Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine
Abstract
1. Introduction
2. Results
2.1. Phenotypic Parameters of Lifecycle
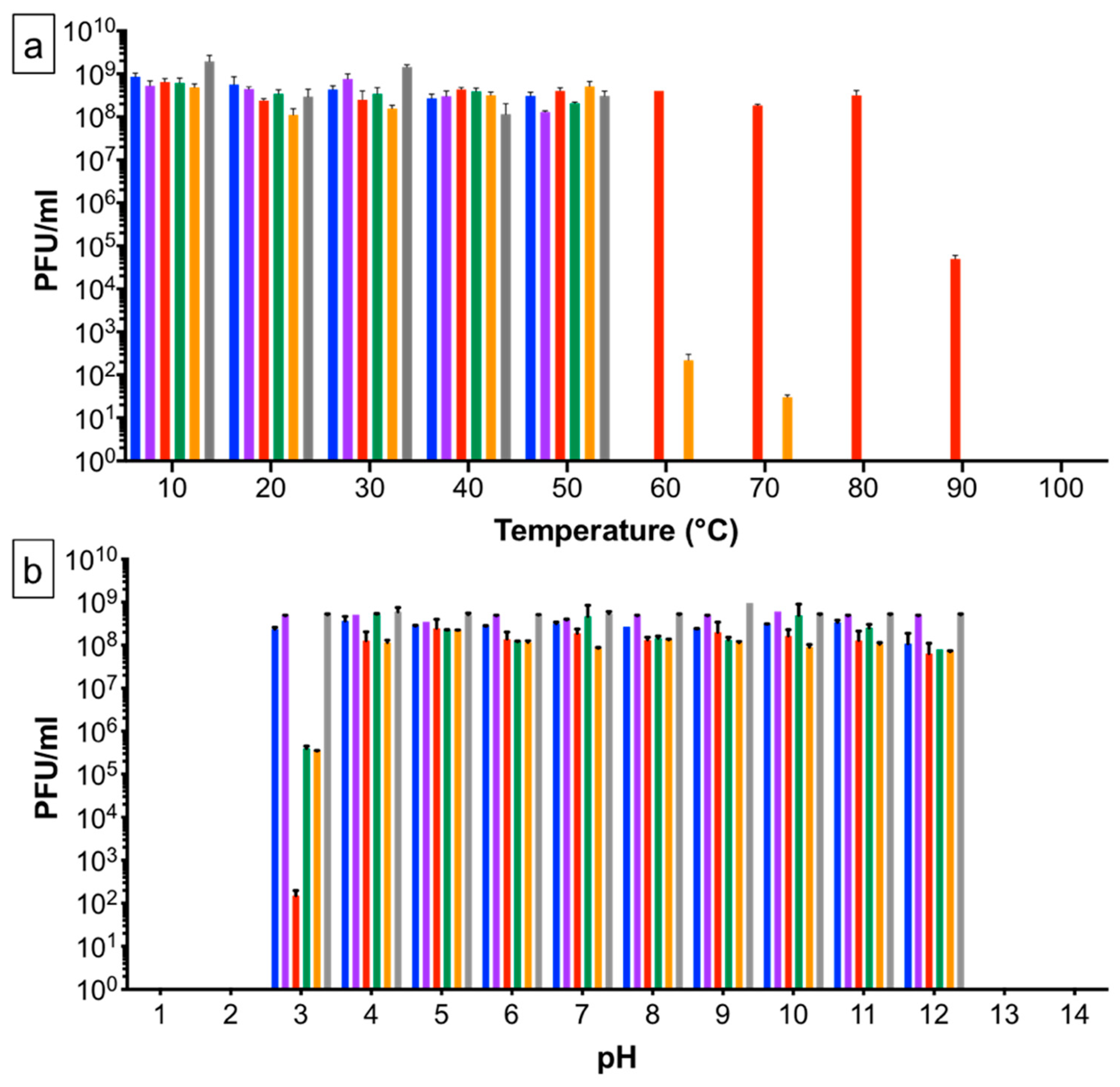
2.2. Temperature and pH Stability of Phages
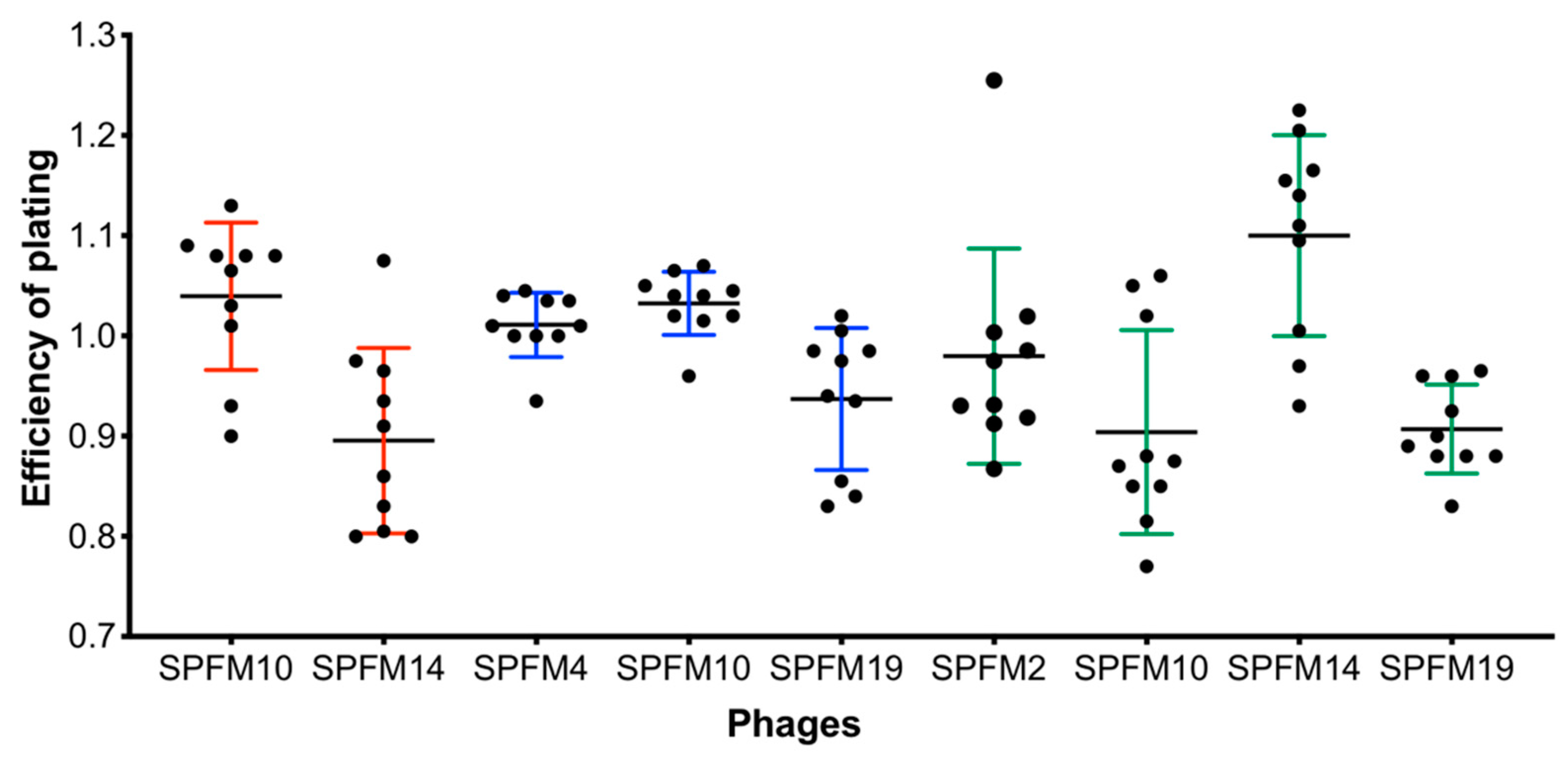
2.3. Identification of the Bacterial Receptor
2.4. Genetic Differences between the Six SPFM Phages
2.5. Infection Dynamics of Six SPFM Phages In Vitro
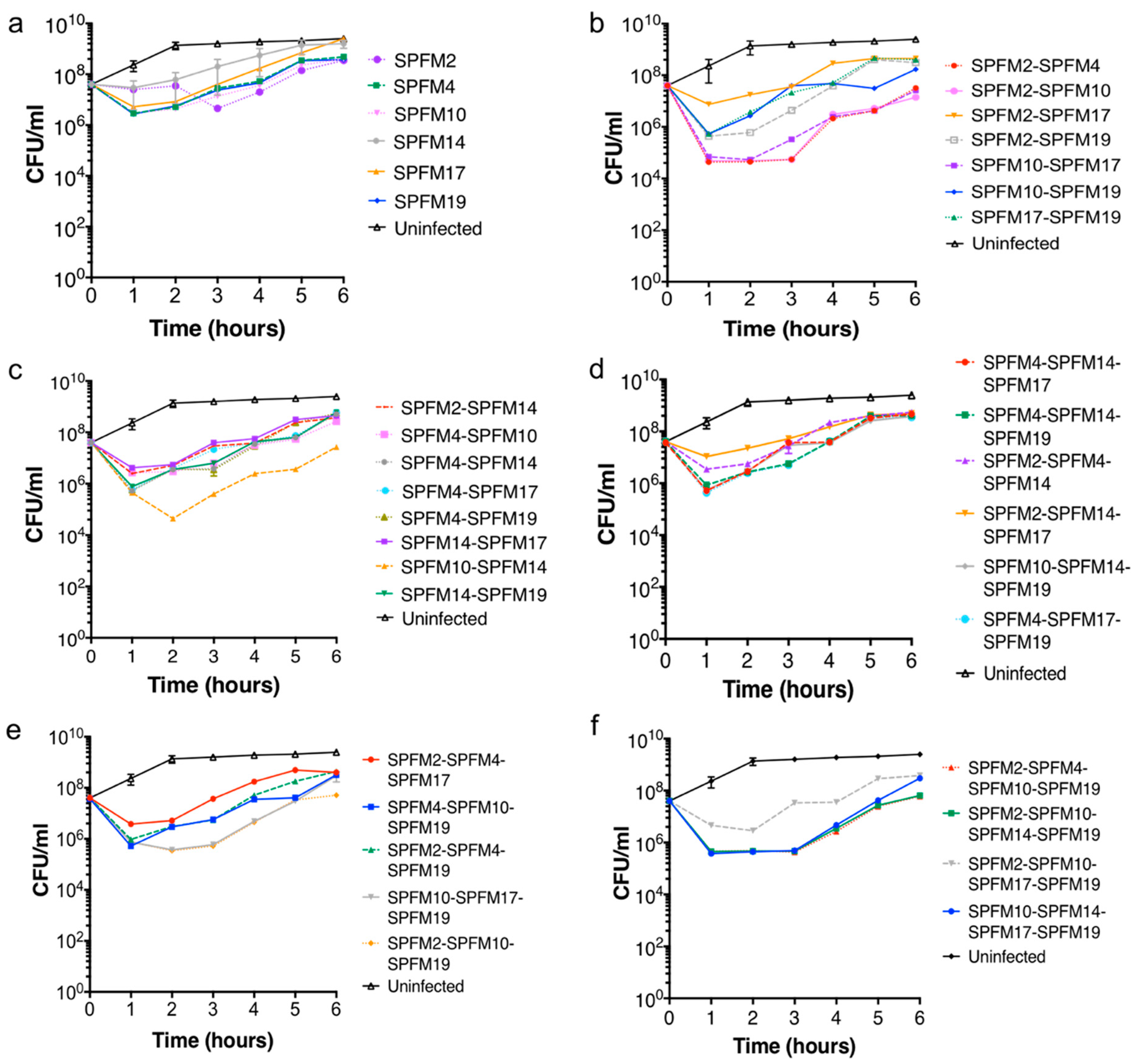
2.5.1. Bacterial killing Assays with Single Phage Suspensions
2.5.2. Efficacy of Multiple Phage Cocktails
2.6. Infection Dynamics of Phage Cocktails In Vivo
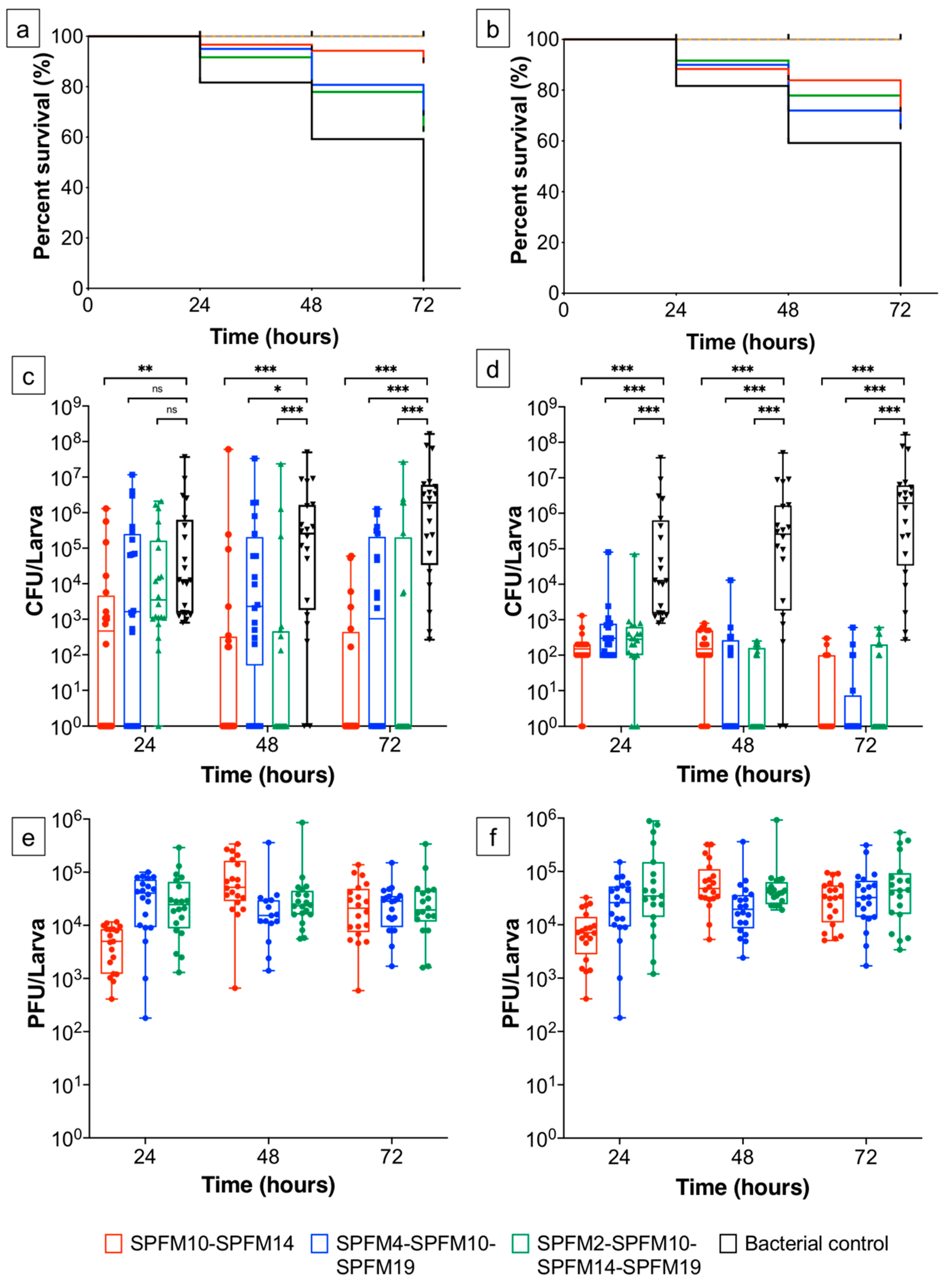
2.6.1. Prophylactic Treatment of Phage Cocktails
2.6.2. Simultaneous Phage Treatment of Infected Larvae
2.6.3. Sensitivity of Recovered Salmonella Colonies after Exposure to Phage Cocktails in a Larvae Model
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Phages and their Genome Accession Numbers
4.3. Phage Propagation and Titration
4.4. One-Step Growth Assay
4.5. Phage Receptor Analysis
4.6. Temperature and pH Stability Assays
4.7. Single-Nucleotide Polymorphism (SNP) and Indels Analysis
4.8. In Vitro Bacterial Killing Assay with Individual Phages and Phage Cocktails
4.9. Testing Efficacy of Phage Cocktails in the In Vivo Galleria Mellonella Infection Model
4.9.1. Preparation of Galleria Mellonella
4.9.2. Salmonella Infected G. mellonella Treated with Phage Cocktails
4.9.3. Larvae Survival Curves and Statistical Analysis
4.9.4. Testing Development of Phage Resistance In Vivo
4.10. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global Burden of Invasive Nontyphoidal. Emerg. Infect. Dis. 2015, 21, 941–949. [Google Scholar] [CrossRef]
- Torgerson, P.R.; Devleesschauwer, B.; Praet, N.; Speybroeck, N.; Willingham, A.L.; Kasuga, F.; Rokni, M.B.; Zhou, X.N.; Fèvre, E.M.; Sripa, B.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001920. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.; Larose, T.; O’Brien, S. Costed Extension to the Second Study of Infectious Intestinal Disease in the Community: Identifying the Proportion of Foodborne Disease in the UK and Attributing Foodborne Disease by Food Commodity; UK Food Standards Agency: London, UK, 2014. [Google Scholar]
- Pires, S.M.; de Knegt, L.; Hald, T. Scientific/technical report submitted to EFSA. Estimation of the relative contribution of different food and animal sources to human Salmonella infections in the European Union. Natl. Food Inst. 2011, 8, 1. [Google Scholar]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef]
- Tassinari, E.; Du, G.; Bawn, M.; Burgess, C.M.; Mccabe, E.M.; Lawlor, P.G.; Gardiner, G.; Kingsley, R.A. Microevolution of antimicrobial resistance and biofilm formation of Salmonella Typhimurium during persistence on pig farms. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered From the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef]
- Campos, J.; Mourao, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the pig production chain: A comprehensive analysis of its impact on human health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, May 2016. [CrossRef]
- Borie, C.; Robeson, J.; Galarce, N. Lytic bacteriophages in Veterinary Medicine: A therapeutic option against bacterial pathogens. Arch. Med. Vet. 2014, 46, 167–179. [Google Scholar] [CrossRef][Green Version]
- Carvalho, C.; Costa, A.R.; Silva, F.; Oliveira, A. Bacteriophages and their derivatives for the treatment and control of food-producing animal infections. Crit. Rev. Microbiol. 2017, 43, 583–601. [Google Scholar] [CrossRef] [PubMed]
- Albino, L.A.A.; Rostagno, M.H.; Hungaro, H.M.; Mendonca, R.C.S. Isolation, characterization, and application of bacteriophages for Salmonella spp. biocontrol in pigs. Foodborne Pathog. Dis. 2014, 11, 602–609. [Google Scholar] [CrossRef]
- Callaway, T.R.; Edrington, T.S.; Brabban, A.; Kutter, B.; Karriker, L.; Stahl, C.; Wagstrom, E.; Anderson, R.; Poole, T.L.; Genovese, K.; et al. Evaluation of Phage Treatment as a Strategy to Reduce Salmonella populations in growing swine. Foodborne Pathog. Dis. 2011, 8, 261–266. [Google Scholar] [CrossRef]
- Saez, A.C.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Direct feeding of microencapsulated bacteriophages to reduce Salmonella colonization in pigs. Foodborne Pathog. Dis. 2011, 8, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Wall, S.K.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl. Environ. Microbiol. 2010, 76, 48–53. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, G.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P. The newly isolated lytic bacteriophages st104a and st104b are highly virulent against Salmonella enterica. J. Appl. Microbiol. 2006, 101, 251–259. [Google Scholar] [CrossRef]
- Lee, N.; Harris, D. The effect of bacteriophage treatment to reduce the rapid dissemination of Salmonella typhimurium in pigs. Swine Res. Report 2001, 50, 196–197. [Google Scholar]
- Seo, B.J.; Song, E.T.; Lee, K.; Kim, J.W.; Jeong, C.G.; Moon, S.H.; Son, J.S.; Kang, S.H.; Cho, H.S.; Jung, B.Y.; et al. Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella Typhimurium. J. Vet. Med. Sci. 2018, 80, 851–860. [Google Scholar] [CrossRef]
- Hooton, S.P.T.; Atterbury, R.J.; Connerton, I.F. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 2011, 151, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Schmidt, K.; Marks, D.; Hatter, S.; Marshall, A.; Albino, L.; Ebner, P. Treatment of Salmonella-Contaminated Eggs and Pork with a Broad-Spectrum, Single Bacteriophage: Assessment of Efficacy and Resistance Development. Foodborne Pathog. Dis. 2016, 13, 679–688. [Google Scholar] [CrossRef]
- Spricigo, D.A.; Bardina, C.; Cortés, P.; Llagostera, M. Use of a bacteriophage cocktail to control Salmonella in food and the food industry. Int. J. Food Microbiol. 2013, 165, 169–174. [Google Scholar] [CrossRef]
- Rosentrater, K.A.; Evers, A.D. Feed and industrial uses for cereals. In Kent’s Technololgy of Cereals, 5th ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 785–837. ISBN 9780081005293. [Google Scholar]
- Henze, L.J.; Koehl, N.J.; Bennett-Lenane, H.; Holm, R.; Grimm, M.; Schneider, F.; Weitschies, W.; Koziolek, M.; Griffin, B.T. Characterization of gastrointestinal transit and luminal conditions in pigs using a telemetric motility capsule. Eur. J. Pharm. Sci. 2021, 156, 105627. [Google Scholar] [CrossRef] [PubMed]
- Thanki, A.M.; Hooton, S.P.T.; Gigante, A.M.; Atterbury, R.J.; Clokie, M.R.J. Potential roles for bacteriophages in reducing Salmonella from poultry and swine. In Salmonella-a Challenge From Farm to Fork; Intechopen: London, UK, 2021. [Google Scholar]
- Ahmad, A.A.; Addy, H.S.; Huang, Q. Biological and Molecular Characterization of a Jumbo Bacteriophage Infecting Plant Pathogenic Ralstonia solanacearum Species Complex Strains. Front. Microbiol. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Wang, L.; Tavanti, A.; Trampuz, A.; Di Luca, M. Antibacterial Efficacy of Two Commercially Available Bacteriophage Formulations, Staphylococcal Bacteriophage and PYO Bacteriophage, Against Methicillin-Resistant Staphylococcus aureus: Prevention and Eradication of Biofilm Formation and Control of a Syst. Front. Microbiol. 2020, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Abbasifar, R.; Kropinski, A.M.; Sabour, P.M.; Chambers, J.R.; MacKinnon, J.; Malig, T.; Griffiths, M.W. Efficiency of bacteriophage therapy against Cronobacter sakazakii in Galleria mellonella (greater wax moth) larvae. Arch. Virol. 2014, 159, 2253–2261. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.M.; Marmo, P.; Henrici De Angelis, L.; Palmieri, M.; Ciacci, N.; Di Lallo, G.; Demattè, E.; Vannuccini, E.; Lupetti, P.; Rossolini, G.M.; et al. φbO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Sci. Rep. 2017, 7, 2614. [Google Scholar] [CrossRef]
- Jeon, J.; Park, J.H.; Yong, D. Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 2019, 19, 70. [Google Scholar] [CrossRef]
- Nale, J.Y.; Vinner, G.K.; Lopez, V.C.; Thanki, A.M.; Phothaworn, P.; Thiennimitr, P.; Garcia, A.; AbuOun, M.; Anjum, M.F.; Korbsrisate, S.; et al. An Optimized Bacteriophage Cocktail Can Effectively Control Salmonella in vitro and in Galleria mellonella. Front. Microbiol. 2021, 11, 11. [Google Scholar] [CrossRef]
- Cools, F.; Torfs, E.; Aizawa, J.; Vanhoutte, B.; Maes, L.; Caljon, G.; Delputte, P.; Cappoen, D.; Cos, P. Optimization and characterization of a Galleria mellonella larval infection model for virulence studies and the evaluation of therapeutics against Streptococcus pneumoniae. Front. Microbiol. 2019, 10, 311. [Google Scholar] [CrossRef]
- Beeton, M.L.; Alves, D.R.; Enright, M.C.; Jenkins, A.T.A. Assessing phage therapy against Pseudomonas aeruginosa using a Galleria mellonella infection model. Int. J. Antimicrob. Agents 2015, 46, 196–200. [Google Scholar] [CrossRef]
- Thanki, A.M.; Brown, N.; Millard, A.D.; Clokie, M.R.J. Genomic Characterization of Jumbo Salmonella Phages That Effectively Target United Kingdom Salmonella Serotypes. 2019, 10, 1419. Front. Microbiol. 2019, 10, 1419. [Google Scholar] [CrossRef]
- Kiarie, E.G.; Mills, A. Role of feed processing on gut health and function in pigs and poultry: Conundrum of optimal particle size and hydrothermal regimens. Front. Vet. Sci. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Lee, J.H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica Serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef]
- Card, R.; Vaughan, K.; Bagnall, M.; Spiropoulos, J.; Cooley, W.; Strickland, T.; Davies, R.; Anjum, M.F. Virulence characterisation of Salmonella enterica isolates of differing antimicrobial resistance recovered from UK livestock and imported meat samples. Front. Microbiol. 2016, 7, 640. [Google Scholar] [CrossRef]
- Topka, G.; Bloch, S.; Jurczak-kurek, A.; Necel, A.; Dydecka, A.; Richert, M. Characterization of Bacteriophage vB-EcoS-95, Isolated From Urban Sewage and Revealing Extremely Rapid Lytic Development. Front. Microbiol. 2018, 9, 3326. [Google Scholar] [CrossRef]
- Yang, H.; Liang, L.; Lin, S.; Jia, S. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010, 10, 131. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages-review. Folia Microbiol. (Praha.) 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.S.Y.; Parumasivam, T.; Gao, F.G.; Carter, E.A.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Britton, W.J.; Kutter, E.; et al. Effects of storage conditions on the stability of spray dried, inhalable bacteriophage powders. Int. J. Pharm. 2017, 521, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.Y.; Wong, J.; Mathai, A.; Morales, S.; Kutter, E.; Britton, W.; Li, J.; Chan, H.K. Production of highly stable spray dried phage formulations for treatment of Pseudomonas aeruginosa lung infection. Eur. J. Pharm. Biopharm. 2017, 121, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vandenheuvel, D.; Singh, A.; Vandersteegen, K.; Klumpp, J.; Lavigne, R.; Van Den Mooter, G. Feasibility of spray drying bacteriophages into respirable powders to combat pulmonary bacterial infections. Eur. J. Pharm. Biopharm. 2013, 84, 578–582. [Google Scholar] [CrossRef]
- Merchant, H.A.; McConnell, E.L.; Liu, F.; Ramaswamy, C.; Kulkarni, R.P.; Basit, A.W.; Murdan, S. Assessment of gastrointestinal pH, fluid and lymphoid tissue in the guinea pig, rabbit and pig, and implications for their use in drug development. Eur. J. Pharm. Sci. 2011, 42, 3–10. [Google Scholar] [CrossRef]
- Vinner, G.K.; Richards, K.; Leppanen, M.; Sagona, A.P.; Malik, D.J. Microencapsulation of enteric bacteriophages in a pH-Responsive solid oral dosage formulation using a scalable membrane emulsification process. Pharmaceutics 2019, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Bardina, C.; Spricigo, D.A.; Cortés, P.; Llagostera, M. Significance of the bacteriophage treatment schedule in reducing salmonella colonization of poultry. Appl. Environ. Microbiol. 2012, 78, 6600–6607. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Chutia, M.; Carr, P.; Hickenbotham, P.T.; Clokie, M.R.J. “Get in early”; Biofilm and wax moth (Galleria mellonella) models reveal new insights into the therapeutic potential of Clostridium difficile bacteriophages. Front. Microbiol. 2016, 7, 1383. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.P.; Pirnay, J.P.; Buckling, A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Im, J.; Lee, J.S.; Jeon, H.J.; Mogeni, O.D.; Kim, J.H.; Rakotozandrindrainy, R.; Baker, S.; Marks, F. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunother. 2019, 15, 1421–1426. [Google Scholar] [CrossRef]
- Cuff, J.A.; Clamp, M.E.; Siddiqui, A.S.; Finlay, M.; Barton, G.J. JPred: A consensus secondary structure prediction server. Bioinformatics 1998, 14, 892–893. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinforma. 2017, 60, 3–15. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. Trabajo práctico No 13. Varianzas en función de variable independiente categórica. Nat. Protoc. 2016, 10, 845–858. [Google Scholar] [CrossRef]
- Bork, P.; Doolittle, R.F. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl. Acad. Sci. 1992, 89, 8990–8994. [Google Scholar] [CrossRef]
- Little, E.; Bork, P.; Doolittle, R.F. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J. Mol. Evol. 1994, 39, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Nair, S.; Pallas, J.; Williams, M.A. Fibronectin: A multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 2011, 35, 147–200. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.S.; Yu, Z.; Maxwell, K.L.; Davidson, A.R. Ig-Like Domains on Bacteriophages: A Tale of Promiscuity and Deceit. J. Mol. Biol. 2006, 359, 496–507. [Google Scholar] [CrossRef]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral Mutation Rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed]
- Kupczok, A.; Neve, H.; Huang, K.D.; Hoeppner, M.P.; Heller, K.J.; Franz, C.M.A.P.; Dagan, T. Rates of mutation and recombination in siphoviridae phage genome evolution over three decades. Mol. Biol. Evol. 2018, 35, 1147–1159. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical advice on the one-step growth curve. Methods Mol. Biol. 2018, 1681, 41–47. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriphages using the small drop plaque assay system. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 81–85. ISBN 978-1-58829-682-5. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
| Efficiency of Plating 1 | |||
|---|---|---|---|
| Phages | SL1344 ΔbutB | SL1344 ΔflgK | SL1344 ΔrfaL |
| SPFM2 | 1.00 | 0.99 | 0 |
| SPFM4 | 0.99 | 1.00 | 0 |
| SPFM10 | 0.98 | 1.00 | 0 |
| SPFM14 | 1.00 | 1.00 | 0 |
| SPFM17 | 0.99 | 1.00 | 0 |
| SPFM19 | 1.00 | 1.00 | 0 |
| Phage | Position in the Genome of SPFM10 (bp) | Locus Tag in SPFM10 1 | Protein Annotation | Base Change | Amino Acid Change | p Value |
|---|---|---|---|---|---|---|
| SPFM2 | 70890 | SPFM10_00081 | Hypothetical protein | A -> T | Thr -> Ser | 1.05 × 10−38 |
| 74294 | SPFM10_00084 | Hypothetical protein | A -> C | Ser -> Leu | 1.70 × 10−43 | |
| 74298 | SPFM10_00084 | Hypothetical protein | G -> T | Ser -> Leu | 4.28 × 10−44 | |
| SPFM4 | 70890 | SPFM10_00081 | Hypothetical protein | A -> T | Thr -> Ser | 2.20 × 10−144 |
| 74291 | SPFM10_00084 | Hypothetical protein | A -> G | Pro -> Leu | 1.34 × 10−238 | |
| 74295 | SPFM10_00084 | Hypothetical protein | G -> T | Ser -> Arg | 3.22 × 10−247 | |
| SPFM14 | 70890 | SPFM10_00081 | Hypothetical protein | A -> T | Thr -> Ser | 5.14 × 10−16 |
| SPFM17 | 57985 | SPFM10_00067 | Chromosome partition protein Smc | A- > G | Gin -> Arg | 1.73 × 10−49 |
| 70890 | SPFM10_00081 | Hypothetical protein | A -> T | Thr -> Ser | 2.77 × 10−66 | |
| 74291 | SPFM10_00084 | Hypothetical protein | G -> A | Pro -> Leu | 6.66 × 10−94 | |
| 74295 | SPFM10_00084 | Hypothetical protein | T -> G | Ser -> Arg | 1.06 × 10−92 | |
| 113292 | SPFM10_00140 | Hypothetical protein | A -> G | Cys -> Cys | 1.10 × 10−65 | |
| SPFM19 | 70890 | SPFM10_00081 | Hypothetical protein | A- > T | Thr -> Ser | 1.55 × 10−50 |
| 72949 | SPFM10_00083 | Hypothetical protein | C -> T | Ser -> Gly | 1.74 × 10−52 |
| Phage or Phages in Cocktail | |
|---|---|
| Single phages | SPFM2 |
| SPFM4 | |
| SPFM10 | |
| SPFM14 | |
| SPFM17 | |
| SPFM19 | |
| Two-phage cocktails | SPFM2-SPFM14 |
| SPFM4-SPFM10 | |
| SPFM4-SPFM14 | |
| SPFM4-SPFM17 | |
| SPFM4-SPFM19 | |
| SPFM10-SPFM14 | |
| SPFM14-SPFM17 | |
| SPFM14-SPFM19 | |
| SPFM2-SPFM4 | |
| SPFM2-SPFM10 | |
| SPFM2-SPFM17 | |
| SPFM2-SPFM19 | |
| SPFM10-SPFM17 | |
| SPFM10-SPFM19 | |
| SPFM17-SPFM19 | |
| Three-phage cocktails | SPFM4-SPFM14-SPFM17 |
| SPFM4-SPFM14-SPFM19 | |
| SPFM2-SPFM4-SPFM14 | |
| SPFM2-SPFM14-SPFM17 | |
| SPFM10-SPFM14-SPFM19 | |
| SPFM4-SPFM17-SPFM19 | |
| SPFM2-SPFM4-SPFM17 | |
| SPFM4-SPFM10-SPFM19 | |
| SPFM2-SPFM4-SPFM19 | |
| SPFM10-SPFM17-SPFM19 | |
| SPFM2-SPFM10-SPFM19 | |
| Four-phage cocktails | SPFM10-SPFM14-SPFM17-SPFM19 |
| SPFM2-SPFM10-SPFM14-SPFM19 | |
| SPFM2-SPFM4-SPFM10-SPFM19 | |
| SPFM2-SPFM10-SPFM17-SPFM19 |
| Groups 1 | Description of Larvae Groups 2 |
|---|---|
| Controls | |
| C-1 | Healthy larvae |
| C-2 | Administered PBS |
| C-3 | Administered the 2-phage cocktail SPFM10-SPFM14 |
| C-4 | Administered the 3-phage cocktail SPFM4-SPFM10-SPFM19 |
| C-5 | Administered the 4-phage cocktail SPFM2-SPFM10-SPFM14-SPFM19 |
| C-6 | Challenged with Salmonella |
| Prophylactic phage treatments | |
| P-2 | Administered 2-phage cocktail SPFM10-SPFM14 one hour prior to challenge with Salmonella |
| P-3 | Administered 3-phage cocktail SPFM4-SPFM10-SPFM19 one hour prior to challenge with Salmonella |
| P-4 | Administered 4-phage cocktail SPFM2-SPFM10-SPFM14-SPFM19 one hour prior to challenge with Salmonella |
| Co-infection studies | |
| CoI-2 | Administered 2-phage cocktail SPFM10-SPFM14 simultaneously with Salmonella |
| CoI-3 | Administered 3-phage cocktail SPFM4-SPFM10-SPFM19 simultaneously with Salmonella |
| CoI-4 | Administered 4-phage cocktail SPFM2-SPFM10-SPFM14-SPFM19 simultaneously with Salmonella |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thanki, A.M.; Clavijo, V.; Healy, K.; Wilkinson, R.C.; Sicheritz-Pontén, T.; Millard, A.D.; Clokie, M.R.J. Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine. Pharmaceuticals 2022, 15, 58. https://doi.org/10.3390/ph15010058
Thanki AM, Clavijo V, Healy K, Wilkinson RC, Sicheritz-Pontén T, Millard AD, Clokie MRJ. Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine. Pharmaceuticals. 2022; 15(1):58. https://doi.org/10.3390/ph15010058
Chicago/Turabian StyleThanki, Anisha M., Viviana Clavijo, Kit Healy, Rachael C. Wilkinson, Thomas Sicheritz-Pontén, Andrew D. Millard, and Martha R. J. Clokie. 2022. "Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine" Pharmaceuticals 15, no. 1: 58. https://doi.org/10.3390/ph15010058
APA StyleThanki, A. M., Clavijo, V., Healy, K., Wilkinson, R. C., Sicheritz-Pontén, T., Millard, A. D., & Clokie, M. R. J. (2022). Development of a Phage Cocktail to Target Salmonella Strains Associated with Swine. Pharmaceuticals, 15(1), 58. https://doi.org/10.3390/ph15010058






