Green Biosynthesis of Silver Nanoparticles Using Annona glabra and Annona squamosa Extracts with Antimicrobial, Anticancer, Apoptosis Potentials, Assisted by In Silico Modeling, and Metabolic Profiling
Abstract
1. Introduction
2. Results
2.1. UPLC-QTOF-MS/MS Metabolic Profiling of the Fruits
2.1.1. Acetogenins
2.1.2. Diterpenes
2.2. Characterization of Silver Nanoparticles
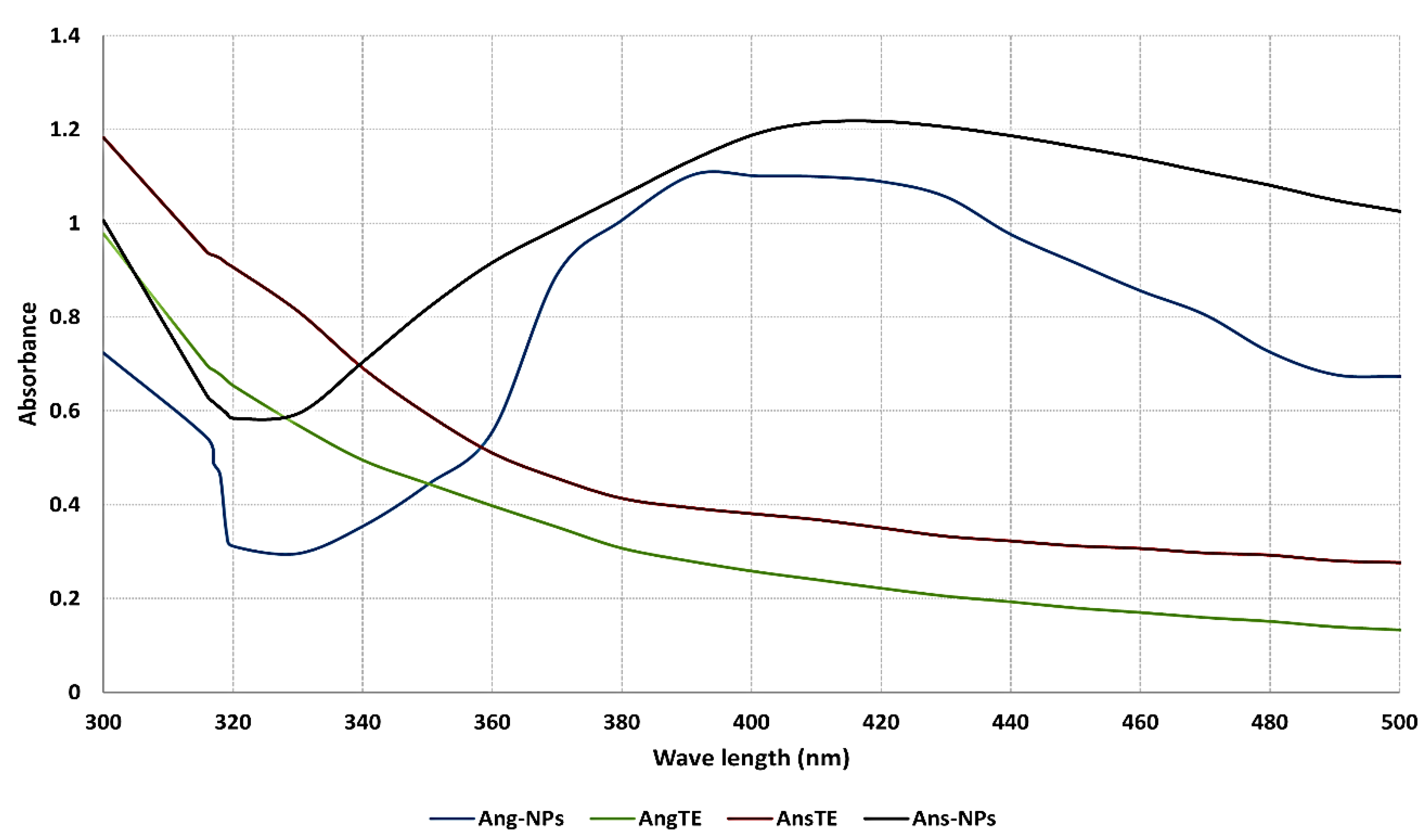
2.2.1. UV Spectroscopy
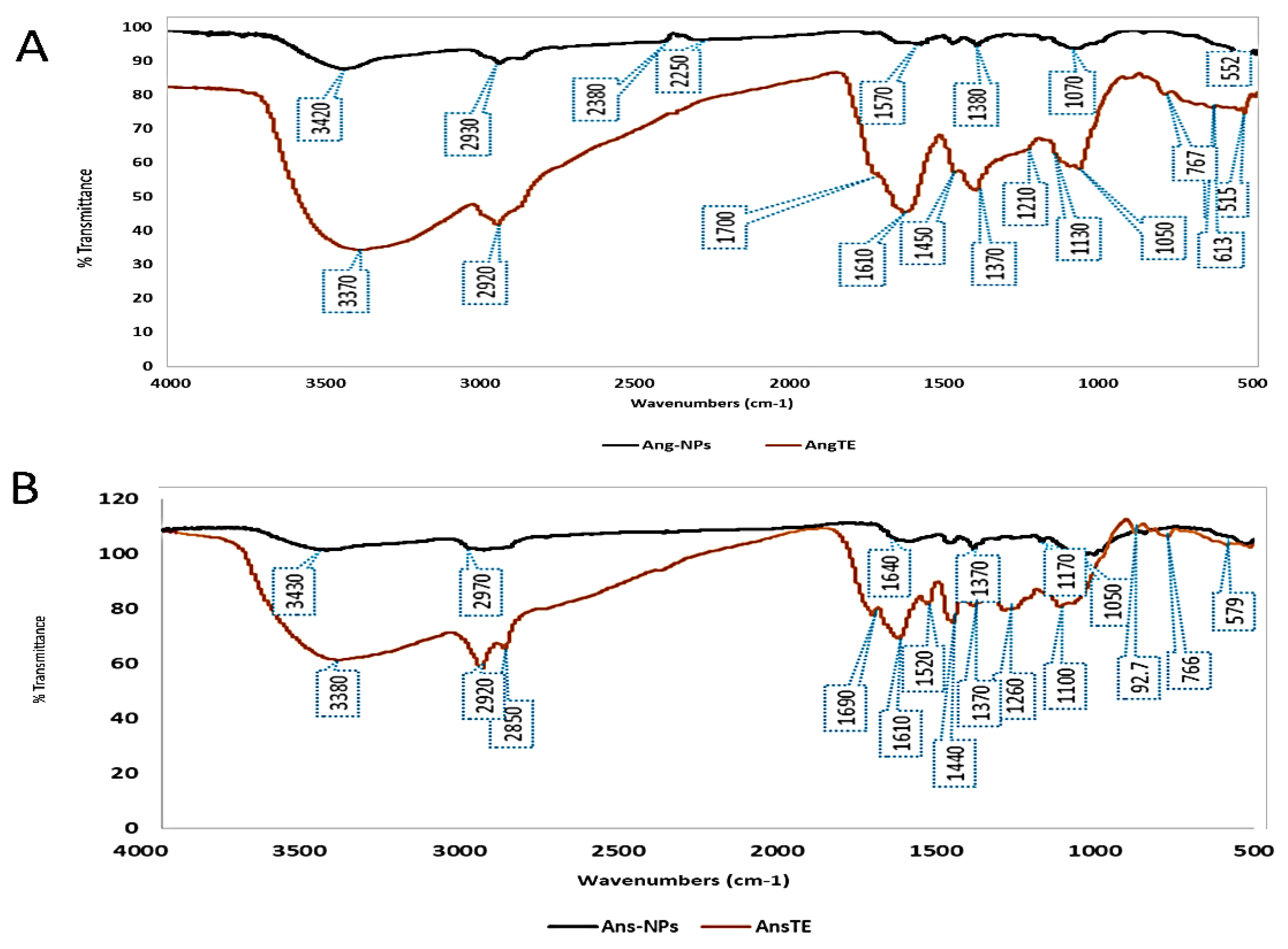
2.2.2. FTIR
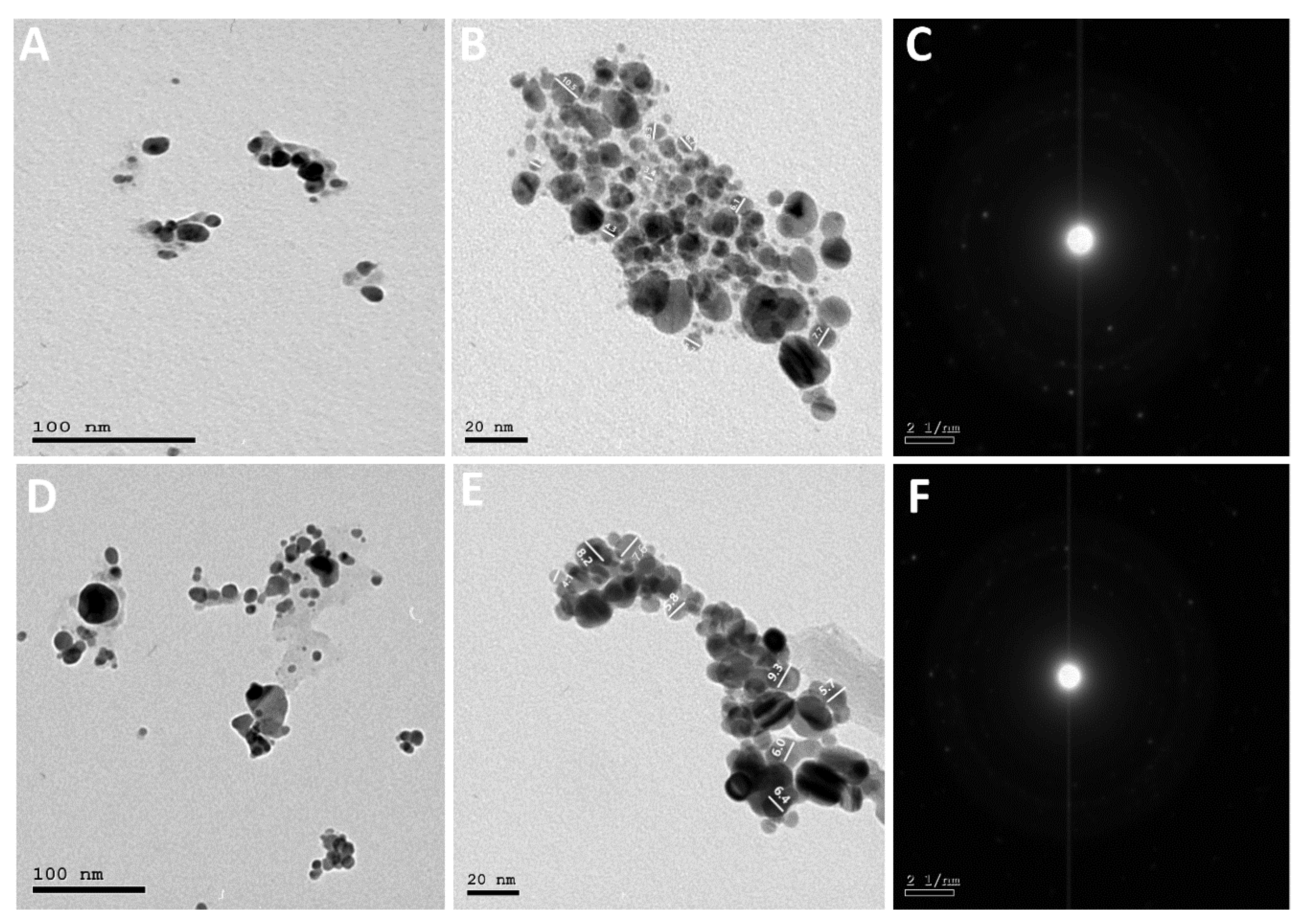
2.2.3. High-Resolution Transmission Electron Microscope (HR-TEM)
2.2.4. Zeta Potential and DLS
2.2.5. X-ray Diffraction (XRD)
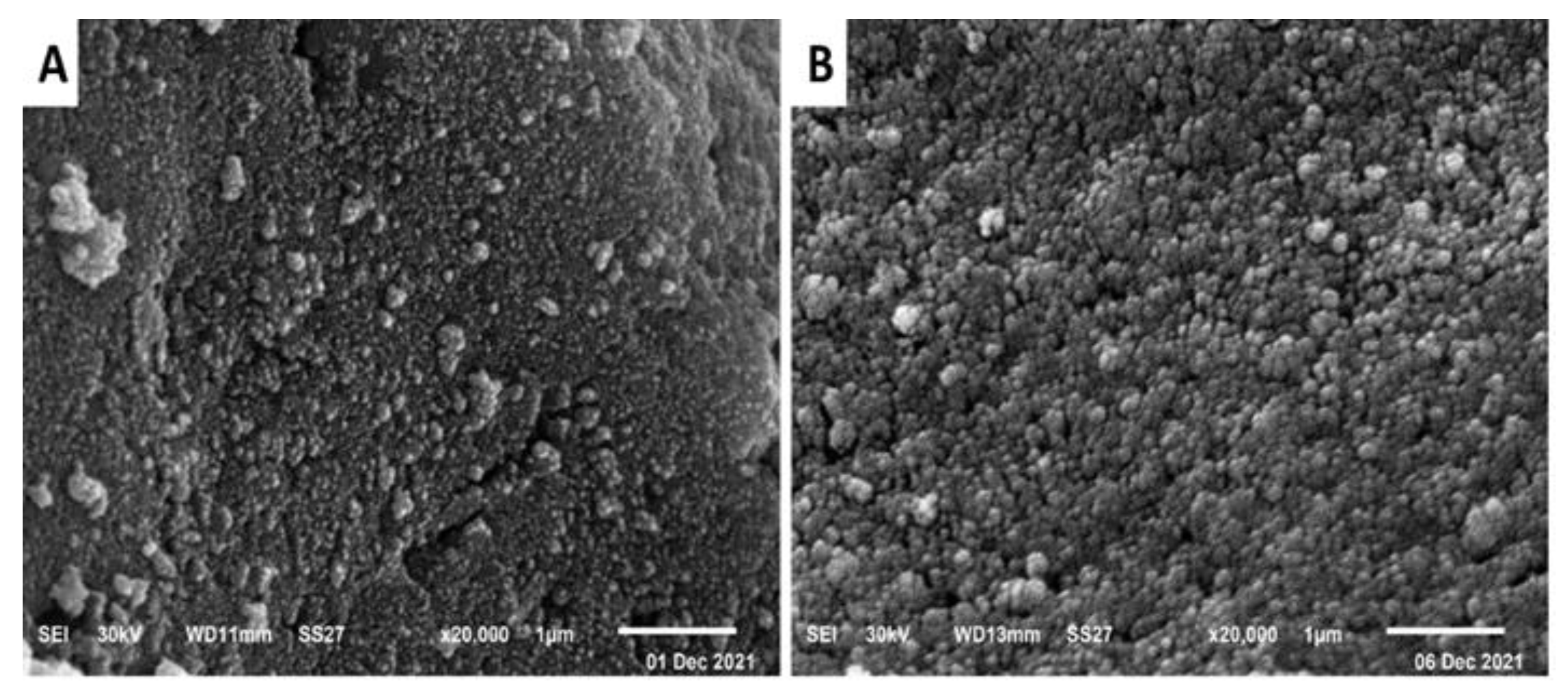
2.2.6. Scanning Electron Microscope (SEM)
2.3. The Biological Study
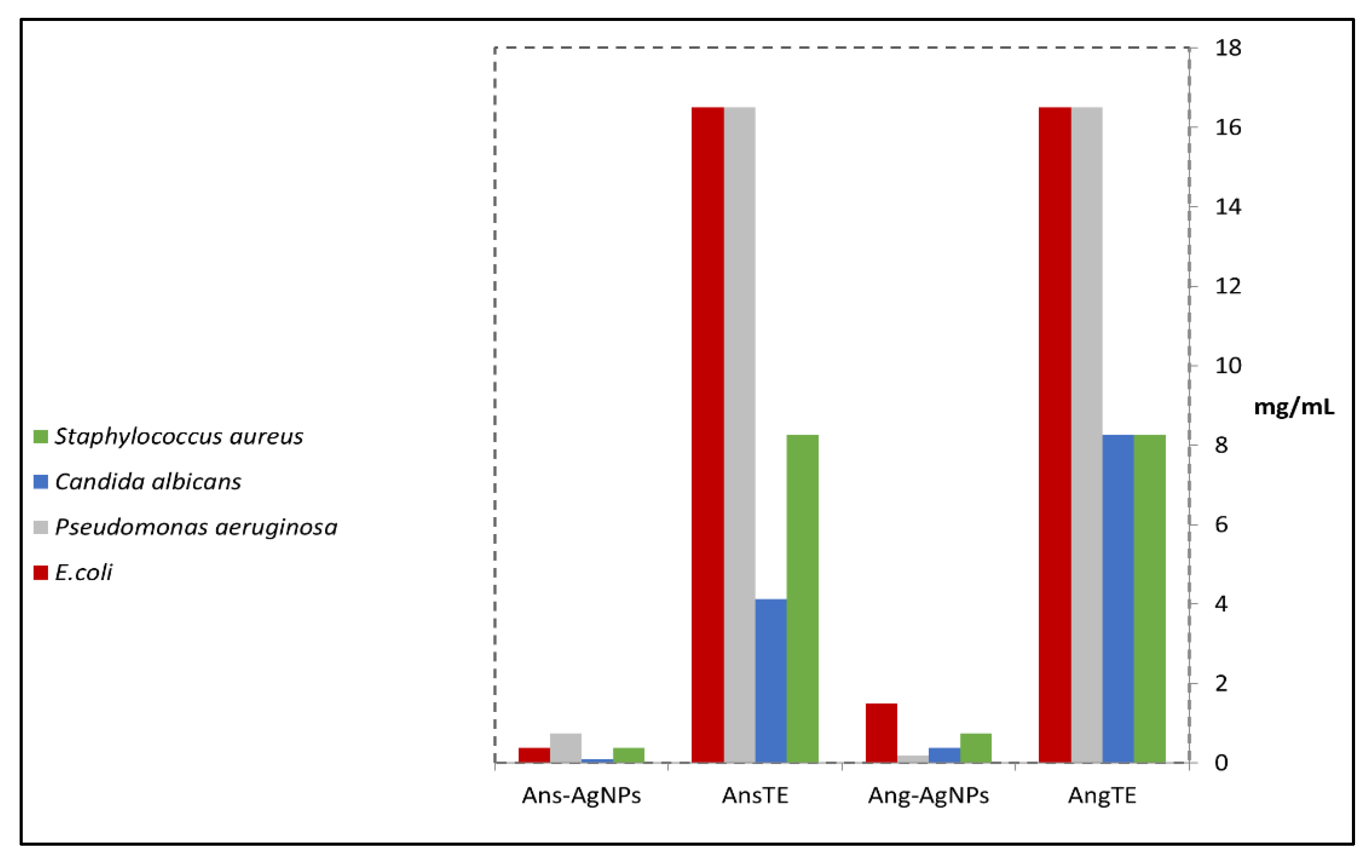
2.3.1. Antimicrobial Evaluation of the Extracts and Their Biogenic Nanoparticles
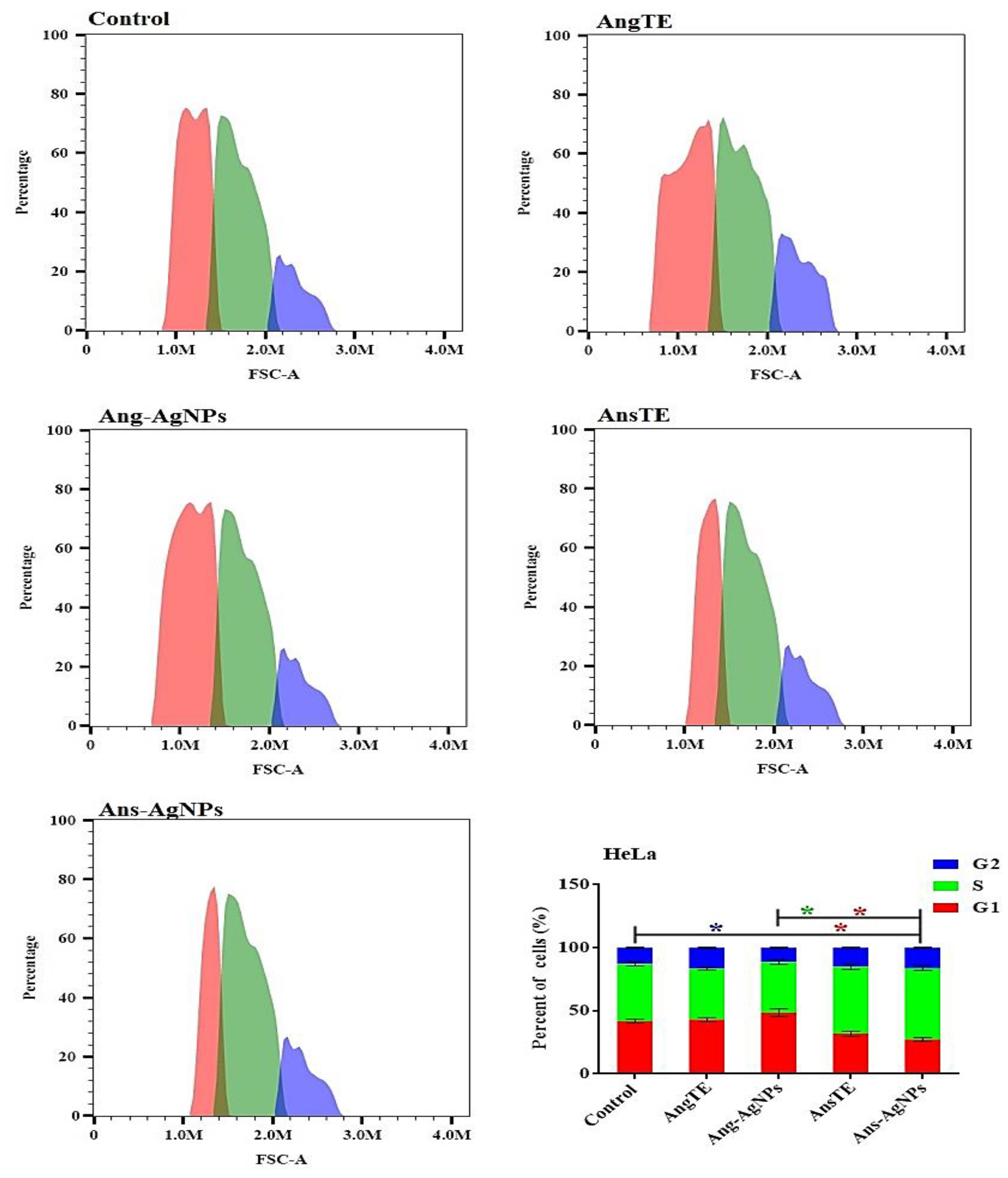
2.3.2. Anticancer Activity
In Vitro Cytotoxic Activity
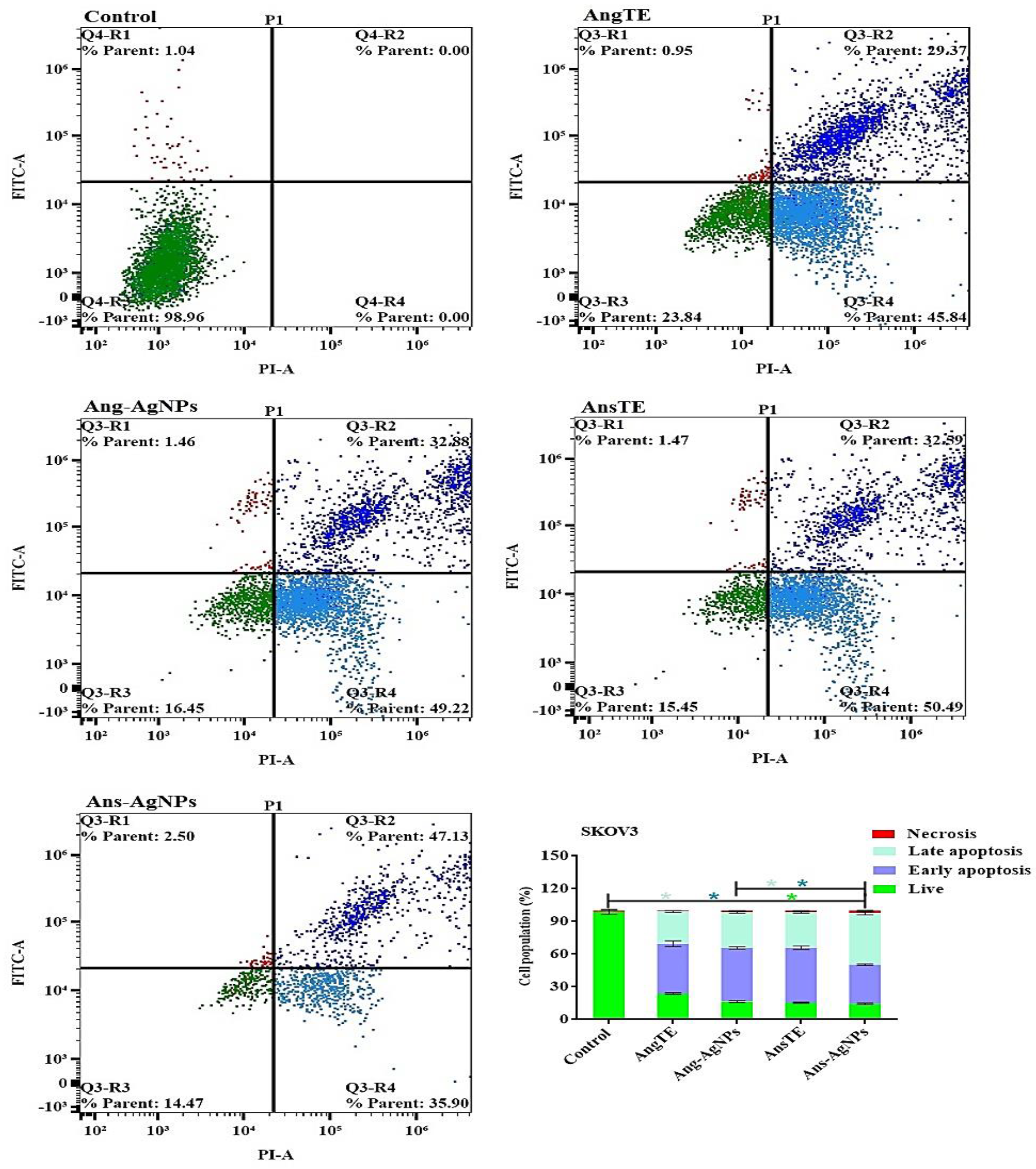
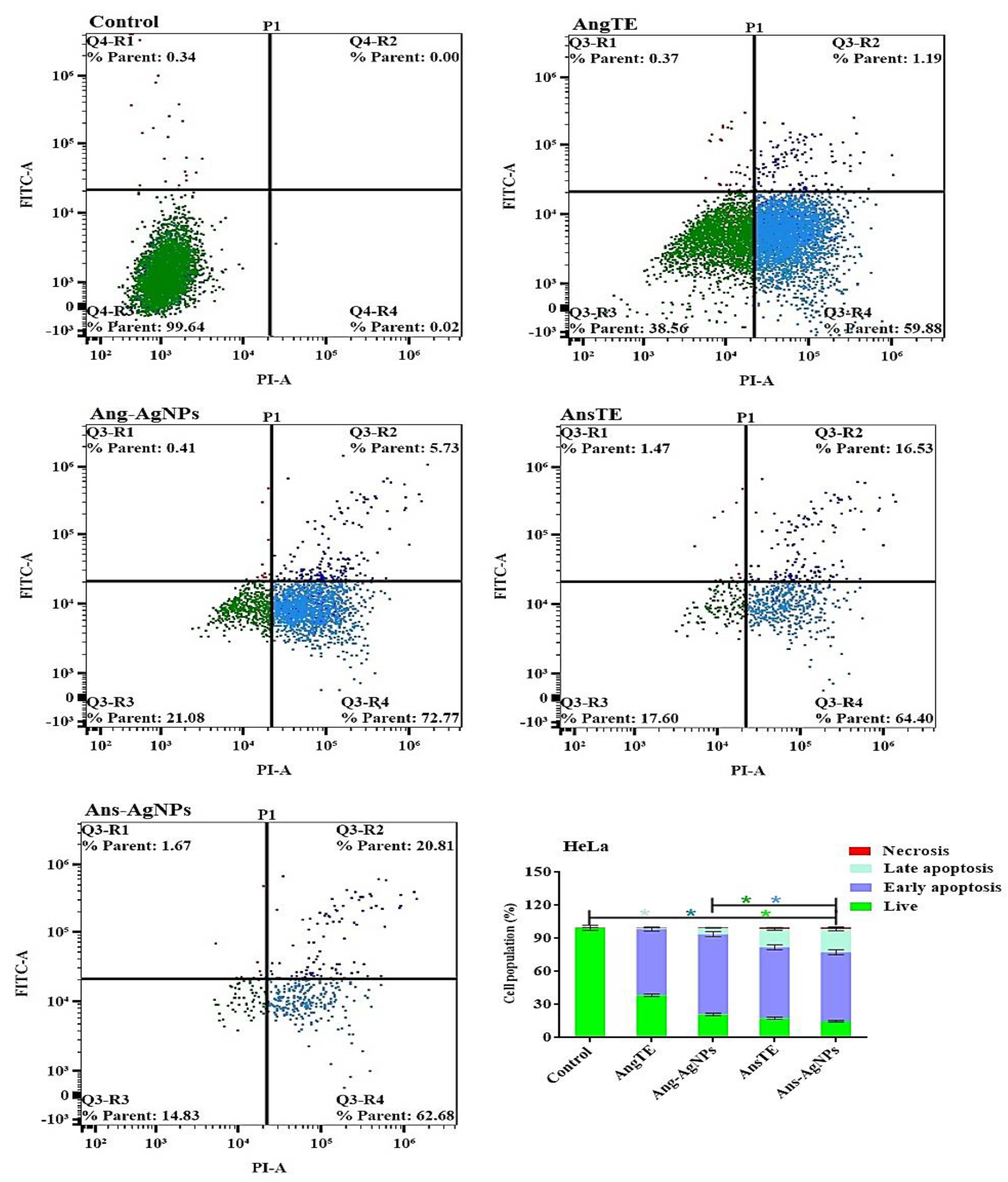
Cell Apoptosis Using Annexin V-FITC
2.4. Docking Study of Bioactive Compounds
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Extract Preparation
4.2. Drugs and Chemicals
4.3. UPLC-QTOF-MS/MS Analysis
4.4. Green Synthesis of Ang-AgNPs and Ans-AgNPs
4.5. Characterization of AgNPs
4.5.1. UV-Vis Spectroscopy
4.5.2. FTIR
4.5.3. HR-TEM
4.5.4. Zeta Potential Analysis
4.5.5. XRD
4.5.6. SEM
4.6. Biological Study
4.6.1. Antibacterial Activity
Determination of Antibacterial Activity of the Extracts
Minimal Inhibitory Concentration (MIC) Determination
Disc Diffusion Assay
4.6.2. In Vitro Cytotoxicity
Cytotoxicity Assay
Cell Cycle Analysis
Apoptosis Analysis
4.7. Molecular Docking Study
4.7.1. Protein Preparation
4.7.2. Validation of the Molecular Docking Method
4.7.3. Ligand Preparation
4.7.4. Molecular Docking Calculations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biba, V.; Amily, A.; Sangeetha, S.; Remani, P. Anticancer, antioxidant and antimicrobial activity of annonaceae family. World J. Pharm. Pharm. Sci. 2014, 3, 1595–1604. [Google Scholar]
- Nugraha, A.S.; Damayanti, Y.D.; Wangchuk, P.; Keller, P.A.J.M. Anti-infective and anti-cancer properties of the Annona species: Their ethnomedicinal uses, alkaloid diversity, and pharmacological activities. Molecules 2019, 24, 4419. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Ma, C.-Y.; Wang, M.-L.; Lu, J.-H.; Hu, P.; Chen, J.-W.; Li, X.; Chen, Y.J.N.P.R. Five new ent-kaurane diterpenes from Annona squamosa L. Pericarps. Nat. Prod. Res. 2020, 34, 2243–2247. [Google Scholar] [CrossRef]
- Dev, A.A.; Joseph, S.M. Anticancer potential of Annona genus: A detailed review. J. Indian Chem. Soc. 2021, 98, 100231. [Google Scholar]
- Feriani, A.; Tir, M.; Gómez-Caravaca, A.M.; del Mar Contreras, M.; Talhaoui, N.; Taamalli, A.; Se-gura-Carretero, A.; Ghazouani, L.; Mufti, A.; Tlili, N.; et al. Hplc-dad-esi-qtof-ms/ms profiling of zy-gophyllum album roots extract and assessment of its cardioprotective effect against deltame-thrin-induced myocardial injuries in rat, by suppression of oxidative stress-related inflammation and apoptosis via nf-κb signaling pathway. J. Ethnopharmacol. 2020, 247, 112266. [Google Scholar]
- Hamed, A.R.; El-Hawary, S.S.; Ibrahim, R.M.; Abdelmohsen, U.R.; El-Halawany, A.M. Identification of chemoprevetive components from halophytes belonging to aizoaceae and cactaceae through lc/ms—Bioassay guided approach. J. Chromatogr. Sci. 2021, 59, 618–626. [Google Scholar] [CrossRef]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.J.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.R.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef] [PubMed]
- Attallah, N.G.; Elekhnawy, E.; Negm, W.A.; Hussein, I.A.; Mokhtar, F.A.; Al-Fakhrany, O.M. In vivo and in vitro antimicrobial activity of biogenic silver nanoparticles against staphylococcus aureus clinical isolates. Pharmaceuticals 2022, 15, 194. [Google Scholar] [CrossRef]
- Thakkar, K.N.; Mhatre, S.S.; Parikh, R.Y. Biological synthesis of metallic nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 257–262. [Google Scholar] [CrossRef]
- Pearce, A.; Haas, M.; Viney, R.; Pearson, S.-A.; Haywood, P.; Brown, C.; Ward, R. Incidence and severity of self-reported chemotherapy side effects in routine care: A prospective cohort study. PLoS ONE 2017, 12, e0184360. [Google Scholar]
- Majeed, H.; Gupta, V. Adverse Effects of Radiation Therapy; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Issa, M.Y.; Mohsen, E.; Younis, I.Y.; Nofal, E.S.; Farag, M.A. Volatiles distribution in jasmine flowers taxa grown in egypt and its commercial products as analyzed via solid-phase microextraction (spme) coupled to chemometrics. Ind. Crops Prod. 2020, 144, 112002. [Google Scholar] [CrossRef]
- Jang, S.J.; Yang, I.J.; Tettey, C.O.; Kim, K.M.; Shin, H.M. In-vitro anticancer activity of green synthesized silver nanoparticles on mcf-7 human breast cancer cells. Mater. Sci. Eng. C 2016, 68, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Langsrud, S.; Sidhu, M.S.; Heir, E.; Holck, A.L. Bacterial disinfectant resistance—A challenge for the food industry. Int. Biodeterior. Biodegrad. 2003, 51, 283–290. [Google Scholar] [CrossRef]
- Ahmed, S.; Ahmad, M.; Swami, B.L.; Ikram, S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: A green expertise. J. Adv. Res. 2016, 7, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Shiekh, K.A.; Olatunde, O.O.; Zhang, B.; Huda, N.; Benjakul, S.J.F.C. Pulsed electric field assisted process for extraction of bioactive compounds from custard apple (Annona squamosa) leaves. Food Chem. 2021, 359, 129976. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Pareek, S.; Sagar, N.; Vyas, N. Bioactive compounds of Annona. In Bioactive Compounds in Underutilized Fruits and Nuts; Springer: Cham, Switzerland, 2020; pp. 37–62. [Google Scholar]
- Ma, C.-Y.; Li, J.-H.; Li, X.; Liu, X.; Chen, J.-W. Eight new cytotoxic annonaceous acetogenins from the seeds of Annona squamosa. Chin. J. Nat. Med. 2019, 17, 291–297. [Google Scholar] [CrossRef]
- Nhiem, N.X.; Hien, N.T.T.; Tai, B.H.; Anh, H.L.T.; Hang, D.T.T.; Quang, T.H.; Van Kiem, P.; Van Minh, C.; Ko, W.; Lee, S.J.B. New ent-kauranes from the fruits of Annona glabra and their inhibitory nitric oxide production in lps-stimulated raw264. 7 macrophages. Bioorganic Med. Chem. Lett. 2015, 25, 254–258. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.-w.; Wang, Y.; Xu, S.-s.; Li, X. Six cytotoxic annonaceous acetogenins from Annona squamosa seeds. Food Chem. 2012, 135, 960–966. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chang, F.-R.; Cho, C.-P.; Wu, Y.-C. Ent-kaurane diterpenoids from Annona glabra. J. Nat. Prod. 2000, 63, 1000–1003. [Google Scholar] [CrossRef]
- Bonneau, N.; Baloul, L.; ba Ndob, I.B.; Senejoux, F.; Champy, P. The fruit of Annona squamosa L. As a source of environmental neurotoxins: From quantification of squamocin to annotation of an-nonaceous acetogenins by lc–ms/ms analysis. Food Chem. 2017, 226, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Avula, B.; Bae, J.-Y.; Majrashi, T.; Wu, T.-Y.; Wang, Y.-H.; Wang, M.; Ali, Z.; Wu, Y.-C.; Khan, I.A. Targeted and non-targeted analysis of annonaceous alkaloids and acetogenins from asimi-na and Annona species using uhplc-qtof-ms. J. Pharm. Biomed. Anal. 2018, 159, 548–566. [Google Scholar] [CrossRef] [PubMed]
- Bykkam, S.; Ahmadipour, M.; Narisngam, S.; Kalagadda, V.R.; Chidurala, S.C. Extensive studies on x-ray diffraction of green synthesized silver nanoparticles. Adv. Nanopart. 2015, 4, 1–10. [Google Scholar] [CrossRef]
- Tarannum, N.; Gautam, Y.K. Facile green synthesis and applications of silver nanoparticles: A state-of-the-art review. RSC Adv. 2019, 9, 34926–34948. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.; Mankad, V.; Gupta, S.K.; Jha, P.K. Size distribution of silver nanoparticles: Uv-visible spectroscopic assessment. Nanosci. Nanotechnol. Lett. 2012, 4, 30–34. [Google Scholar] [CrossRef]
- Varadavenkatesan, T.; Vinayagam, R.; Selvaraj, R. Structural characterization of silver nanoparticles phyto-mediated by a plant waste, seed hull of vigna mungo and their biological applications. J. Mol. Struct. 2017, 1147, 629–635. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (mic) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Burmeister, L.; Bartlett, M.; Rinaldi, M. Multicenter evaluation of four methods of yeast inoculum preparation. J. Clin. Microbiol. 1988, 26, 1437–1441. [Google Scholar] [CrossRef]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. Clsi methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Al-Abd, A.M.; Lightfoot, D.A.; El-Shemy, H.A. Anti-cancer characteristics of mevinolin against three different solid tumor cell lines was not solely p53-dependent. J. Enzyme Inhib. Med. Chem. 2012, 27, 673–679. [Google Scholar] [CrossRef]
- Nunez, R. DNA measurement and cell cycle analysis by flow cytometry. Curr. Issues Mol. Biol. 2001, 3, 67–70. [Google Scholar]
- Vafaei, S.; Sadat Shandiz, S.A.; Piravar, Z. Zinc-phosphate nanoparticles as a novel anticancer agent: An in vitro evaluation of their ability to induce apoptosis. Biol. Trace Elem. Res. 2020, 198, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Wakui, N.; Yoshino, R.; Yasuo, N.; Ohue, M.; Sekijima, M. Exploring the selectivity of inhibitor complexes with bcl-2 and bcl-xl: A molecular dynamics simulation approach. J. Mol. Graph. Model. 2018, 79, 166–174. [Google Scholar] [CrossRef] [PubMed]






| Zone of Inhibition | ||||
|---|---|---|---|---|
| Ang-AgNPs (mm) | Ang-AgNPs + Gentamicin (mm) | Gentamicin (mm) | Reference Zone of Inhibition | |
| Pseudomonas aeruginosa | 10 | 20 | 14 | 15 |
| E. coli | 10 | 19 | 14 | 15 |
| Zone of Inhibition | ||||
|---|---|---|---|---|
| Ans-AgNPs (mm) | Ans-AgNPs + Gentamicin (mm) | Gentamicin | Reference Zone of Inhibition in CLSI | |
| Pseudomonas aeruginosa | 13 | 20 | 12 | 15 |
| E. coli | 12 | 15 | 15 | 15 |
| Zone of Inhibition | |||
|---|---|---|---|
| Ans-AgNPs | Ans-AgNPs + Vancomycin | Vancomycin | |
| Staphylococcus aureus | 15 | 12 | 17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokhtar, F.A.; Selim, N.M.; Elhawary, S.S.; Abd El Hadi, S.R.; Hetta, M.H.; Albalawi, M.A.; Shati, A.A.; Alfaifi, M.Y.; Elbehairi, S.E.I.; Fahmy, L.I.; et al. Green Biosynthesis of Silver Nanoparticles Using Annona glabra and Annona squamosa Extracts with Antimicrobial, Anticancer, Apoptosis Potentials, Assisted by In Silico Modeling, and Metabolic Profiling. Pharmaceuticals 2022, 15, 1354. https://doi.org/10.3390/ph15111354
Mokhtar FA, Selim NM, Elhawary SS, Abd El Hadi SR, Hetta MH, Albalawi MA, Shati AA, Alfaifi MY, Elbehairi SEI, Fahmy LI, et al. Green Biosynthesis of Silver Nanoparticles Using Annona glabra and Annona squamosa Extracts with Antimicrobial, Anticancer, Apoptosis Potentials, Assisted by In Silico Modeling, and Metabolic Profiling. Pharmaceuticals. 2022; 15(11):1354. https://doi.org/10.3390/ph15111354
Chicago/Turabian StyleMokhtar, Fatma A., Nabil M. Selim, Seham S. Elhawary, Soha R. Abd El Hadi, Mona H. Hetta, Marzough A. Albalawi, Ali A. Shati, Mohammad Y. Alfaifi, Serag Eldin I. Elbehairi, Lamiaa I. Fahmy, and et al. 2022. "Green Biosynthesis of Silver Nanoparticles Using Annona glabra and Annona squamosa Extracts with Antimicrobial, Anticancer, Apoptosis Potentials, Assisted by In Silico Modeling, and Metabolic Profiling" Pharmaceuticals 15, no. 11: 1354. https://doi.org/10.3390/ph15111354
APA StyleMokhtar, F. A., Selim, N. M., Elhawary, S. S., Abd El Hadi, S. R., Hetta, M. H., Albalawi, M. A., Shati, A. A., Alfaifi, M. Y., Elbehairi, S. E. I., Fahmy, L. I., & Ibrahim, R. M. (2022). Green Biosynthesis of Silver Nanoparticles Using Annona glabra and Annona squamosa Extracts with Antimicrobial, Anticancer, Apoptosis Potentials, Assisted by In Silico Modeling, and Metabolic Profiling. Pharmaceuticals, 15(11), 1354. https://doi.org/10.3390/ph15111354






