar] [CrossRef][Green Version]
- Takeda, K.I.; Minato, H.; Hamamoto, K.; Horibe, I.; Nagasaki, T.; Ikuta, M. Studies on sesquiterpenoids. Part VII. Isolation and synthesis of ujacazulene. J. Chem. Soc. 1964, 692, 3577–3583. [Google Scholar] [CrossRef]
- Souza, A.D.L.; Rodrigues-Filho, E.; Souza, A.Q.L.; Henrique-Silva, F.; Pereira, J.O. A new guaiane mannoside from a Eutypa-like fungus isolated from Murraya paniculata in Brazil. J. Braz. Chem. Soc. 2008, 19, 1321–1325. [Google Scholar] [CrossRef]
- Edathara, P.M.; Gorre, M.; Kagita, S.; Vuree, S.; Cingeetham, A.; Nanchari, S.R.; Annamaneni, S.; Digumarthi, R.R.; Satti, V. Association of promoter polymorphisms of Fas-FasL genes with development of chronic myeloid leukemia. Tumor Biol. 2016, 37, 5475–5484. [Google Scholar] [CrossRef]
- Burin, S.M.; Cacemiro, M.D.; Cominal, J.G.; Grandis, R.A.; Machado, A.R.; Donaires, F.S.; Cintra, A.C.; Ambrosio, L.; Antunes, L.M.; Sampaio, S.V.; et al. Bothrops moojeni L-amino acid oxidase induces apoptosis and epigenetic modulation on Bcr-Abl(+) cells. J. Venom. Anim. Toxins 2020, 26, e20200123. [Google Scholar] [CrossRef]
- Triana, J.; Eiroa, J.L.; Morales, M.; Perez, F.J.; Brouard, I.; Quintana, J.; Ruiz-Estévez, M.; Estévez, F.; León, F. Sesquiterpenoids Isolated from two species of the Asteriscus Alliance. J. Nat. Prod. 2016, 79, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Yang, Y.; Ren, D.; Long, S.; Qin, X.; Liu, J.; Zeng, Y.; Lan, W.; Ma, W.; Liu, W. Hirsutanol A inhibits T-acute lymphocytic leukemia Jurkat cell viability through cell cycle arrest and p53-dependent induction of apoptosis. Exp. Ther. Med. 2021, 22, 741. [Google Scholar] [CrossRef]
- Masuda, Y.; Kadokura, T.; Ishii, M.; Takada, K.; Kitajima, J. Hinesol, a compound isolated from the essential oils of Atractylodes lancea rhizome, inhibits cell growth and induces apoptosis in human leukemia HL-60 cells. J. Nat. Med.-Tokyo 2015, 69, 332–339. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, M.; Nie, H.; Yuan, Y. PD-1 and PD-L1 in cancer immunotherapy: Clinical implications and future considerations. Hum. Vaccines Immunother. 2019, 15, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Basso, I.N.; Kim, D.D.H. Target spectrum of the BCR-ABL tyrosine kinase inhibitors in chronic myeloid leukemia. Int. J. Hematol. 2021, 113, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Bagg, A.; Pear, W.; Nowell, P.C.; Hess, J.L. Chronic myelogenous leukemia: Laboratory diagnosis and monitoring. Gene Chromosome Canc. 2001, 32, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Su, E.; Han, X.; Jiang, G. The transforming growth factor beta 1/SMAD signaling pathway involved in human chronic myeloid leukemia. Tumori 2010, 96, 659–666. [Google Scholar] [CrossRef]
- Muselli, F.; Peyron, J.F.; Mary, D. Druggable biochemical pathways and potential therapeutic alternatives to target leukemic stem cells and eliminate the residual disease in chronic myeloid leukemia. Int. J. Mol. Sci. 2019, 20, 5616. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Ji, C. The emerging roles of Notch signaling in leukemia and stem cells. Biomark. Res. 2013, 1, 23. [Google Scholar] [CrossRef]
- Pal, P.; Kanaujiya, J.K.; Lochab, S.; Tripathi, S.B.; Bhatt, M.L.; Singh, P.K.; Sanyal, S.; Trivedi, A.K. 2-D gel electrophoresis-based proteomic analysis reveals that ormeloxifen induces G0-G1 growth arrest and ERK-mediated apoptosis in chronic myeloid leukemia cells K562. Proteomics 2011, 11, 1517–1529. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Hamosh, A. Searching online, endelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinform. 2017, 58, 1.2.1–1.2.12. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef]
- Li, Y.H.; Yu, C.Y.; Li, X.X.; Zhang, P.; Tang, J.; Yang, Q.; Fu, T.; Zhang, X.; Cui, X.; Tu, G.; et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018, 46, D1121–D1127. [Google Scholar] [CrossRef]
- Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 8 May 2022).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease ontology: A backbone for disease semantic integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef]
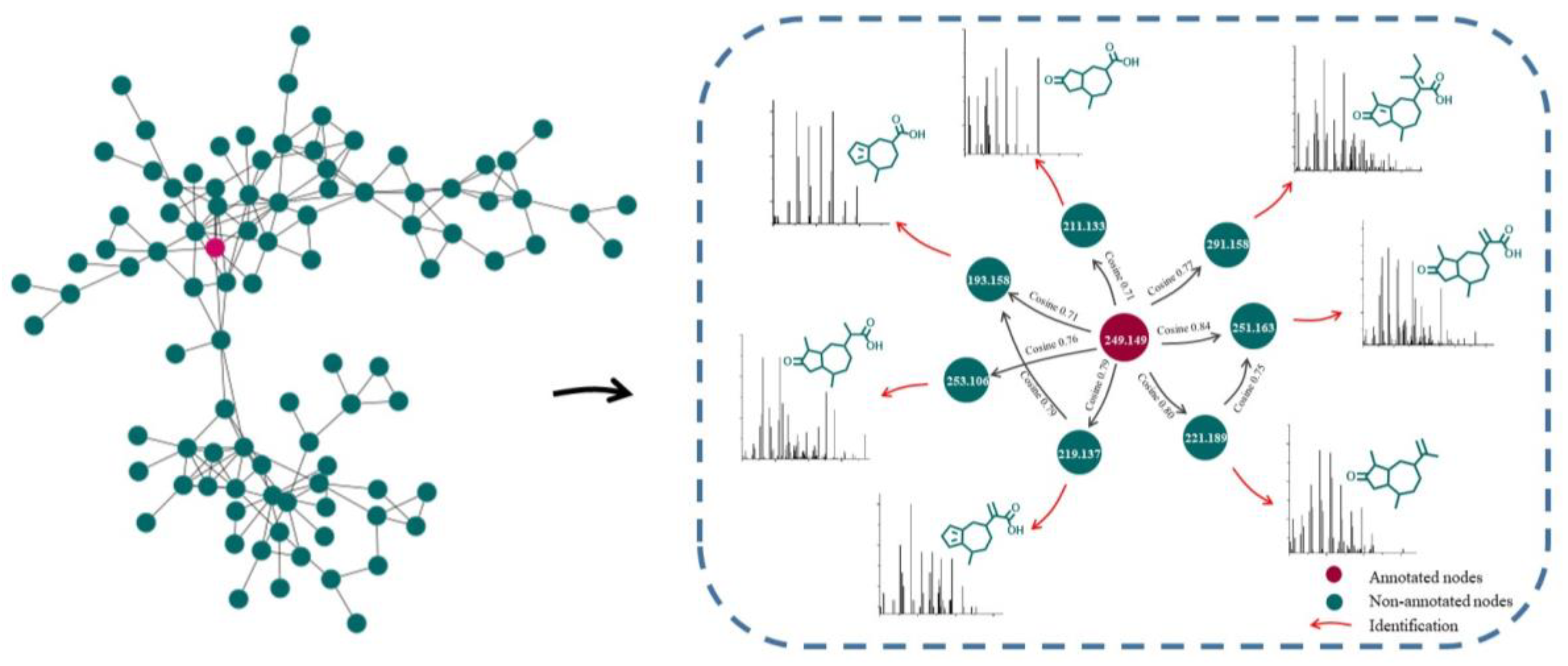
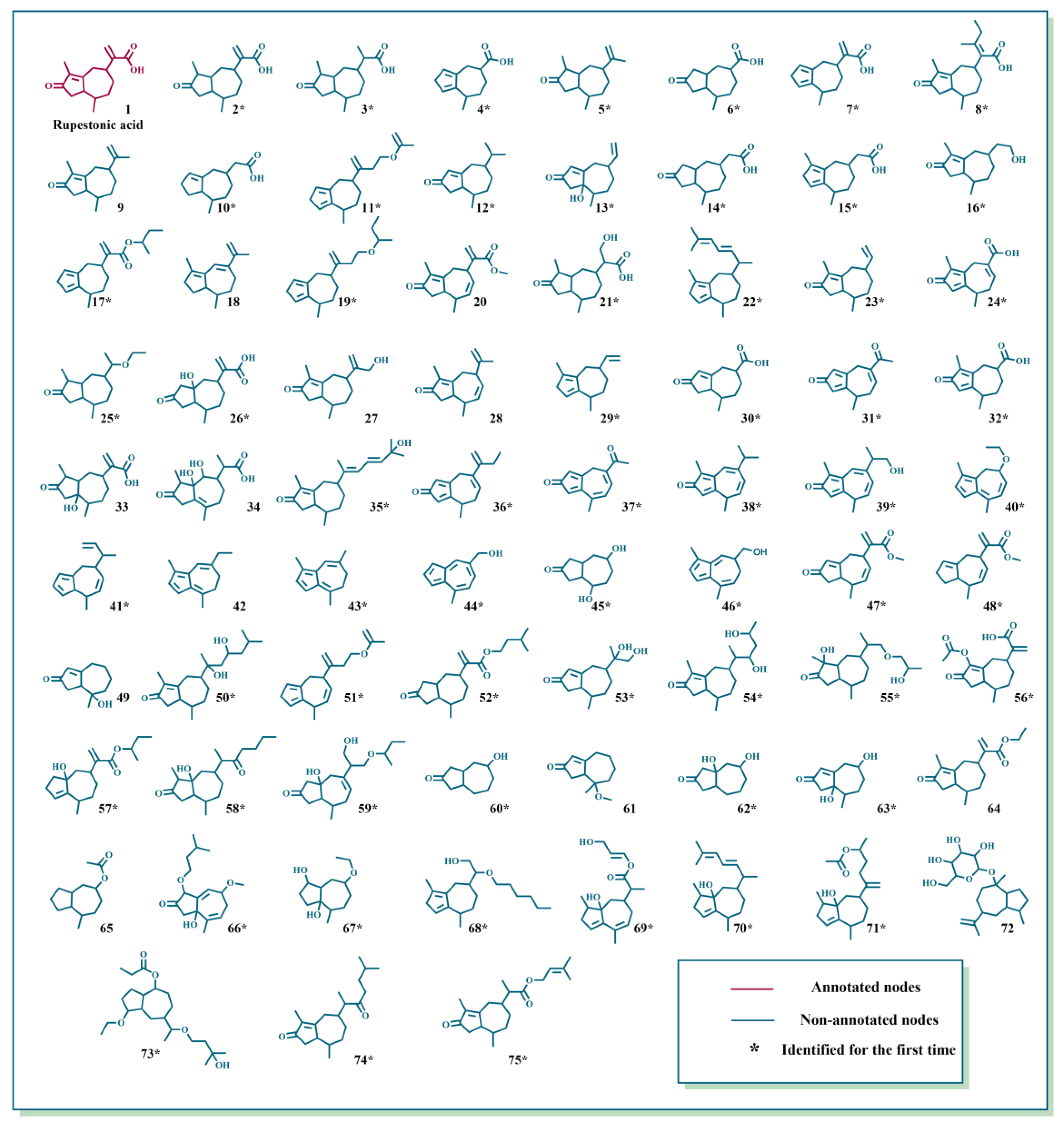

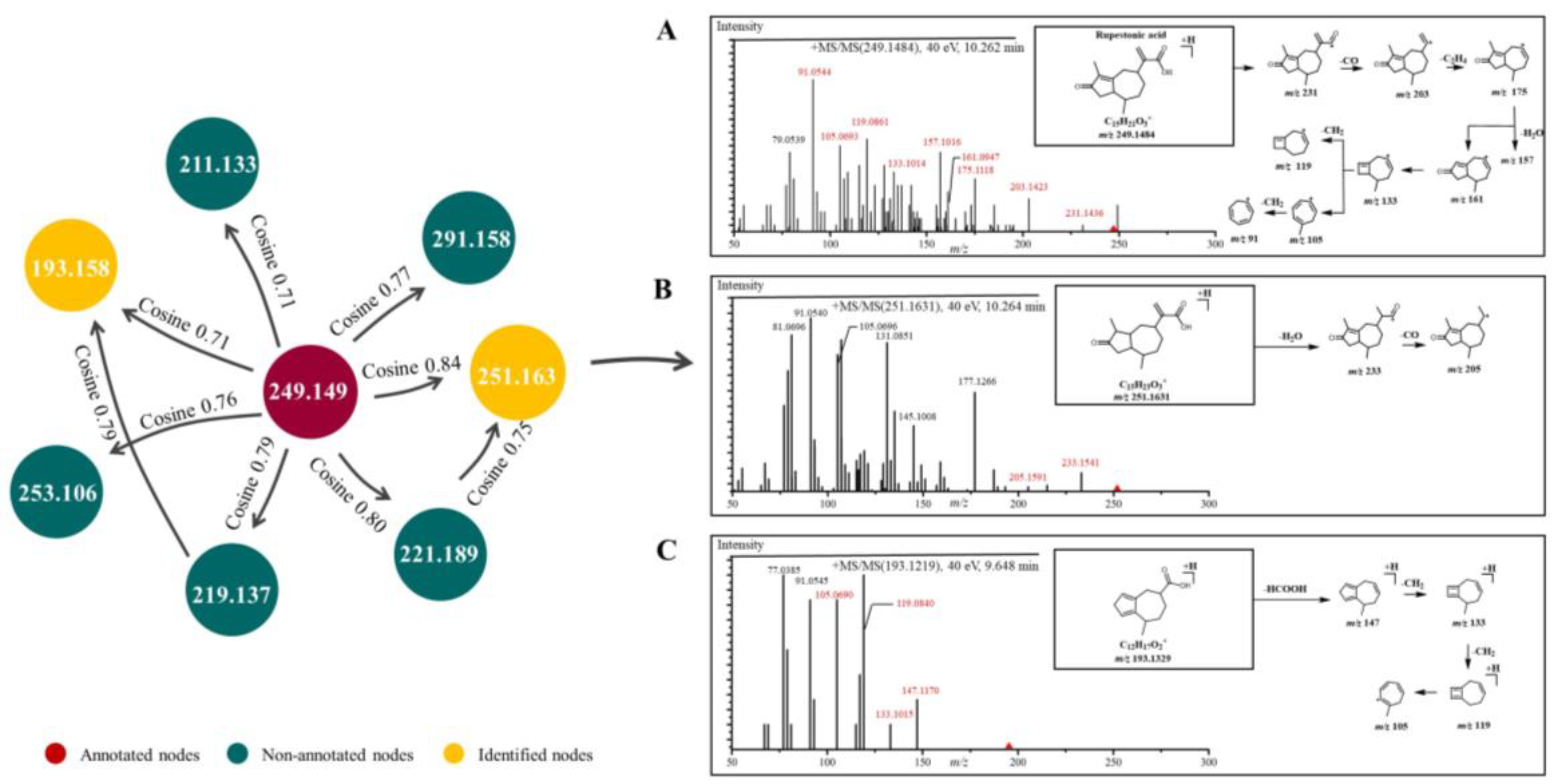
| Compound | Molecular Formula | Molecular Ion | Retention Time (min) | Error (ppm) | Fragment Ions (m/z) | References |
|---|---|---|---|---|---|---|
| 1 | C15H20O3 | 249.1484 | 10.262 | −0.5 | 231.1436, 203.1423, 175.1118, 161.0947, 157.1016, 133.1014, 119.0861, 105.0693, 91.0544, 81.0697, 79.0539 | [27] |
| 2 * | C15H22O3 | 251.1631 | 10.264 | −4.3 | 233.1541, 205.1591, 177.1266, 131.0851, 105.0696 | / |
| 3 * | C15H24O3 | 253.1788 | 8.585 | −4.0 | 217.1578, 177.1263, 133.1007, 119.0850, 105.0694 | / |
| 4 * | C12H16O2 | 193.1219 | 9.648 | −2.1 | 147.1170, 133.1015, 119.0840, 105.0690 | / |
| 5 * | C15H24O | 221.1898 | 12.214 | −0.9 | 203.1736, 161.1327, 147.1157, 119.0846, 105.0694 | / |
| 6 * | C12H18O3 | 211.1325 | 9.607 | −1.2 | 175.1098, 147.1166, 119.0850, 105.0688 | / |
| 7 * | C14H18O2 | 219.1373 | 10.458 | −3.0 | 145.1006, 131.0843, 119.0852, 105.0691, 91.0539, 77.0391 | / |
| 8 * | C18H26O3 | 291.1955 | 11.031 | 0.1 | 245.1901,147.0808, 133.1008, 119.0847, 105.0698, 69.0692 | / |
| 9 | C15H22O | 219.1739 | 11.779 | −3.4 | 161.0961, 105.0695 | [28] |
| 10 * | C13H20O2 | 209.1531 | 10.107 | −2.4 | 191.1505, 147.1169, 133.1007, 105.0695 | / |
| 11 * | C18H26O | 259.2059 | 14.156 | 1.0 | 199.1510, 173.1326, 131.0853, 105.0695, 91.0539 | / |
| 12 * | C14H22O | 207.1737 | 10.085 | −3.1 | 189.1274, 147.0812, 105.0697, 91.0542 | / |
| 13 * | C13H18O2 | 207.1378 | 9.958 | −0.8 | 189.1270, 105.0697, 91.0540 | / |
| 14 * | C13H20O3 | 225.1487 | 8.473 | 0.8 | 165.1263, 147.1164, 133.1000, 105.0692, 91.0539 | / |
| 15 * | C14H20O2 | 221.1532 | 12.627 | −1.8 | 161.1313. 147.1179, 133.1023, 119.0851, 105.0690 | / |
| 16 * | C14H22O2 | 223.1693 | 10.367 | 0.2 | 205.1580, 190.1361, 161.0962, 119.0846, 105.0706, 91.0535, 79.0557, 67.0544 | / |
| 17 * | C18H26O2 | 275.1996 | 12.681 | −3.5 | 173.1320, 159.1161, 131.0852, 105.0693, 91.0539 | / |
| 18 | C15H22 | 203.1791 | 9.823 | −1.6 | 161.1317, 145.0994, 133.1016, 119.0845, 105.0695 | / |
| 19 * | C19H30O | 275.2363 | 13.171 | −0.5 | 199.1488, 173.1340, 159.1173, 133.1012, 119.0854, 105.0693 | / |
| 20 | C16H20O3 | 261.1482 | 11.287 | −1.2 | 229.1236, 201.1268, 187.1121, 147.0811, 105.0698, 91.0541 | [29] |
| 21 * | C15H24O4 | 269.1742 | 9.915 | −2.0 | 233.1571, 215.1345, 119.0853, 105.0699 | / |
| 22 * | C20H30 | 271.2412 | 11.057 | −3.1 | 215.1827, 159.1165, 145.1002, 133.1016, 119.0852, 105.0690 | / |
| 23 * | C14H20O | 205.1582 | 15.026 | −2.4 | 190.1352, 175.1107, 105.0695, 91.0535 | / |
| 24 * | C13H14O3 | 219.1007 | 9.609 | −4.0 | 177.0539, 149.0589, 141.0700, 115.0532, 105.0699 | / |
| 25 * | C16H28O2 | 253.2153 | 9.538 | −3.6 | 189.1270, 177.1274, 159.1167, 145.1010, 133.1005, 105.0696 | / |
| 26 * | C14H20O4 | 253.1428 | 10.433 | −2.5 | 189.1267, 133.1010, 119.0848, 105.0696 | / |
| 27 | C15H22O2 | 235.1688 | 7.907 | −1.9 | 189.1260, 177.1272, 159.1157, 142.0774, 133.1009, 105.0695 | / |
| 28 | C15H20O | 217.1584 | 11.009 | −1.3 | 159.0803, 145.0647, 141.0701, 119.0853, 105.0699 | [30] |
| 29 * | C14H20 | 189.1643 | 11.308 | 2.8 | 145.1002, 129.0695, 119.0850, 105.0687 | / |
| 30 * | C12H16O3 | 209.1168 | 9.778 | −2.0 | 151.1111, 105.0687 | / |
| 31 * | C13H14O2 | 203.1063 | 13.237 | −1.8 | 161.0964, 147.0798, 105.0699 | / |
| 32 * | C13H16O3 | 221.1162 | 9.961 | −4.6 | 206.0918, 161.0944, 128.0612, 105.0691 | / |
| 33 | C15H22O4 | 267.1592 | 7.603 | 0.8 | 249.1479, 189.1270, 161.0957, 105.0689 | [31] |
| 34 | C15H22O5 | 283.1534 | 5.029 | −2.1 | 265.1420, 247.1323, 235.1325, 189.1267, 105.0693 | [32] |
| 35 * | C20H30O2 | 303.231 | 13.414 | −2.8 | 285.2205, 267.2112, 173.1315, 105.0696 | / |
| 36 * | C15H18O | 215.1426 | 10.391 | −2.1 | 185.0959, 167.0689, 145.0998, 129.0668, 115.0359, 105.0708 | / |
| 37 * | C13H12O2 | 201.0908 | 9.560 | −1.0 | 129.0694, 115.0545, 105.0702, 91.0535 | / |
| 38 * | C15H18O | 215.1427 | 11.434 | −1.6 | 199.1099, 185.0961, 115.0540, 105.0698 | / |
| 39 * | C15H18O2 | 231.1377 | 11.625 | −1.1 | 213.1222, 195.1155, 156.0938, 143.0857, 105.0698, 91.0540 | / |
| 40 * | C14H18O | 203.1427 | 10.986 | −1.7 | 174.0656, 157.1011, 141.0695, 128.0624, 115.0539, 105.0694 | / |
| 41 * | C15H20 | 201.1637 | 11.243 | −0.4 | 173.1302, 145.1007, 105.0693 | / |
| 42 | C14H18 | 187.1481 | 10.345 | −0.1 | 157.1007, 145.1008, 91.0545 | [33] |
| 43 * | C13H16 | 173.1317 | 9.893 | −0.1 | 157.0644, 143.0846, 115.0533, 105.0687 | / |
| 44 * | C12H12O | 173.0959 | 10.455 | −1.1 | 143.0856, 105.0697 | / |
| 45 * | C10H16O3 | 185.1174 | 10.304 | 1.0 | 91.0543, 79.0538 | / |
| 46 * | C13H16O | 189.1271 | 7.903 | −1.5 | 156.0941, 141.0696, 128.0615, 105.0713, 91.0543 | / |
| 47 * | C15H18O3 | 247.1326 | 9.406 | −1.1 | 187.1121, 145.1008, 128.0628, 105.0684 | / |
| 48 * | C15H20O2 | 233.1533 | 9.898 | −1.3 | 173.1322, 145.1009, 105.0705 | / |
| 49 | C11H16O2 | 181.1219 | 11.453 | −2.2 | 163.1114, 135.1163, 105.0702, 91.0540 | [34] |
| 50 * | C20H34O3 | 323.2573 | 13.420 | −2.4 | 287.2362, 269.2266, 177.1264, 145.1004 | / |
| 51 * | C18H24O | 257.1902 | 12.629 | 0.8 | 199.1520, 173.1322, 128.0623, 105.0702 | / |
| 52 * | C19H30O3 | 307.2268 | 10.024 | 0.1 | 179.0969, 165.1260, 119.0852, 105.0701 | / |
| 53 * | C14H22O3 | 239.1634 | 9.166 | −3.2 | 193.1672, 161.0953, 145.1013, 105.0699, 91.0532 | / |
| 54 * | C18H30O3 | 295.2260 | 10.115 | −2.6 | 259.2019, 179.1432, 151.1113, 105.0697 | / |
| 55 * | C18H32O4 | 313.2371 | 12.458 | −0.8 | 233.1538, 201.1292, 133.1018, 105.0702 | / |
| 56 * | C16H20O5 | 293.1380 | 9.655 | −1.2 | 257.1157, 173.1339, 149.0960, 145.0997, 105.0697 | / |
| 57 * | C18H28O3 | 293.2103 | 14.021 | −2.8 | 275.2009, 173.1320, 159.1171, 133.1008, 105.0699 | / |
| 58 * | C19H32O3 | 309.2414 | 10.632 | −3.3 | 291.2270, 205.1568, 191.1436, 161.0951, 119.0848, 105.0702 | / |
| 59 * | C18H30O4 | 311.2212 | 9.305 | −1.6 | 275.2006, 163.1111, 145.1008, 119.0858, 105.0695 | / |
| 60 * | C10H16O2 | 169.1223 | 8.886 | −3.0 | 151.1131, 123.1164, 91.0538 | / |
| 61 | C12H18O2 | 195.1375 | 13.301 | −2.3 | 131.0857, 105.0697 | [35] |
| 62 * | C10H16O3 | 185.1174 | 10.304 | 1.0 | 97.0642, 91.0543, 79.0538, 69.0695, 55.0538 | / |
| 63 * | C11H16O3 | 197.1109 | 8.471 | −1.6 | 161.0944, 133.1008, 105.0691, 91.0539 | / |
| 64 | C17H24O3 | 277.1790 | 14.188 | −3.0 | 203.1430, 189.1272, 175.1113, 135.1163, 105.0695 | [36] |
| 65 | C13H22O2 | 211.1687 | 9.454 | −2.6 | 133.1016, 105.0707 | [37] |
| 66 * | C17H26O4 | 295.1899 | 9.349 | −1.6 | 247.1677, 119.0847 | / |
| 67 * | C13H24O3 | 229.1798 | 9.432 | −0.1 | 149.1330, 131.1009, 121.1008, 105.0696, 91.0542 | / |
| 68 * | C20H34O2 | 307.2623 | 12.592 | −2.8 | 289.2502, 187.1477, 161.1326, 147.1170 | / |
| 69 * | C18H26O4 | 307.1897 | 13.979 | −2.2 | 207.1726, 161.1313, 149.1321, 105.0694 | / |
| 70 * | C20H32O | 289.2522 | 9.255 | −1.4 | 271.2408, 215.1786, 161.1322, 119.0848, 105.0692 | / |
| 71 * | C20H32O3 | 321.2420 | 11.191 | −1.3 | 303.2310, 243.2109, 203.1786, 189.1640, 175.1477, 147.1167, 119.0852 | / |
| 72 | C21H36O6 | 385.2579 | 15.588 | −1.5 | 321.2033, 293.1721, 221.1901, 135.1154, 105.0695 | [38] |
| 73 * | C22H40O5 | 385.2937 | 16.090 | −3.8 | 340.2603, 209.1631, 153.1260, 135.1168, 105.0702, 95.0858 | / |
| 74 * | C20H32O2 | 305.2469 | 9.411 | −2.0 | 206.1653, 191.1429, 173.1327, 133.1009, 107.0855 | / |
| 75 * | C20H30O3 | 319.2258 | 12.104 | −3.0 | 206.1662, 191.1428, 173.1327, 135.0799, 119.0850 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, X.; Wang, L.; Yang, X.; Yang, J.; Zhou, J.; Lan, C.; Kantawong, F.; Kumsaiyai, W.; Wu, J.; Zeng, J. Integrated Chemical Characterization, Network Pharmacology and Transcriptomics to Explore the Mechanism of Sesquiterpenoids Isolated from Gynura divaricata (L.) DC. against Chronic Myelogenous Leukemia. Pharmaceuticals 2022, 15, 1435. https://doi.org/10.3390/ph15111435
Ye X, Wang L, Yang X, Yang J, Zhou J, Lan C, Kantawong F, Kumsaiyai W, Wu J, Zeng J. Integrated Chemical Characterization, Network Pharmacology and Transcriptomics to Explore the Mechanism of Sesquiterpenoids Isolated from Gynura divaricata (L.) DC. against Chronic Myelogenous Leukemia. Pharmaceuticals. 2022; 15(11):1435. https://doi.org/10.3390/ph15111435
Chicago/Turabian StyleYe, Xinyuan, Long Wang, Xin Yang, Jie Yang, Jie Zhou, Cai Lan, Fahsai Kantawong, Warunee Kumsaiyai, Jianming Wu, and Jing Zeng. 2022. "Integrated Chemical Characterization, Network Pharmacology and Transcriptomics to Explore the Mechanism of Sesquiterpenoids Isolated from Gynura divaricata (L.) DC. against Chronic Myelogenous Leukemia" Pharmaceuticals 15, no. 11: 1435. https://doi.org/10.3390/ph15111435
APA StyleYe, X., Wang, L., Yang, X., Yang, J., Zhou, J., Lan, C., Kantawong, F., Kumsaiyai, W., Wu, J., & Zeng, J. (2022). Integrated Chemical Characterization, Network Pharmacology and Transcriptomics to Explore the Mechanism of Sesquiterpenoids Isolated from Gynura divaricata (L.) DC. against Chronic Myelogenous Leukemia. Pharmaceuticals, 15(11), 1435. https://doi.org/10.3390/ph15111435






