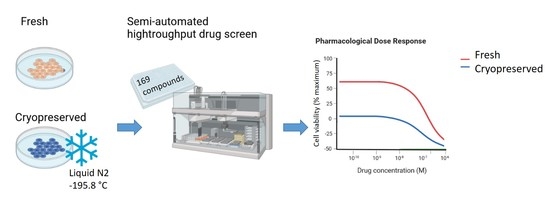
Testing the Stability of Drug Resistance on Cryopreserved, Gene-Engineered Human Induced Pluripotent Stem Cells
Abstract
:1. Introduction
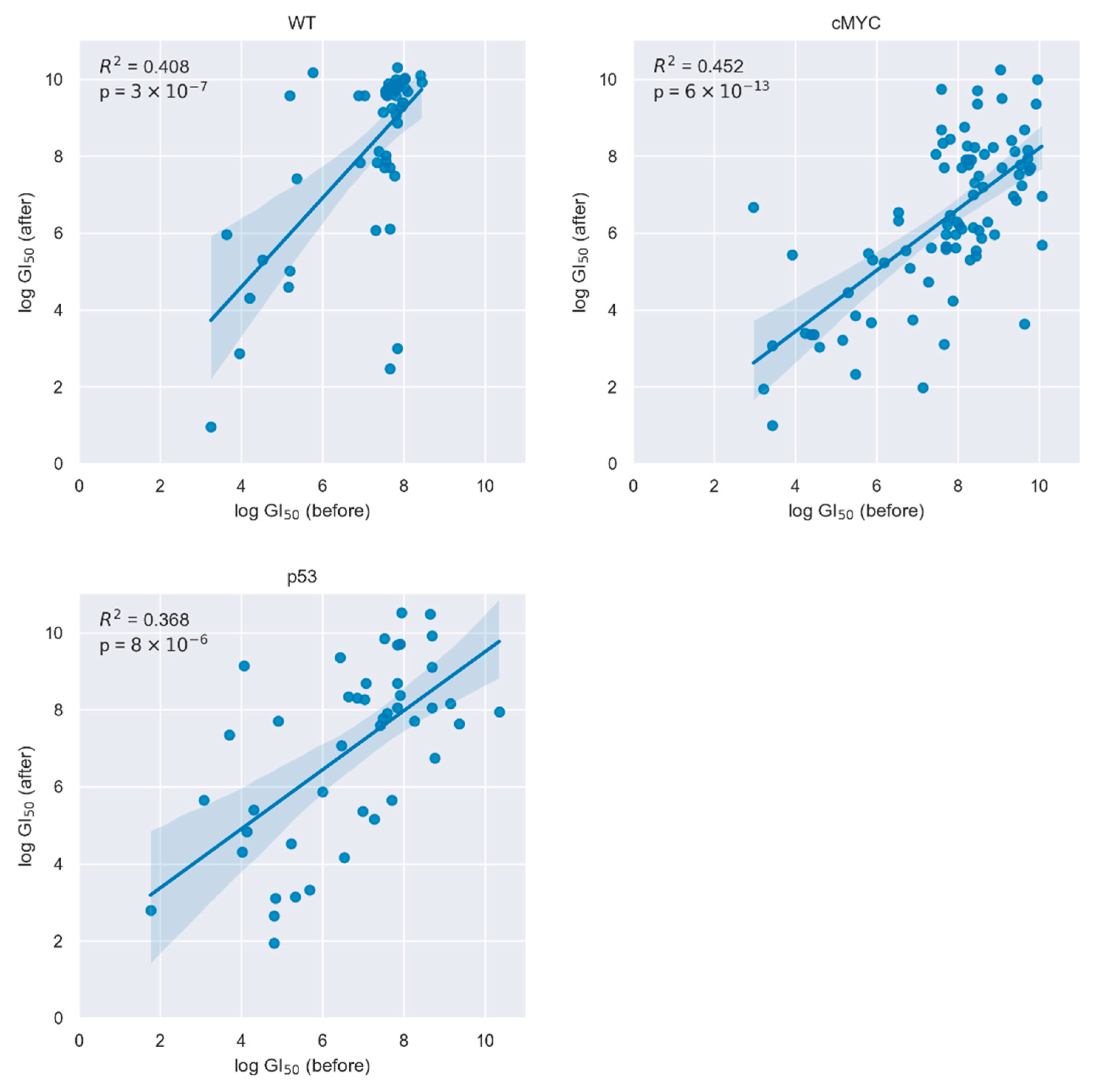
2. Results
3. Discussion
4. Material and Methods
4.1. Cell Growth and Cell Models
4.2. Cryopreservation Procedure
4.3. In Vitro Pharmacology Testing
4.4. Statistical Evaluation of Repeatability of Drug Responses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prinz, F.; Schlange, T.; Asadullah, K. Believe it or not: How much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 2011, 10, 712. [Google Scholar] [CrossRef] [Green Version]
- Haibe-Kains, B.; El-Hachem, N.; Birkbak, N.J.; Jin, A.C.; Beck, A.H.; Aerts, H.J.W.L.; Quackenbush, J. Inconsistency in large pharmacogenomic studies. Nature 2013, 504, 389–393. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Koga, T.; Chaim, I.A.; Benitez, J.A.; Markmiller, S.; Parisian, A.D.; Hevner, R.F.; Turner, K.M.; Hessenauer, F.M.; D’Antonio, M.; Nguyen, N.D.; et al. Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat. Commun. 2020, 11, 550. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Martinez, I.; Nivet, E.; Xia, Y.; Hishida, T.; Aguirre, A.; Ocampo, A.; Ma, L.; Morey, R.; Krause, M.N.; Zembrzycki, A.; et al. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat. Commun. 2016, 7, 10743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrjardi, N.Z.; Hänggi, D.; Kahlert, U.D. Current biomarker-associated procedures of cancer modeling-a reference in the context of IDH1 mutant glioma. Cell Death Dis. 2020, 11, 998. [Google Scholar] [CrossRef]
- De Sousa, P.A.; Steeg, R.; Wachter, E.; Bruce, K.; King, J.; Hoeve, M.; Khadun, S.; McConnachie, G.; Holder, J.; Kurtz, A.; et al. Rapid establishment of the European Bank for induced Pluripotent Stem Cells (EBiSC)-the Hot Start experience. Stem Cell Res. 2017, 20, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Wakeman, D.R.; Hiller, B.M.; Marmion, D.J.; McMahon, C.W.; Corbett, G.T.; Mangan, K.P.; Ma, J.; Little, L.E.; Xie, Z.; Perez-Rosello, T.; et al. Cryopreservation Maintains Functionality of Human iPSC Dopamine Neurons and Rescues Parkinsonian Phenotypes In Vivo. Stem Cell Rep. 2017, 9, 149–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, H.K.; Faubion, M.G.; Hjortness, M.K.; Palecek, S.P.; Shusta, E.V. Cryopreservation of Brain Endothelial Cells Derived from Human Induced Pluripotent Stem Cells Is Enhanced by Rho-Associated Coiled Coil-Containing Kinase Inhibition. Tissue Eng. Part C Methods 2016, 22, 1085–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, N.J.; Singh Dolt, K.; Canham, M.A.; Kilbride, P.; Morris, G.J.; Kunath, T. Cryopreservation of Human Midbrain Dopaminergic Neural Progenitor Cells Poised for Neuronal Differentiation. Front. Cell Dev. Biol. 2020, 8, 1279. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; DeStefano, J.G.; Nerenberg, R.F.; Grifno, G.N.; Ye, R.; Gallagher, E.; Searson, P.C. Long-Term Cryopreservation Preserves Blood–Brain Barrier Phenotype of iPSC-Derived Brain Microvascular Endothelial Cells and Three-Dimensional Microvessels. Mol. Pharm. 2020, 17, 3425–3434. [Google Scholar] [CrossRef] [PubMed]
- Van den Brink, L.; Brandão, K.O.; Yiangou, L.; Mol, M.P.H.; Grandela, C.; Mummery, C.L.; Verkerk, A.O.; Davis, R.P. Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Res. 2020, 43, 101698. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.J. Technical Considerations in the Freezing, Low-Temperature Storage and Thawing of Stem Cells for Cellular Therapies. Transfus. Med. Hemotherapy 2019, 46, 134–150. [Google Scholar] [CrossRef] [PubMed]
- Marotta, D.; Rao, C.; Fossati, V. Human Induced Pluripotent Stem Cell (iPSC) Handling Protocols: Maintenance, Expansion, and Cryopreservation. In Methods in Molecular Biology; Springer: New York, NY, USA; pp. 1–15.
- Horiguchi, I.; Kino-oka, M. Current Developments in the Stable Production of Human Induced Pluripotent Stem Cells. Engineering 2021, 7, 144–152. [Google Scholar] [CrossRef]
- Shafa, M.; Walsh, T.; Panchalingam, K.M.; Richardson, T.; Menendez, L.; Tian, X.; Suresh Babu, S.; Dadgar, S.; Beller, J.; Yang, F.; et al. Long-Term Stability and Differentiation Potential of Cryopreserved cGMP-Compliant Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2019, 21, 108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Z.; Belbachir, N.; Zhang, T.; Liu, Y.; Shrestha, R.; Wu, J.C. Effects of Cryopreservation on Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Assessing Drug Safety Response Profiles. Stem Cell Rep. 2021, 16, 168–181. [Google Scholar] [CrossRef]
- Lee, J.-K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- Chen, Y.; Tristan, C.A.; Chen, L.; Jovanovic, V.M.; Malley, C.; Chu, P.-H.; Ryu, S.; Deng, T.; Ormanoglu, P.; Tao, D.; et al. A versatile polypharmacology platform promotes cytoprotection and viability of human pluripotent and differentiated cells. Nat. Methods 2021, 18, 528–541. [Google Scholar] [CrossRef]
- Işbilir, A.; Möller, J.; Arimont, M.; Bobkov, V.; Perpiñá-Viciano, C.; Hoffmann, C.; Inoue, A.; Heukers, R.; De Graaf, C.; Smit, M.J.; et al. Advanced fluorescence microscopy reveals disruption of dynamic CXCR4 dimerization by subpocket-specific inverse agonists. Proc. Natl. Acad. Sci. USA 2020, 117, 29144–29154. [Google Scholar] [CrossRef]
- Hewera, M.; Nickel, A.-C.; Knipprath, N.; Muhammad, S.; Fan, X.; Steiger, H.-J.; Hänggi, D.; Kahlert, U.D. An inexpensive and easy-to-implement approach to a Quality Management System for an academic research lab. F1000Research 2020, 9, 660. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, C.; Kuhn, L.-M.; Tigges, J.; Fritsche, E.; Kahlert, U.D. Efficient Modulation of TP53 Expression in Human Induced Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2020, 52, e102. [Google Scholar] [CrossRef] [PubMed]
- Hewera, M.; Hänggi, D.; Gerlach, B.; Kahlert, U.D. eLabFTW as an Open Science tool to improve the quality and translation of preclinical research. F1000Research 2021, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Toscano, A.; Khan, D.; Nickel, A.-C.; Hewera, M.; Kamp, M.A.; Fischer, I.; Steiger, H.-J.; Zhang, W.; Muhammad, S.; Hänggi, D.; et al. Robot technology identifies a Parkinsonian therapeutics repurpose to target stem cells of glioblastoma. CNS Oncol. 2020, 9, CNS58. [Google Scholar] [CrossRef]
- Skipper, S.; Perktold, J. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 92–96. [Google Scholar]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
| Stable (Drugs That Show up in Top 20 List before and after Freezing) | Unstable (Drugs That Show up as Top 20 Performer Only before Freezing Whereas after That They Fall behind the Threshold) | |||
|---|---|---|---|---|
| Drug Name | Rank before Freezing | Rank after Freezing | Drug | Rank before Freezing |
| Cabazitaxel | 3 | 3 | Ixabepilone | 1 |
| Docetaxel | 4 | 12 | Decitabine | 2 |
| Topotecan hydrochloride | 5 | 5 | Gemcitabine | 6 |
| Erlotinib | 7 | 7 | Nintedanib Ethanesulfonate Salt | 9 |
| Homoharringtonine | 8 | 11 | Mitoxantrone hydrochloride | 12 |
| Podophyllotoxin | 10 | 8 | Vinblastine sulfate | 13 |
| Imiquimod | 11 | 10 | Paclitaxel | 15 |
| Acalabrutinib | 14 | 15 | Romidepsin | 16 |
| Nintedanib | 17 | |||
| Carfilzomib | 18 | |||
| Lenalidomide | 19 | |||
| Sumatriptan succinate | 20 | |||
| Stable (Drugs That Show up in Top 20 List before and after Freezing) | Unstable (Drugs That Show up as Top 20 Performer Only before Freezing Whereas after That They Fall behind the Threshold) | |||
|---|---|---|---|---|
| Drug Name | Rank before Freezing | Rank after Freezing | Drug | Rank before Freezing |
| Ixabepilone | 1 | 4 | Ibrutinib | 2 |
| Cabazitaxel | 3 | 17 | Docetaxel | 4 |
| Podophyllotoxin | 5 | 10 | Vinblastine sulfate | 6 |
| Homoharringtonine | 7 | 13 | Decitabine | 12 |
| Doxorubicin hydrochloride | 8 | 16 | Nintedanib | 13 |
| Paclitaxel | 9 | 1 | Erlotinib | 18 |
| Panobinostat | 10 | 3 | Topotecan hydrochloride | 19 |
| Bortezomib | 11 | 5 | ||
| Carfilzomib | 14 | 12 | ||
| Nintedanib Ethanesulfonate Salt | 15 | 6 | ||
| Romidepsin | 16 | 8 | ||
| Acalabrutinib | 17 | 19 | ||
| Mitoxantrone hydrochloride | 20 | 9 | ||
| Stable (Drugs That Show up in Top 20 List before and after Freezing) | (Drugs That Show up as Top 20 Performer Only before Freezing Whereas after That They Fall behind the Threshold) | |||
|---|---|---|---|---|
| Drug Name | Rank before Freezing | Rank after Freezing | Drug | Rank before Freezing |
| Doxorubicin hydrochloride | 3 | 3 | Nintedanib Ethanesulfonate Salt | 1 |
| Ixabepilone | 4 | 2 | Cabazitaxel | 2 |
| Mitoxantrone hydrochloride | 5 | 8 | Nintedanib | 6 |
| Homoharringtonine | 7 | 14 | Acalabrutinib | 15 |
| Podophyllotoxin | 8 | 12 | Paclitaxel | 16 |
| Topotecan hydrochloride | 9 | 13 | Erlotinib | 18 |
| Vinblastine sulfate | 10 | 7 | Palbociclib Isethionate | 19 |
| Docetaxel | 11 | 11 | Dasatinib | 20 |
| Panobinostat | 12 | 20 | ||
| Bortezomib | 13 | 18 | ||
| Carfilzomib | 14 | 5 | ||
| Romidepsin | 17 | 16 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, D.; Nickel, A.-C.; Jeising, S.; Uhlmann, C.; Muhammad, S.; Hänggi, D.; Fischer, I.; Kahlert, U.D. Testing the Stability of Drug Resistance on Cryopreserved, Gene-Engineered Human Induced Pluripotent Stem Cells. Pharmaceuticals 2021, 14, 919. https://doi.org/10.3390/ph14090919
Khan D, Nickel A-C, Jeising S, Uhlmann C, Muhammad S, Hänggi D, Fischer I, Kahlert UD. Testing the Stability of Drug Resistance on Cryopreserved, Gene-Engineered Human Induced Pluripotent Stem Cells. Pharmaceuticals. 2021; 14(9):919. https://doi.org/10.3390/ph14090919
Chicago/Turabian StyleKhan, Dilaware, Ann-Christin Nickel, Sebastian Jeising, Constanze Uhlmann, Sajjad Muhammad, Daniel Hänggi, Igor Fischer, and Ulf Dietrich Kahlert. 2021. "Testing the Stability of Drug Resistance on Cryopreserved, Gene-Engineered Human Induced Pluripotent Stem Cells" Pharmaceuticals 14, no. 9: 919. https://doi.org/10.3390/ph14090919
APA StyleKhan, D., Nickel, A.-C., Jeising, S., Uhlmann, C., Muhammad, S., Hänggi, D., Fischer, I., & Kahlert, U. D. (2021). Testing the Stability of Drug Resistance on Cryopreserved, Gene-Engineered Human Induced Pluripotent Stem Cells. Pharmaceuticals, 14(9), 919. https://doi.org/10.3390/ph14090919