The STING in Non-Alcoholic Fatty Liver Diseases: Potential Therapeutic Targets in Inflammation-Carcinogenesis Pathway
Abstract
1. Introduction
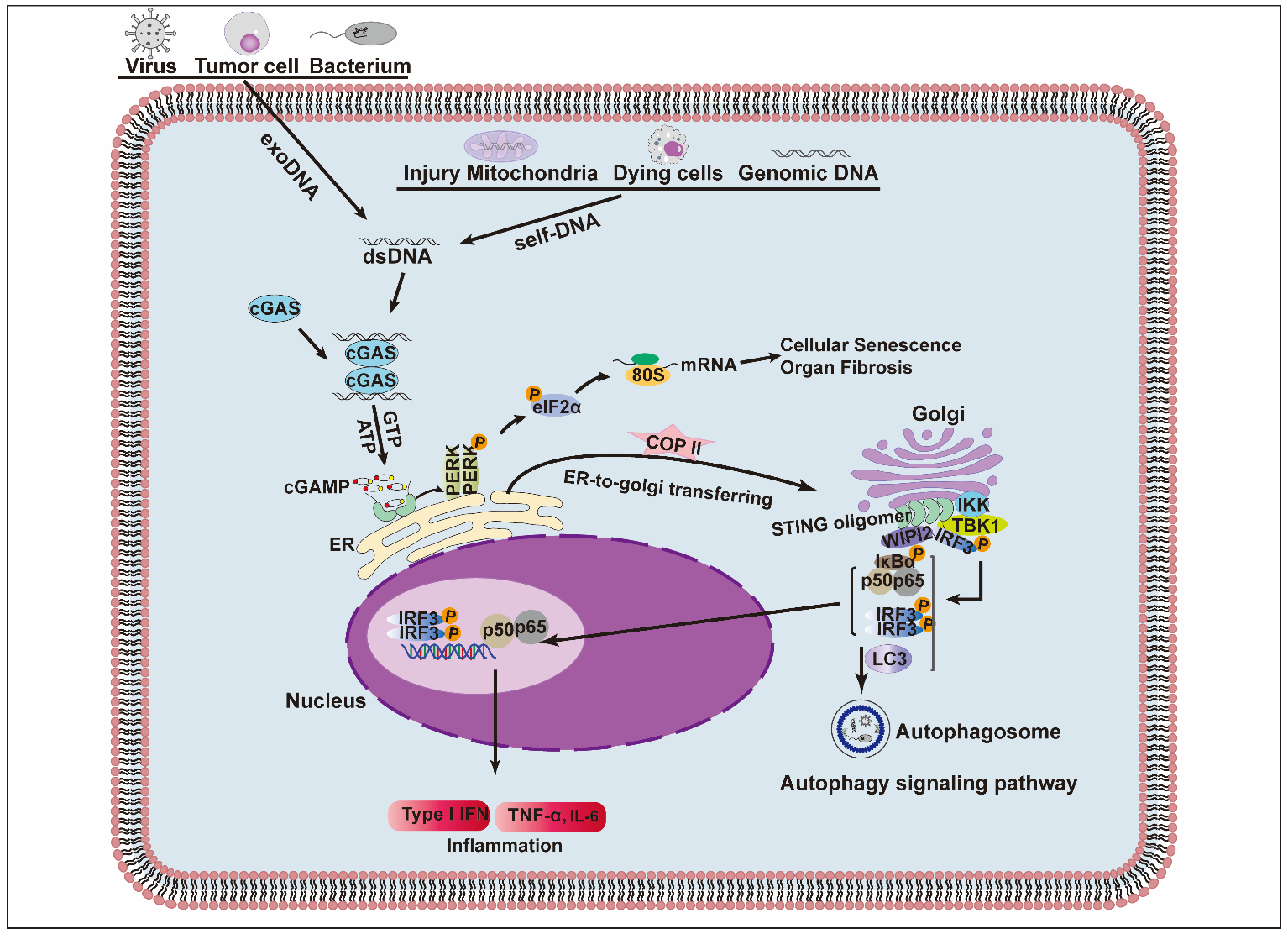
2. Structure and Signaling Pathway of STING
3. NAFL/NASH and STING
4. NASH-Associated HCC and STING
5. STING Inhibitors and Agonists
5.1. Covalent Inhibitors
5.2. Noncovalent Inhibitors
5.3. CDNs Agonists
5.4. Non-CDN Agonists
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, H.W.; Wong, V.W. Changing NAFLD Epidemiology in China. Hepatology 2019, 70, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Scorletti, E.; Carr, R.M. A new perspective on NAFLD: Focusing on lipid droplets. J. Hepatol. 2022, 76, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.A.; Patil, R.; Harrison, S.A. NAFLD-related hepatocellular carcinoma: The growing challenge. Hepatology 2022. [Google Scholar] [CrossRef]
- Eslam, M.; Sanyal, A.J.; George, J.; on behalf of theInternational Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1991. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, H.; Bu, Q.; Wei, S.; Li, L.; Zhou, J.; Zhou, S.; Su, W.; Liu, M.; Liu, Z.; et al. Role of XBP1 in regulating the progression of non-alcoholic steatohepatitis. J. Hepatol. 2022, 77, 312–325. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, J.; Man, K.; Chu, E.S.; Yau, T.O.; Sung, J.C.; Go, M.Y.; Deng, J.; Lu, L.; Wong, V.W.; et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J. Hepatol. 2014, 61, 1365–1375. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Yang, X.; Yang, Z.; Li, J.W.; Xu, M.Q.; Qu, Y.X.; Tang, J.J.; Li, Y.F.; Wang, L.; Shao, Y.W.; et al. Discovery of an insulin-induced gene binding compound that ameliorates nonalcoholic steatohepatitis by inhibiting sterol regulatory element-binding protein-mediated lipogenesis. Hepatology 2022, Volume, page. [Google Scholar] [CrossRef]
- Mohs, A.; Otto, T.; Schneider, K.M.; Peltzer, M.; Boekschoten, M.; Holland, C.H.; Hudert, C.A.; Kalveram, L.; Wiegand, S.; Saez-Rodriguez, J.; et al. Hepatocyte-specific NRF2 activation controls fibrogenesis and carcinogenesis in steatohepatitis. J. Hepatol. 2021, 74, 638–648. [Google Scholar] [CrossRef]
- Garcia-Monzon, C.; Lo Iacono, O.; Mayoral, R.; Gonzalez-Rodriguez, A.; Miquilena-Colina, M.E.; Lozano-Rodriguez, T.; Garcia-Pozo, L.; Vargas-Castrillon, J.; Casado, M.; Bosca, L.; et al. Hepatic insulin resistance is associated with increased apoptosis and fibrogenesis in nonalcoholic steatohepatitis and chronic hepatitis C. J. Hepatol. 2011, 54, 142–152. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, J.; Wang, L.; Tang, Y.; Zhou, Q.; Ji, S.; Wang, Y.; Santos, L.; Haeusler, R.A.; Que, J.; et al. Inhibition of PU.1 ameliorates metabolic dysfunction and non-alcoholic steatohepatitis. J. Hepatol. 2020, 73, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.; Mackey, J.B.G.; Jamieson, T.; Ramon-Gil, E.; Drake, T.M.; Fercoq, F.; Clark, W.; Gilroy, K.; Hedley, A.; Nixon, C.; et al. CXCR2 inhibition enables NASH-HCC immunotherapy. Gut 2022, 71, 1937–1938. [Google Scholar] [CrossRef]
- Muzurovic, E.; Peng, C.C.; Belanger, M.J.; Sanoudou, D.; Mikhailidis, D.P.; Mantzoros, C.S. Nonalcoholic Fatty Liver Disease and Cardiovascular Disease: A Review of Shared Cardiometabolic Risk Factors. Hypertension 2022, 79, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Cholankeril, G.; Kanwal, F. NAFLD and HCC: Time to Bridge the Gap. Hepatology 2021, 74, 2336–2338. [Google Scholar] [CrossRef] [PubMed]
- Le, M.H.; Yeo, Y.H.; Li, X.; Li, J.; Zou, B.; Wu, Y.; Ye, Q.; Huang, D.Q.; Zhao, C.; Zhang, J.; et al. 2019 Global NAFLD Prevalence: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2021. [Google Scholar] [CrossRef]
- Huang, X.; Yao, Y.; Hou, X.; Wei, L.; Rao, Y.; Su, Y.; Zheng, G.; Ruan, X.Z.; Li, D.; Chen, Y. Macrophage SCAP Contributes to Metaflammation and Lean NAFLD by Activating STING-NF-kappaB Signaling Pathway. Cell. Mol. Gastroenterol. Hepatol. 2022, 14, 1–26. [Google Scholar] [CrossRef]
- Qiao, J.T.; Cui, C.; Qing, L.; Wang, L.S.; He, T.Y.; Yan, F.; Liu, F.Q.; Shen, Y.H.; Hou, X.G.; Chen, L. Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism 2018, 81, 13–24. [Google Scholar] [CrossRef]
- Xu, D.; Tian, Y.; Xia, Q.; Ke, B. The cGAS-STING Pathway: Novel Perspectives in Liver Diseases. Front. Immunol. 2021, 12, 682736. [Google Scholar] [CrossRef]
- Mukai, K.; Konno, H.; Akiba, T.; Uemura, T.; Waguri, S.; Kobayashi, T.; Barber, G.N.; Arai, H.; Taguchi, T. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 2016, 7, 11932. [Google Scholar] [CrossRef]
- Zhao, B.; Du, F.; Xu, P.; Shu, C.; Sankaran, B.; Bell, S.L.; Liu, M.; Lei, Y.; Gao, X.; Fu, X.; et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 2019, 569, 718–722. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, X.C.; Chen, Z.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, H.; Ma, L.; Zhou, J.; Guo, X.; Woo, S.L.; Pei, Y.; Knight, L.R.; Deveau, M.; Chen, Y.; et al. Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 2018, 155, 1971–1984.e1974. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liu, Y.; An, W.; Song, J.; Zhang, Y.; Zhao, X. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Investig. 2019, 129, 546–555. [Google Scholar] [CrossRef]
- Wang, X.; Rao, H.; Zhao, J.; Wee, A.; Li, X.; Fei, R.; Huang, R.; Wu, C.; Liu, F.; Wei, L. STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Lab. Investig. 2020, 100, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Cen, D.; Ge, Q.; Xie, C.; Zheng, Q.; Guo, J.; Zhang, Y.; Wang, Y.; Li, X.; Gu, Z.; Cai, X. ZnS@BSA Nanoclusters Potentiate Efficacy of Cancer Immunotherapy. Adv. Mater. 2021, 33, e2104037. [Google Scholar] [CrossRef]
- Sen, T.; Rodriguez, B.L.; Chen, L.; Corte, C.M.D.; Morikawa, N.; Fujimoto, J.; Cristea, S.; Nguyen, T.; Diao, L.; Li, L.; et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019, 9, 646–661. [Google Scholar] [CrossRef]
- Jing, W.; McAllister, D.; Vonderhaar, E.P.; Palen, K.; Riese, M.J.; Gershan, J.; Johnson, B.D.; Dwinell, M.B. STING agonist inflames the pancreatic cancer immune microenvironment and reduces tumor burden in mouse models. J. Immunother. Cancer 2019, 7, 115. [Google Scholar] [CrossRef]
- Wang-Bishop, L.; Wehbe, M.; Shae, D.; James, J.; Hacker, B.C.; Garland, K.; Chistov, P.P.; Rafat, M.; Balko, J.M.; Wilson, J.T. Potent STING activation stimulates immunogenic cell death to enhance antitumor immunity in neuroblastoma. J. Immunother. Cancer 2020, 8, e000282. [Google Scholar] [CrossRef]
- Haag, S.M.; Gulen, M.F.; Reymond, L.; Gibelin, A.; Abrami, L.; Decout, A.; Heymann, M.; van der Goot, F.G.; Turcatti, G.; Behrendt, R.; et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018, 559, 269–273. [Google Scholar] [CrossRef]
- Li, S.; Hong, Z.; Wang, Z.; Li, F.; Mei, J.; Huang, L.; Lou, X.; Zhao, S.; Song, L.; Chen, W.; et al. The Cyclopeptide Astin C Specifically Inhibits the Innate Immune CDN Sensor STING. Cell Rep. 2018, 25, 3405–3421.e3407. [Google Scholar] [CrossRef]
- Gao, J.; Zheng, M.; Wu, X.; Zhang, H.; Su, H.; Dang, Y.; Ma, M.; Wang, F.; Xu, J.; Chen, L.; et al. CDK inhibitor Palbociclib targets STING to alleviate autoinflammation. EMBO Rep. 2022, 23, e53932. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Mann, C.C.; Orzalli, M.H.; King, D.S.; Kagan, J.C.; Lee, A.S.Y.; Kranzusch, P.J. Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappaB Signaling Adaptation. Cell Rep. 2019, 27, 1165–1175.e1165. [Google Scholar] [CrossRef]
- Fang, R.; Jiang, Q.; Guan, Y.; Gao, P.; Zhang, R.; Zhao, Z.; Jiang, Z. Golgi apparatus-synthesized sulfated glycosaminoglycans mediate polymerization and activation of the cGAMP sensor STING. Immunity 2021, 54, 962–975.e968. [Google Scholar] [CrossRef]
- Gui, X.; Yang, H.; Li, T.; Tan, X.; Shi, P.; Li, M.; Du, F.; Chen, Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019, 567, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, Y.; Zhu, Y.; Zhang, Q.; Guan, H.; Liu, S.; Chen, S.; Mei, C.; Chen, C.; Liao, Z.; et al. A non-canonical cGAS-STING-PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat. Cell Biol. 2022, 24, 766–782. [Google Scholar] [CrossRef] [PubMed]
- Doria, A.; Zen, M.; Bettio, S.; Gatto, M.; Bassi, N.; Nalotto, L.; Ghirardello, A.; Iaccarino, L.; Punzi, L. Autoinflammation and autoimmunity: Bridging the divide. Autoimmun. Rev. 2012, 12, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, X.; Zheng, S.; Khanabdali, R.; Kalionis, B.; Wu, J.; Wan, W.; Tai, X. An Update on Inflamm-Aging: Mechanisms, Prevention, and Treatment. J. Immunol. Res. 2016, 2016, 8426874. [Google Scholar] [CrossRef]
- Chen, Q.; Boire, A.; Jin, X.; Valiente, M.; Er, E.E.; Lopez-Soto, A.; Jacob, L.; Patwa, R.; Shah, H.; Xu, K.; et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 2016, 533, 493–498. [Google Scholar] [CrossRef]
- Ahn, J.; Konno, H.; Barber, G.N. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene 2015, 34, 5302–5308. [Google Scholar] [CrossRef]
- Powell, E.E.; Wong, V.W.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef]
- Shindo, N.; Fujisawa, T.; Sugimoto, K.; Nojima, K.; Oze-Fukai, A.; Yoshikawa, Y.; Wang, X.; Yasuda, O.; Ikegami, H.; Rakugi, H. Involvement of microsomal triglyceride transfer protein in nonalcoholic steatohepatitis in novel spontaneous mouse model. J. Hepatol. 2010, 52, 903–912. [Google Scholar] [CrossRef]
- Wu, J.W.; Wang, S.P.; Alvarez, F.; Casavant, S.; Gauthier, N.; Abed, L.; Soni, K.G.; Yang, G.; Mitchell, G.A. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology 2011, 54, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.A.; Lee, H.C.; Choe, J.; Kim, M.J.; Lee, M.J.; Chang, H.S.; Bae, I.Y.; Kim, H.K.; An, J.; Shim, J.H.; et al. Association between non-alcoholic fatty liver disease and cancer incidence rate. J. Hepatol. 2017, 68, 140–146. [Google Scholar] [CrossRef]
- Gardin, A.; Remih, K.; Gonzales, E.; Andersson, E.R.; Strnad, P. Modern therapeutic approaches to liver-related disorders. J. Hepatol. 2022, 76, 1392–1409. [Google Scholar] [CrossRef]
- Adams, D.H. Sinusoidal Endothelial Cells as Orchestrators of the Gut Liver Immune Axis. Hepatology 2021, 74, 1690–1691. [Google Scholar] [CrossRef]
- Wang, S.; Wu, Q.; Chen, T.; Su, R.; Pan, C.; Qian, J.; Huang, H.; Yin, S.; Xie, H.; Zhou, L.; et al. Blocking CD47 promotes anti-tumor immunity through CD103+ dendritic cell-NK cell axis in murine hepatocellular carcinoma model. J. Hepatol. 2022, 77, 467–478. [Google Scholar] [CrossRef]
- Kim, K.E.; Lee, J.; Shin, H.J.; Jeong, E.A.; Jang, H.M.; Ahn, Y.J.; An, H.S.; Lee, J.Y.; Shin, M.C.; Kim, S.K.; et al. Lipocalin-2 Activates Hepatic stellate cells and Promotes Non-alcoholic Steatohepatitis in High-Fat Diet-Fed ob/ob mice. Hepatology 2022. [Google Scholar] [CrossRef]
- Yang, W.; He, H.; Wang, T.; Su, N.; Zhang, F.; Jiang, K.; Zhu, J.; Zhang, C.; Niu, K.; Wang, L.; et al. Single-Cell Transcriptomic Analysis Reveals a Hepatic Stellate Cell-Activation Roadmap and Myofibroblast Origin During Liver Fibrosis in Mice. Hepatology 2021, 74, 2774–2790. [Google Scholar] [CrossRef]
- Wang, S.S.; Tang, X.T.; Lin, M.; Yuan, J.; Peng, Y.J.; Yin, X.; Shang, G.; Ge, G.; Ren, Z.; Zhou, B.O. Perivenous Stellate Cells Are the Main Source of Myofibroblasts and Cancer-Associated Fibroblasts Formed After Chronic Liver Injuries. Hepatology 2021, 74, 1578–1594. [Google Scholar] [CrossRef]
- Moreno-Fernandez, M.E.; Miraldi, E.R.; Divanovic, S. Not Chopped Liver-A Careful, Fate-Mapping Study of Macrophages in NASH. Cell Metab. 2020, 32, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Zapotoczny, B.; Szafranska, K.; Kus, E.; Braet, F.; Wisse, E.; Chlopicki, S.; Szymonski, M. Tracking Fenestrae Dynamics in Live Murine Liver Sinusoidal Endothelial Cells. Hepatology 2019, 69, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, K.K.; Simon-Santamaria, J.; McCuskey, R.S.; Smedsrod, B. Liver Sinusoidal Endothelial Cells. Compr. Physiol. 2015, 5, 1751–1774. [Google Scholar] [CrossRef] [PubMed]
- Gracia-Sancho, J.; Caparros, E.; Fernandez-Iglesias, A.; Frances, R. Role of liver sinusoidal endothelial cells in liver diseases. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 411–431. [Google Scholar] [CrossRef]
- Hammoutene, A.; Rautou, P.E. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J. Hepatol. 2019, 70, 1278–1291. [Google Scholar] [CrossRef]
- Desroches-Castan, A.; Tillet, E.; Ricard, N.; Ouarne, M.; Mallet, C.; Belmudes, L.; Coute, Y.; Boillot, O.; Scoazec, J.Y.; Bailly, S.; et al. Bone Morphogenetic Protein 9 Is a Paracrine Factor Controlling Liver Sinusoidal Endothelial Cell Fenestration and Protecting Against Hepatic Fibrosis. Hepatology 2019, 70, 1392–1408. [Google Scholar] [CrossRef]
- Garcia-Martinez, I.; Santoro, N.; Chen, Y.; Hoque, R.; Ouyang, X.; Caprio, S.; Shlomchik, M.J.; Coffman, R.L.; Candia, A.; Mehal, W.Z. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J. Clin. Investig. 2016, 126, 859–864. [Google Scholar] [CrossRef]
- Ahn, J.; Xia, T.; Rabasa Capote, A.; Betancourt, D.; Barber, G.N. Extrinsic Phagocyte-Dependent STING Signaling Dictates the Immunogenicity of Dying Cells. Cancer Cell 2018, 33, 862–873.e865. [Google Scholar] [CrossRef]
- Cai, B.; Dongiovanni, P.; Corey, K.E.; Wang, X.; Shmarakov, I.O.; Zheng, Z.; Kasikara, C.; Davra, V.; Meroni, M.; Chung, R.T.; et al. Macrophage MerTK Promotes Liver Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab. 2020, 31, 406–421.e407. [Google Scholar] [CrossRef]
- Pradere, J.P.; Kluwe, J.; De Minicis, S.; Jiao, J.J.; Gwak, G.Y.; Dapito, D.H.; Jang, M.K.; Guenther, N.D.; Mederacke, I.; Friedman, R.; et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology 2013, 58, 1461–1473. [Google Scholar] [CrossRef]
- Shmuel-Galia, L.; Humphries, F.; Lei, X.; Ceglia, S.; Wilson, R.; Jiang, Z.; Ketelut-Carneiro, N.; Foley, S.E.; Pechhold, S.; Houghton, J.; et al. Dysbiosis exacerbates colitis by promoting ubiquitination and accumulation of the innate immune adaptor STING in myeloid cells. Immunity 2021, 54, 1137–1153.e1138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tang, K.; Zhang, Y.; Ma, R.; Ma, J.; Li, Y.; Luo, S.; Liang, X.; Ji, T.; Gu, Z.; et al. Cell-free tumor microparticle vaccines stimulate dendritic cells via cGAS/STING signaling. Cancer Immunol. Res. 2015, 3, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.S.; Park, H.W.; Ho, A.; Semple, I.A.; Kim, B.; Jang, I.; Park, H.; Reilly, S.; Saltiel, A.R.; Lee, J.H. Lipotoxicity induces hepatic protein inclusions through TANK binding kinase 1-mediated p62/sequestosome 1 phosphorylation. Hepatology 2018, 68, 1331–1346. [Google Scholar] [CrossRef]
- Akhmetova, K.; Balasov, M.; Chesnokov, I. Drosophila STING protein has a role in lipid metabolism. Elife 2021, 10, e67358. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Xu, E.; Zheng, R.; Zhangchen, Z.; Zhong, R.; Huang, F.; Ye, J.; Sun, H.; Fan, Y.; Xie, S.; et al. Traditional Chinese medicine Lingguizhugan decoction ameliorate HFD-induced hepatic-lipid deposition in mice by inhibiting STING-mediated inflammation in macrophages. Chin. Med. 2022, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, Q.; Yan, C.; Niu, C.; Zhou, J.; Liu, J.; Song, Y.; Zhou, F.; Fan, Y.; Ren, J.; et al. The Absence of STING Ameliorates Non-Alcoholic Fatty Liver Disease and Reforms Gut Bacterial Community. Front. Immunol. 2022, 13, 931176. [Google Scholar] [CrossRef]
- Maher, J.J. Macrophages Steal STING From the Infectious Disease Playbook to Promote Nonalcoholic Fatty Liver Disease. Gastroenterology 2018, 155, 1687–1688. [Google Scholar] [CrossRef]
- Karim, M.A.; Singal, A.G.; Kum, H.C.; Lee, Y.T.; Park, S.; Rich, N.E.; Noureddin, M.; Yang, J.D. Clinical Characteristics and Outcomes of Nonalcoholic Fatty Liver Disease-Associated Hepatocellular Carcinoma in the United States. Clin. Gastroenterol. Hepatol. 2022. [Google Scholar] [CrossRef]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef]
- Claveria-Cabello, A.; Avila, M.A. HIF2alpha Activation in NASH: A New Force Pushing Toward HCC. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 678–680. [Google Scholar] [CrossRef]
- Gabbia, D.; Cannella, L.; De Martin, S. The Role of Oxidative Stress in NAFLD-NASH-HCC Transition-Focus on NADPH Oxidases. Biomedicines 2021, 9, 687. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Alvarez, M.I.; Sebastian, D.; Vives, S.; Ivanova, S.; Bartoccioni, P.; Kakimoto, P.; Plana, N.; Veiga, S.R.; Hernandez, V.; Vasconcelos, N.; et al. Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease. Cell 2019, 177, 881–895.e817. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Q.; Teoh, N.; Xu, L.; Pok, S.; Li, X.; Chu, E.S.H.; Chiu, J.; Dong, L.; Arfianti, E.; Haigh, W.G.; et al. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat. Commun. 2018, 9, 4490. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Herceg, Z. From hepatitis to hepatocellular carcinoma: A proposed model for cross-talk between inflammation and epigenetic mechanisms. Genome Med. 2012, 4, 8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yan, C.; Huo, X.; Wang, S.; Feng, Y.; Gong, Z. Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J. Hepatol. 2015, 63, 420–428. [Google Scholar] [CrossRef]
- Thomsen, M.K.; Skouboe, M.K.; Boularan, C.; Vernejoul, F.; Lioux, T.; Leknes, S.L.; Berthelsen, M.F.; Riedel, M.; Cai, H.; Joseph, J.V.; et al. The cGAS-STING pathway is a therapeutic target in a preclinical model of hepatocellular carcinoma. Oncogene 2020, 39, 1652–1664. [Google Scholar] [CrossRef]
- Li, A.; Yi, M.; Qin, S.; Song, Y.; Chu, Q.; Wu, K. Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 35. [Google Scholar] [CrossRef]
- Bolhaqueiro, A.C.F.; Ponsioen, B.; Bakker, B.; Klaasen, S.J.; Kucukkose, E.; van Jaarsveld, R.H.; Vivie, J.; Verlaan-Klink, I.; Hami, N.; Spierings, D.C.J.; et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019, 51, 824–834. [Google Scholar] [CrossRef]
- Venkatesan, S.; Angelova, M.; Puttick, C.; Zhai, H.; Caswell, D.R.; Lu, W.T.; Dietzen, M.; Galanos, P.; Evangelou, K.; Bellelli, R.; et al. Induction of APOBEC3 Exacerbates DNA Replication Stress and Chromosomal Instability in Early Breast and Lung Cancer Evolution. Cancer Discov. 2021, 11, 2456–2473. [Google Scholar] [CrossRef]
- Watkins, T.B.K.; Lim, E.L.; Petkovic, M.; Elizalde, S.; Birkbak, N.J.; Wilson, G.A.; Moore, D.A.; Gronroos, E.; Rowan, A.; Dewhurst, S.M.; et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 2020, 587, 126–132. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wei, L.; Xu, F.; Zhao, F.; Huang, Y.; Fan, Z.; Mei, S.; Hu, Y.; Zhai, L.; Guo, J.; et al. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci. Signal. 2022, 15, eabg8744. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Wu, X.; Ma, D.; Wu, J.; Wang, L.; Jiang, Y.; Fei, Y.; Zhu, C.; Tan, R.; et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 2018, 563, 131–136. [Google Scholar] [CrossRef]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.C.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef] [PubMed]
- Lemos, H.; Mohamed, E.; Huang, L.; Ou, R.; Pacholczyk, G.; Arbab, A.S.; Munn, D.; Mellor, A.L. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016, 76, 2076–2081. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Jang, G.Y.; Lee, J.W.; Park, S.H.; Han, H.D.; Park, Y.M.; Kang, T.H. Improvement of STING-mediated cancer immunotherapy using immune checkpoint inhibitors as a game-changer. Cancer Immunol. Immunother. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, F.; Cao, Y.; Dang, Y.; Ge, B. The multifaceted functions of cGAS. J. Mol. Cell Biol. 2022, 14, mjac031. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Gong, Y.; Qiu, D.; Tang, H.; Zhang, J.; Yuan, Z.; Huang, Y.; Qin, Y.; Ye, L.; Yang, Y. Hyperbaric oxygen facilitates teniposide-induced cGAS-STING activation to enhance the antitumor efficacy of PD-1 antibody in HCC. J. Immunother. Cancer 2022, 10, e004006. [Google Scholar] [CrossRef]
- Lee, K.M.; Lin, C.C.; Servetto, A.; Bae, J.; Kandagatla, V.; Ye, D.; Kim, G.; Sudhan, D.R.; Mendiratta, S.; Gonzalez Ericsson, P.I.; et al. Epigenetic Repression of STING by MYC Promotes Immune Evasion and Resistance to Immune Checkpoint Inhibitors in Triple-Negative Breast Cancer. Cancer Immunol. Res. 2022, 10, 829–843. [Google Scholar] [CrossRef]
- Bu, Y.; Liu, F.; Jia, Q.A.; Yu, S.N. Decreased Expression of TMEM173 Predicts Poor Prognosis in Patients with Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0165681. [Google Scholar] [CrossRef]
- Hong, Z.; Mei, J.; Li, C.; Bai, G.; Maimaiti, M.; Hu, H.; Yu, W.; Sun, L.; Zhang, L.; Cheng, D.; et al. STING inhibitors target the cyclic dinucleotide binding pocket. Proc. Natl. Acad. Sci. USA 2021, 118, e2105465118. [Google Scholar] [CrossRef] [PubMed]
- Siu, T.; Altman, M.D.; Baltus, G.A.; Childers, M.; Ellis, J.M.; Gunaydin, H.; Hatch, H.; Ho, T.; Jewell, J.; Lacey, B.M.; et al. Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING. ACS Med. Chem. Lett. 2019, 10, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.L.; Buchan, G.J.; Ruhl, M.; Mukai, K.; Salvatore, S.R.; Ogawa, E.; Andersen, S.D.; Iversen, M.B.; Thielke, A.L.; Gunderstofte, C.; et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E7768–E7775. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Kong, D.; Guo, H.; Xing, C.; Lv, J.; Bian, H.; Lv, N.; Zhang, C.; Chen, D.; Liu, M.; et al. Gelsevirine improves age-related and surgically induced osteoarthritis in mice by reducing STING availability and local inflammation. BioChem. Pharmacol. 2022, 198, 114975. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Goldeck, M.; Cavlar, T.; Deimling, T.; Witte, G.; Rohl, I.; Hopfner, K.P.; Ludwig, J.; Hornung, V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 2013, 498, 380–384. [Google Scholar] [CrossRef]
- Andrade, W.A.; Firon, A.; Schmidt, T.; Hornung, V.; Fitzgerald, K.A.; Kurt-Jones, E.A.; Trieu-Cuot, P.; Golenbock, D.T.; Kaminski, P.A. Group B Streptococcus Degrades Cyclic-di-AMP to Modulate STING-Dependent Type I Interferon Production. Cell Host Microbe 2016, 20, 49–59. [Google Scholar] [CrossRef]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef]
- Wu, J.J.; Zhao, L.; Hu, H.G.; Li, W.H.; Li, Y.M. Agonists and inhibitors of the STING pathway: Potential agents for immunotherapy. Med. Res. Rev. 2020, 40, 1117–1141. [Google Scholar] [CrossRef]
- Huang, Y.H.; Liu, X.Y.; Du, X.X.; Jiang, Z.F.; Su, X.D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 2012, 19, 728–730. [Google Scholar] [CrossRef]
- Bowie, A.G. Innate sensing of bacterial cyclic dinucleotides: More than just STING. Nat. Immunol. 2012, 13, 1137–1139. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Shi, H.; Wu, J.; Zhang, X.; Sun, L.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 2013, 51, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Kranzusch, P.J.; Wilson, S.C.; Lee, A.S.; Berger, J.M.; Doudna, J.A.; Vance, R.E. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2′,3′ cGAMP Signaling. Mol. Cell 2015, 59, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Benkovics, T.; Peng, F.; Phillips, E.M.; An, C.; Bade, R.S.; Chung, C.K.; Dance, Z.E.X.; Fier, P.S.; Forstater, J.H.; Liu, Z.; et al. Diverse Catalytic Reactions for the Stereoselective Synthesis of Cyclic Dinucleotide MK-1454. J. Am. Chem. Soc. 2022, 144, 5855–5863. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Altman, M.D.; Lesburg, C.A.; Perera, S.A.; Piesvaux, J.A.; Schroeder, G.K.; Wyss, D.F.; Cemerski, S.; Chen, Y.; DiNunzio, E.; et al. Discovery of MK-1454: A Potent Cyclic Dinucleotide Stimulator of Interferon Genes Agonist for the Treatment of Cancer. J. Med. Chem. 2022, 65, 5675–5689. [Google Scholar] [CrossRef]
- Kim, S.; Li, L.; Maliga, Z.; Yin, Q.; Wu, H.; Mitchison, T.J. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem. Biol. 2013, 8, 1396–1401. [Google Scholar] [CrossRef]
- A Developed STING Agonist Has Systemic Antitumor Activity. Cancer Discov. 2019, 9, 12. [CrossRef]
- Cavlar, T.; Deimling, T.; Ablasser, A.; Hopfner, K.P.; Hornung, V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013, 32, 1440–1450. [Google Scholar] [CrossRef]
- Gao, P.; Zillinger, T.; Wang, W.; Ascano, M.; Dai, P.; Hartmann, G.; Tuschl, T.; Deng, L.; Barchet, W.; Patel, D.J. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Rep. 2014, 8, 1668–1676. [Google Scholar] [CrossRef]
- Pan, B.S.; Perera, S.A.; Piesvaux, J.A.; Presland, J.P.; Schroeder, G.K.; Cumming, J.N.; Trotter, B.W.; Altman, M.D.; Buevich, A.V.; Cash, B.; et al. An orally available non-nucleotide STING agonist with antitumor activity. Science 2020, 369, eaba6098. [Google Scholar] [CrossRef]
- Liu, J.; Huang, X.; Ding, J. Identification of MSA-2: An oral antitumor non-nucleotide STING agonist. Signal. Transduct. Target. Ther 2021, 6, 18. [Google Scholar] [CrossRef]
- Chin, E.N.; Yu, C.; Vartabedian, V.F.; Jia, Y.; Kumar, M.; Gamo, A.M.; Vernier, W.; Ali, S.H.; Kissai, M.; Lazar, D.C.; et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science 2020, 369, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tang, M.; Yuan, Z.; Li, Z.; Hu, B.; Bai, X.; Chu, J.; Xu, X.; Zhang, X.Q. Targeted delivery of a STING agonist to brain tumors using bioengineered protein nanoparticles for enhanced immunotherapy. Bioact. Mater. 2022, 16, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Luo, M.; Wang, Z.; Feng, Q.; Wilhelm, J.; Wang, X.; Li, W.; Wang, J.; Cholka, A.; Fu, Y.X.; et al. Prolonged activation of innate immune pathways by a polyvalent STING agonist. Nat. Biomed. Eng. 2021, 5, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Pei, J.; Luo, Q.; Zeng, X.; Li, Q.; Yang, Z.; Quan, J. Identification of alpha-Mangostin as an Agonist of Human STING. ChemMedChem 2018, 13, 2057–2064. [Google Scholar] [CrossRef]
- Luo, M.; Liu, Z.; Zhang, X.; Han, C.; Samandi, L.Z.; Dong, C.; Sumer, B.D.; Lea, J.; Fu, Y.X.; Gao, J. Synergistic STING activation by PC7A nanovaccine and ionizing radiation improves cancer immunotherapy. J. Control. Release 2019, 300, 154–160. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, J.; Zheng, X.; Liu, Z.; Zhang, X.; Li, Y.; Wilhelm, J.; Cao, J.; Huang, G.; Zhang, J.; et al. Intratumoral administration of STING-activating nanovaccine enhances T cell immunotherapy. J. Immunother. Cancer 2022, 10, e003960. [Google Scholar] [CrossRef]
- Boudreau, C.E.; Najem, H.; Ott, M.; Horbinski, C.; Fang, D.; DeRay, C.M.; Levine, J.M.; Curran, M.A.; Heimberger, A.B. Intratumoral Delivery of STING Agonist Results in Clinical Responses in Canine Glioblastoma. Clin. Cancer Res. 2021, 27, 5528–5535. [Google Scholar] [CrossRef]
- Luo, Z.; Ji, Y.; Zhang, D.; Gao, H.; Jin, Z.; Yang, M.; Ying, W. Microbial DNA enrichment promotes liver steatosis and fibrosis in the course of non-alcoholic steatohepatitis. Acta Physiol. 2022, 235, e13827. [Google Scholar] [CrossRef]
- Ma, H.; Kang, Z.; Foo, T.K.; Shen, Z.; Xia, B. Disrupted BRCA1-PALB2 interaction induces tumor immunosuppression and T-lymphocyte infiltration in HCC through cGAS-STING pathway. Hepatology 2022. [Google Scholar] [CrossRef]


| Compound | Mechanism | Reference |
|---|---|---|
| Tetrahydroisoquinoline derivatives (Compound 18) | Target CBD binding pocket | [92] |
| Astin C | Inhibits STING-IRF3 interaction via the binding of the C-terminal domain of STING | [30] |
| SN-011 | Target cyclic dinucleotide binding pocket | [91] |
| NO2-FAs | Suppresses STING palmitoylation | [93] |
| C-176, C-178 | Suppresses murine STING palmitoylation | [29] |
| C170, C-171 | Suppresses murine STING palmitoylation | |
| H-151 | Suppresses human STING palmitoylation | |
| Gelsevirine | Targeting STING for K48 ubiquitination and degradation | [94] |
| Palbociclib | Directly targets STING Y167 | [31] |
| Compound | Phase; Start Time; Notes | Reference/Clinical Trials ID |
|---|---|---|
| MK-1454 | Ⅰ (2017): Intratumoral(IT); solid tumors or lymphomas | NCT03010176; NCT04220866; [103,104] |
| Ⅱ (2020): IT; HNSCC | ||
| MK-2118 | Ⅰ (2017): IT/SQ; solid tumors or lymphomas | NCT03249792 |
| BMS-986301 | Ⅰ (2019): IV; advanced solid cancers | NCT03956680 |
| BI-STING (BI1387446) | Ⅰ (2020): IT; advanced solid cancers | NCT04147234 |
| SB 11285 | Ⅰ (2019): IV; advanced solid tumors: melanoma, HNSCC, and solid tumor | NCT04096638 |
| IMSA-101(GB492) | Ⅰ/Ⅱ (2019): IV; solid tumor | NCT04020185; CTR20211689 |
| Ⅰ (2021): I.V.; adult advanced malignant tumor | ||
| E7766 | Ⅰ (2020): IT; advanced solid tumors or lymphomas | NCT04144140; NCT04109092 |
| Withdrawn (Ⅰ(2020): urinary bladder neoplasms) | ||
| MIW815(ADU-S100) | Terminated: Ⅰ (2016): advanced/metastatic solid tumors or lymphomas | NCT02675439; NCT03172936; NCT03937141 |
| Terminated: Ⅰ(2017): advanced/metastatic solid tumors or lymphomas) | ||
| Terminated: Ⅱ (2019: head and neck cancer) | ||
| Compound 3 | Preclinical: Support IV | [106] |
| a-Mangostin | Preclinical | [114] |
| GSK3745417 | Ⅰ (2019): IV; advanced solid tumors | NCT03843359 |
| BMS-986301 | Ⅰ (2019): IT/IM; advanced solid cancers | NCT03956680 |
| SYNB1891 | Ⅰ (2019): IT; metastatic solid neoplasm lymphoma | NCT04167137 |
| TAK-676 | Ⅰ (2020): IV; advanced solid tumors | NCT04420884; NCT04879849; NCT04541108 |
| Early phase Ⅰ (2021): solid tumor | ||
| SNX281 | Ⅰ (2020): IV; solid tumors and lymphoma | NCT04609579 |
| MSA-1 | Preclinical | No data |
| MSA-2 | Preclinical: orally available; antitumor | |
| SR-001 | Preclinical: pro-drug | [111,112] |
| SR717 | Preclinical: The compound was modified from the structure of SR-012 and was combined with cGAMP at the same position on STING; it could induce the same conformational change of STING and had a stronger response when combined with STING | |
| HG381 | Ⅰ (2021): advanced solid tumors | NCT04998422; CTR20211765; |
| Ⅰ (2021): advanced solid tumors Report: colorectal cancer; a better efficacy than ADB-S100 | ||
| KL340399 | IND (2022: NMPA): IV; advanced solid tumor | CXHL2200223; CXHL2200224 |
| NOX-66 | Completed: Ⅰ/Ⅱ (2017): Cancer | NCT02941523; NCT04957290 |
| Ⅰ/Ⅱ (2021): metastatic castration-resistant prostate cancer and other solid tumors | ||
| PC7A | Preclinical; form biomolecular condensates; intratumoral administration | [113,115,116] |
| IACS-8779 | Preclinical (2021); IT; Canine glioblastoma | [117] |
| TAK-500 | Recruiting:Ⅰ(2022), IV, HCC/Pancreatic cancer | NCT05070247 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, J.; Xing, C.; Chen, Y.; Bian, H.; Lv, N.; Wang, Z.; Liu, M.; Su, L. The STING in Non-Alcoholic Fatty Liver Diseases: Potential Therapeutic Targets in Inflammation-Carcinogenesis Pathway. Pharmaceuticals 2022, 15, 1241. https://doi.org/10.3390/ph15101241
Lv J, Xing C, Chen Y, Bian H, Lv N, Wang Z, Liu M, Su L. The STING in Non-Alcoholic Fatty Liver Diseases: Potential Therapeutic Targets in Inflammation-Carcinogenesis Pathway. Pharmaceuticals. 2022; 15(10):1241. https://doi.org/10.3390/ph15101241
Chicago/Turabian StyleLv, Juan, Chunlei Xing, Yuhong Chen, Huihui Bian, Nanning Lv, Zhibin Wang, Mingming Liu, and Li Su. 2022. "The STING in Non-Alcoholic Fatty Liver Diseases: Potential Therapeutic Targets in Inflammation-Carcinogenesis Pathway" Pharmaceuticals 15, no. 10: 1241. https://doi.org/10.3390/ph15101241
APA StyleLv, J., Xing, C., Chen, Y., Bian, H., Lv, N., Wang, Z., Liu, M., & Su, L. (2022). The STING in Non-Alcoholic Fatty Liver Diseases: Potential Therapeutic Targets in Inflammation-Carcinogenesis Pathway. Pharmaceuticals, 15(10), 1241. https://doi.org/10.3390/ph15101241







