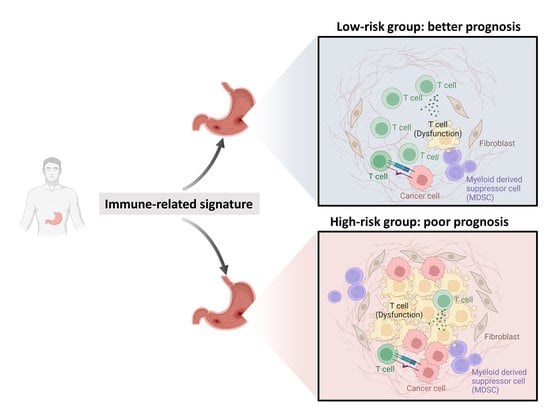
A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer
Abstract
1. Introduction
2. Results
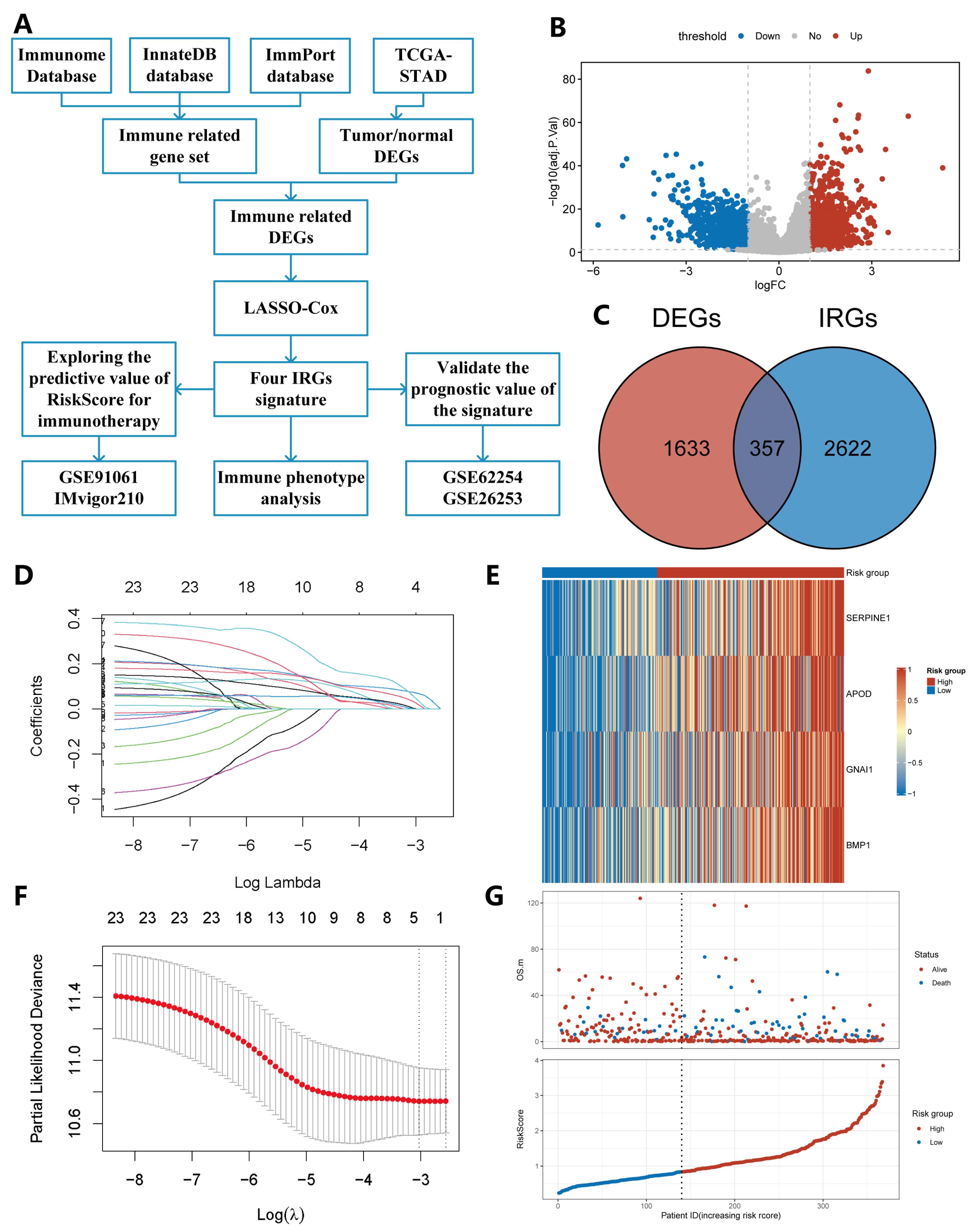
2.1. Construction of IRS
2.2. The Prognostic Value of the IRS
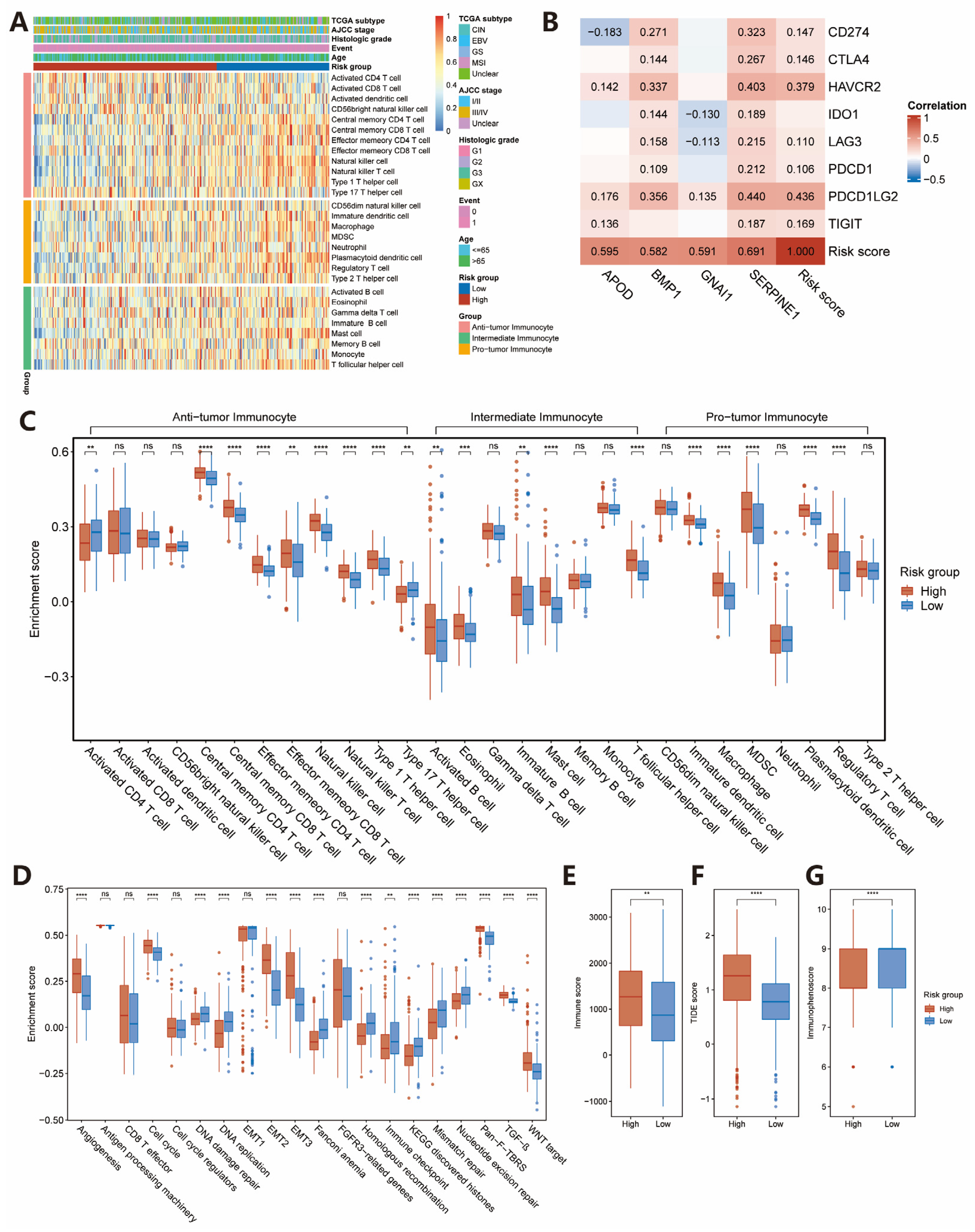
2.3. Estimation of TME Immune Infiltration and ICIs Response
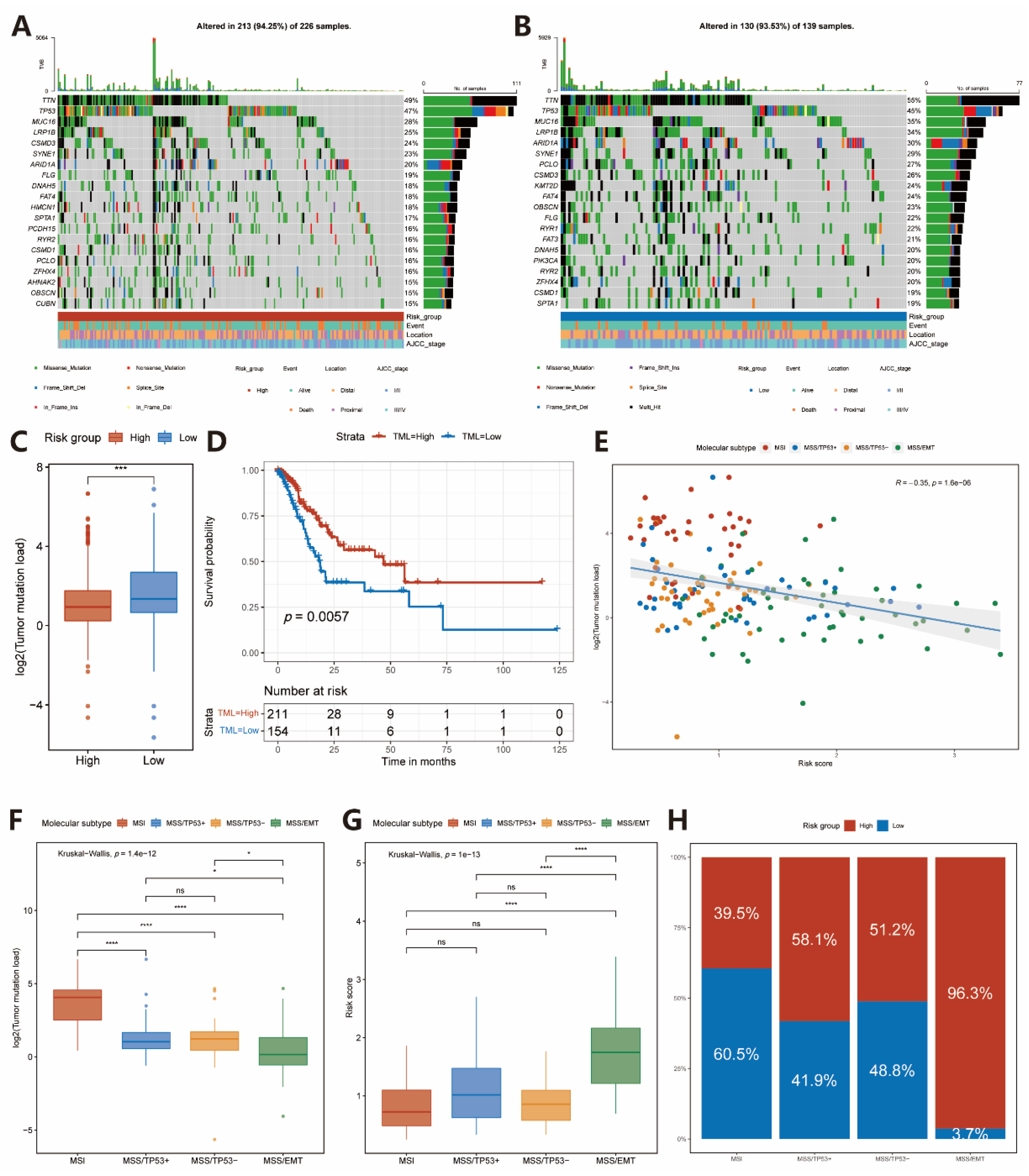
2.4. Somatic Variations in Two Subgroups
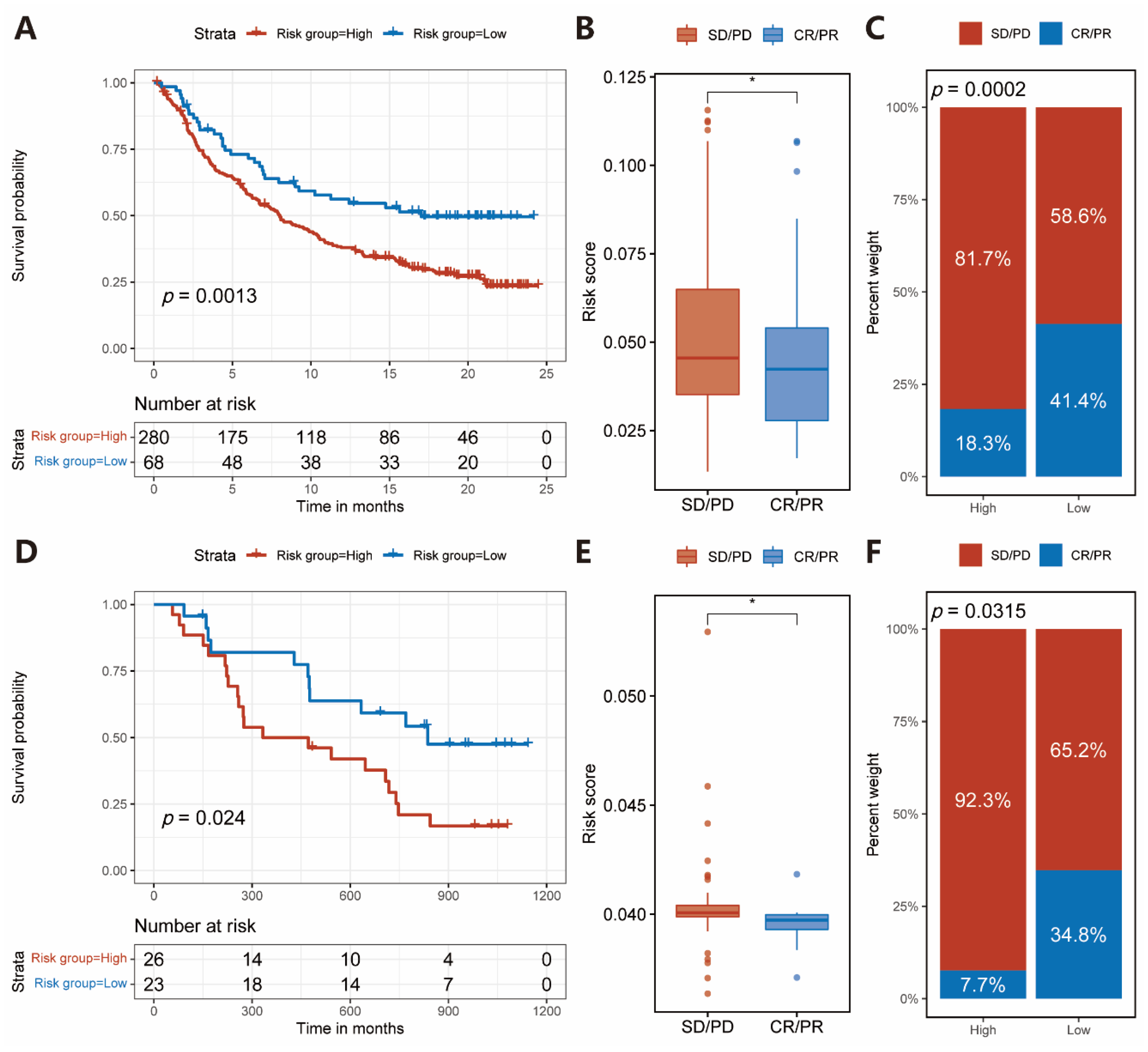
2.5. Immunotherapeutic Benefits Predicted by the IRS
2.6. Construction and Verification of Nomogram
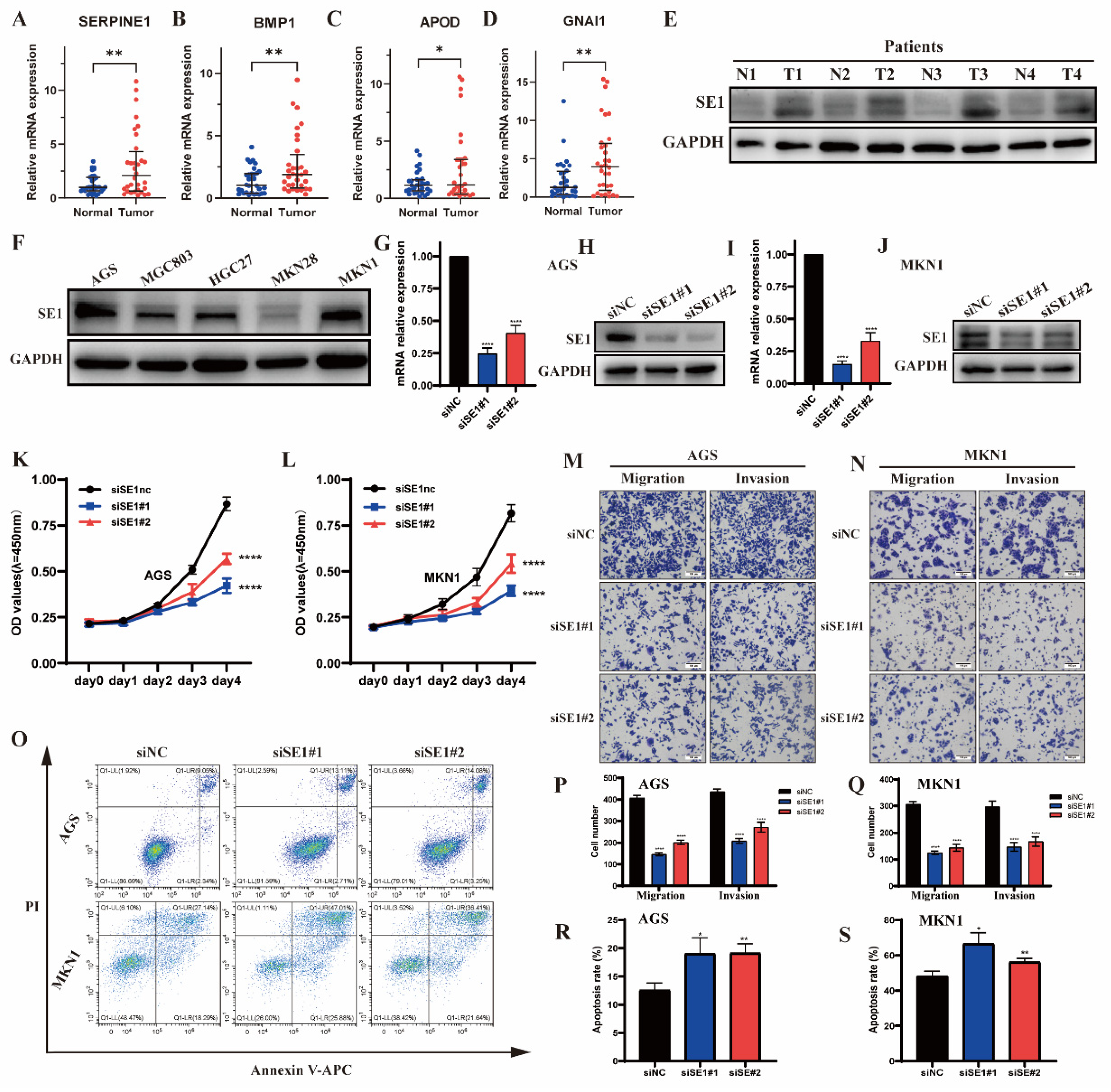
2.7. Validation of the IRGs in Clinical Tissue Samples
2.8. Silencing of SERPINE1 Inhibited the Proliferation, Invasion, Metastasis but Promoted the Apoptosis of GC Cells
3. Discussion
4. Materials and Methods
4.1. Study Design and Data Collection
4.2. Establishment and Validation of IRS
4.3. Estimation of TME Immune Infiltration and Functional Annotation
4.4. Quantification of the Immunotherapy Response
4.5. Correlation between the IRS and Somatic Variants
4.6. IRS Predicting ICIs Therapy Benefits
4.7. Construction of Nomogram
4.8. Cell Lines and Cell Culture
4.9. Small Interfering (si)RNA Transfection
4.10. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
4.11. Western Blot Analysis
4.12. Cell Proliferation, Migration, Invasion, and Cell Apoptosis Assays
4.13. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wilke, H.; Muro, K.; Van Cutsem, E.; Oh, S.-C.; Bodoky, G.; Shimada, Y.; Hironaka, S.; Sugimoto, N.; Lipatov, O.; Kim, T.-Y.; et al. Ramucirumab plus Paclitaxel versus Placebo plus Paclitaxel in Patients with Previously Treated Advanced Gastric or Gastro-Oesophageal Junction Adenocarcinoma (RAINBOW): A Double-Blind, Randomised Phase 3 Trial. Lancet Oncol. 2014, 15, 1224–1235. [Google Scholar] [CrossRef]
- Koizumi, W.; Narahara, H.; Hara, T.; Takagane, A.; Akiya, T.; Takagi, M.; Miyashita, K.; Nishizaki, T.; Kobayashi, O.; Takiyama, W.; et al. S-1 plus Cisplatin versus S-1 Alone for First-Line Treatment of Advanced Gastric Cancer (SPIRITS Trial): A Phase III Trial. Lancet Oncol. 2008, 9, 215–221. [Google Scholar] [CrossRef]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in Combination with Chemotherapy versus Chemotherapy Alone for Treatment of HER2-Positive Advanced Gastric or Gastro-Oesophageal Junction Cancer (ToGA): A Phase 3, Open-Label, Randomised Controlled Trial. Lancet Lond. Engl. 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Kawazoe, A.; Yañez, P.; Li, N.; Lonardi, S.; Kolesnik, O.; Barajas, O.; Bai, Y.; Shen, L.; Tang, Y.; et al. The KEYNOTE-811 Trial of Dual PD-1 and HER2 Blockade in HER2-Positive Gastric Cancer. Nature 2021, 600, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-K.; Boku, N.; Satoh, T.; Ryu, M.-H.; Chao, Y.; Kato, K.; Chung, H.C.; Chen, J.-S.; Muro, K.; Kang, W.K.; et al. Nivolumab in Patients with Advanced Gastric or Gastro-Oesophageal Junction Cancer Refractory to, or Intolerant of, at Least Two Previous Chemotherapy Regimens (ONO-4538-12, ATTRACTION-2): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Lond. Engl. 2017, 390, 2461–2471. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.-P.; et al. Safety and Efficacy of Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 Clinical KEYNOTE-059 Trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for Patients with PD-L1-Positive Advanced Gastric Cancer (KEYNOTE-012): A Multicentre, Open-Label, Phase 1b Trial. Lancet Oncol. 2016, 17, 717–726. [Google Scholar] [CrossRef]
- Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [CrossRef]
- AlMusawi, S.; Ahmed, M.; Nateri, A.S. Understanding Cell-Cell Communication and Signaling in the Colorectal Cancer Microenvironment. Clin. Transl. Med. 2021, 11, e308. [Google Scholar] [CrossRef]
- Choi, H.; Na, K.J. Integrative Analysis of Imaging and Transcriptomic Data of the Immune Landscape Associated with Tumor Metabolism in Lung Adenocarcinoma: Clinical and Prognostic Implications. Theranostics 2018, 8, 1956–1965. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T Cell Dysfunction and Exclusion Predict Cancer Immunotherapy Response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling Pathways in Cancer-Associated Fibroblasts and Targeted Therapy for Cancer. Signal Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef]
- Chen, X.; Song, E. Turning Foes to Friends: Targeting Cancer-Associated Fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Enomoto, A.; Woods, S.L.; Burt, A.D.; Takahashi, M.; Worthley, D.L. Cancer-Associated Fibroblasts in Gastrointestinal Cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 282–295. [Google Scholar] [CrossRef]
- Wu, F.; Li, G.-Z.; Liu, H.-J.; Zhao, Z.; Chai, R.-C.; Liu, Y.-Q.; Jiang, H.-Y.; Zhai, Y.; Feng, Y.-M.; Li, R.-P.; et al. Molecular Subtyping Reveals Immune Alterations in IDH Wild-Type Lower-Grade Diffuse Glioma. J. Pathol. 2020, 251, 272–283. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and Clinical Activity of Pembrolizumab for Treatment of Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck (KEYNOTE-012): An Open-Label, Multicentre, Phase 1b Trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-Tumor Genomic Biomarkers for PD-1 Checkpoint Blockade-Based Immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The History and Advances in Cancer Immunotherapy: Understanding the Characteristics of Tumor-Infiltrating Immune Cells and Their Therapeutic Implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Bejarano, L.; Jordāo, M.J.C.; Joyce, J.A. Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov. 2021, 11, 933–959. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.R.; Yang, M.H.; Lim, Y.J.; Lee, J.H.; Chang, D.K.; Kim, Y.-H.; Son, H.J.; Kim, J.J.; Rhee, J.C.; Kim, J.Y. Association between Plasma Levels of Plasminogen Activator Inhibitor-1 and Colorectal Neoplasms. Gut Liver 2013, 7, 519–523. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Teng, F.; Zhang, J.; Chen, Y.; Shen, X.; Su, C.; Guo, Y.; Wang, P.; Shi, C.; Lei, M.; Cao, Y.; et al. LncRNA NKX2-1-AS1 Promotes Tumor Progression and Angiogenesis via Upregulation of SERPINE1 Expression and Activation of the VEGFR-2 Signaling Pathway in Gastric Cancer. Mol. Oncol. 2021, 15, 1234–1255. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.D.; Cheng, H.; Martin, S.E.; Si, H.; Ormanoglu, P.; Carlson, S.; Clavijo, P.E.; Yang, X.; Das, R.; Cornelius, S.; et al. Integrated Genomic and Functional MicroRNA Analysis Identifies MiR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.C.Y.; Lawrie, A.; Demaries, S.; Massaeli, H.; Burry, A.; Yablonsky, S.; Sarjeant, J.M.; Fera, E.; Rassart, E.; Pickering, J.G.; et al. Apolipoprotein D and Platelet-Derived Growth Factor-BB Synergism Mediates Vascular Smooth Muscle Cell Migration. Circ. Res. 2004, 95, 179–186. [Google Scholar] [CrossRef]
- Bajo-Grañeras, R.; Crespo-Sanjuan, J.; García-Centeno, R.M.; Garrote-Adrados, J.A.; Gutierrez, G.; García-Tejeiro, M.; Aguirre-Gervás, B.; Calvo-Nieves, M.D.; Bustamante, R.; Ganfornina, M.D.; et al. Expression and Potential Role of Apolipoprotein D on the Death–Survival Balance of Human Colorectal Cancer Cells under Oxidative Stress Conditions. Int. J. Colorectal Dis. 2013, 28, 751–766. [Google Scholar] [CrossRef]
- Søiland, H.; Skaland, I.; Varhaug, J.E.; Kørner, H.; Janssen, E.A.M.; Gudlaugsson, E.; Baak, J.P.A.; Søreide, J.A. Co-Expression of Estrogen Receptor α and Apolipoprotein D in Node Positive Operable Breast Cancer–Possible Relevance for Survival and Effects of Adjuvant Tamoxifen in Postmenopausal Patients. Acta Oncol. 2009, 48, 514–521. [Google Scholar] [CrossRef]
- Garrigue-Antar, L.; Hartigan, N.; Kadler, K.E. Post-Translational Modification of Bone Morphogenetic Protein-1 Is Required for Secretion and Stability of the Protein. J. Biol. Chem. 2002, 277, 43327–43334. [Google Scholar] [CrossRef]
- Ge, G.; Greenspan, D.S. BMP1 Controls TGFbeta1 Activation via Cleavage of Latent TGFbeta-Binding Protein. J. Cell Biol. 2006, 175, 111–120. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, X.; Wang, T.; Xing, J. Overexpression of BMP1 Reflects Poor Prognosis in Clear Cell Renal Cell Carcinoma. Cancer Gene Ther. 2020, 27, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-Y.; Tung, S.-Y.; Pan, H.-Y.; Yen, C.-W.; Xu, H.-W.; Deng, Y.-F.; Lin, Y.-J.; Hsu, W.-T.; Wu, C.-S.; Li, C. Upregulation of Bone Morphogenetic Protein 1 Is Associated with Poor Prognosis of Late-Stage Gastric Cancer Patients. BMC Cancer 2018, 18, 508. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, B.; Yi, M.; Qiu, H.; Yuan, X. A Prognostic Nomogram Model Based on MRNA Expression of DNA Methylation-Driven Genes for Gastric Cancer. Front. Oncol. 2020, 10, 584733. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.-Y.; Dong, Z.-N.; Fu, X.-T.; Lin, L.-Y.; Wang, L.; Ye, G.-D.; Luo, Q.-C.; Chen, Y.-C. Identification of a Tumor Microenvironment-Relevant Gene Set-Based Prognostic Signature and Related Therapy Targets in Gastric Cancer. Theranostics 2020, 10, 8633–8647. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Yang, Z.; Fu, Y.; Zheng, L.; Liu, F.; Chai, L.; Jia, J. The Pyroptosis-Related Signature Predicts Prognosis and Indicates Immune Microenvironment Infiltration in Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 676485. [Google Scholar] [CrossRef]
- Qiu, J.; Sun, M.; Wang, Y.; Chen, B. Identification and Validation of an Individualized Autophagy-Clinical Prognostic Index in Gastric Cancer Patients. Cancer Cell Int. 2020, 20, 178. [Google Scholar] [CrossRef]
- Verdeil, G.; Fuertes Marraco, S.A.; Murray, T.; Speiser, D.E. From T Cell “Exhaustion” to Anti-Cancer Immunity. Biochim. Biophys. Acta 2016, 1865, 49–57. [Google Scholar] [CrossRef]
- Wherry, E.J.; Kurachi, M. Molecular and Cellular Insights into T Cell Exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ Attenuates Tumour Response to PD-L1 Blockade by Contributing to Exclusion of T Cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFβ Drives Immune Evasion in Genetically Reconstituted Colon Cancer Metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef]
- Holmgaard, R.B.; Schaer, D.A.; Li, Y.; Castaneda, S.P.; Murphy, M.Y.; Xu, X.; Inigo, I.; Dobkin, J.; Manro, J.R.; Iversen, P.W.; et al. Targeting the TGFβ Pathway with Galunisertib, a TGFβRI Small Molecule Inhibitor, Promotes Anti-Tumor Immunity Leading to Durable, Complete Responses, as Monotherapy and in Combination with Checkpoint Blockade. J. Immunother. Cancer 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- de Streel, G.; Bertrand, C.; Chalon, N.; Liénart, S.; Bricard, O.; Lecomte, S.; Devreux, J.; Gaignage, M.; De Boeck, G.; Mariën, L.; et al. Selective Inhibition of TGF-Β1 Produced by GARP-Expressing Tregs Overcomes Resistance to PD-1/PD-L1 Blockade in Cancer. Nat. Commun. 2020, 11, 4545. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.A.; Riskin, J.; Hempel, H.A.; Fousek, K.; Lind, H.; Hamilton, D.H.; McCampbell, K.K.; Maeda, D.Y.; Zebala, J.A.; Su, Z.; et al. Simultaneous Inhibition of CXCR1/2, TGF-β, and PD-L1 Remodels the Tumor and Its Microenvironment to Drive Antitumor Immunity. J. Immunother. Cancer 2020, 8, e000326. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, L.; Jiang, X.; Li, Q. Mismatch Repair Deficiency/Microsatellite Instability-High as a Predictor for Anti-PD-1/PD-L1 Immunotherapy Efficacy. J. Hematol. Oncol.J Hematol Oncol 2019, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.C.; Sohn, B.H.; Cheong, J.-H.; Kim, S.-B.; Lee, J.E.; Park, K.C.; Lee, S.H.; Park, J.-L.; Park, Y.-Y.; Lee, H.-S.; et al. Clinical and Genomic Landscape of Gastric Cancer with a Mesenchymal Phenotype. Nat. Commun. 2018, 9, 1777. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture from Expression Data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer Immunology. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non-Small Cell Lung Cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhang, L.; Qian, Y.; Fang, Q.; Xiao, Y.; Chen, G.; Cai, G.; Abula, A.; Wang, Z.; Zhai, E.; et al. A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer. Pharmaceuticals 2022, 15, 1401. https://doi.org/10.3390/ph15111401
Xu X, Zhang L, Qian Y, Fang Q, Xiao Y, Chen G, Cai G, Abula A, Wang Z, Zhai E, et al. A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer. Pharmaceuticals. 2022; 15(11):1401. https://doi.org/10.3390/ph15111401
Chicago/Turabian StyleXu, Xiang, Lipeng Zhang, Yan Qian, Qian Fang, Yongbiao Xiao, Guizeng Chen, Guojing Cai, Alimujiang Abula, Zhao Wang, Ertao Zhai, and et al. 2022. "A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer" Pharmaceuticals 15, no. 11: 1401. https://doi.org/10.3390/ph15111401
APA StyleXu, X., Zhang, L., Qian, Y., Fang, Q., Xiao, Y., Chen, G., Cai, G., Abula, A., Wang, Z., Zhai, E., Chen, J., Cai, S., & Wu, H. (2022). A SERPINE1-Based Immune Gene Signature Predicts Prognosis and Immunotherapy Response in Gastric Cancer. Pharmaceuticals, 15(11), 1401. https://doi.org/10.3390/ph15111401