Abstract
This study aimed to investigate the effects of genetic variants and haplotypes in the renin–angiotensin system (RAS) on the risk of warfarin-induced bleeding complications at therapeutic international normalized ratios (INRs). Four single nucleotide polymorphisms (SNPs) of AGT, two SNPs of REN, three SNPs of ACE, four SNPs of AGTR1, and one SNP of AGTR2, in addition to VKORC1 and CYP2C9 variants, were investigated. We utilized logistic regression and several machine learning methods for bleeding prediction. The study included 142 patients, among whom 21 experienced bleeding complications. We identified a haplotype, H2 (TCG), carrying three single nucleotide polymorphisms (SNPs) of ACE (rs1800764, rs4341, and rs4353), which showed a significant relation with bleeding complications. After adjusting covariates, patients with H2/H2 experienced a 0.12-fold (95% CI 0.02–0.99) higher risk of bleeding complications than the others. In addition, G allele carriers of AGT rs5050 and A allele carriers of AGTR1 rs2640543 had 5.0- (95% CI 1.8–14.1) and 3.2-fold (95% CI 1.1–8.9) increased risk of bleeding complications compared with the TT genotype and GG genotype carriers, respectively. The AUROC values (mean, 95% CI) across 10 random iterations using five-fold cross-validated multivariate logistic regression, elastic net, random forest, support vector machine (SVM)–linear kernel, and SVM–radial kernel models were 0.732 (0.694–0.771), 0.741 (0.612–0.870), 0.723 (0.589–0.857), 0.673 (0.517–0.828), and 0.680 (0.528–0.832), respectively. The highest quartile group (≥75th percentile) of weighted risk score had approximately 12.0 times (95% CI 3.1–46.7) increased risk of bleeding, compared to the 25–75th percentile group, respectively. This study demonstrated that RAS-related polymorphisms, including the H2 haplotype of the ACE gene, could affect bleeding complications during warfarin treatment for patients with mechanical heart valves. Our results could be used to develop individually tailored intervention strategies to prevent warfarin-induced bleeding.
1. Introduction
Warfarin has been one of the most widely used oral anticoagulants since its approval [1]. Although direct oral anticoagulants have become popular for patients who need anticoagulation therapy, warfarin remains the first-line anticoagulant for patients with heart valve prostheses [2]. Nevertheless, warfarin has several limitations, including a narrow therapeutic range and wide inter- and intra-individual variabilities [1].
Bleeding is the most serious complication of warfarin treatment [3]. Although close monitoring based on the international normalized ratio (INR) is known to be effective for evaluating the efficacy and safety of warfarin, it has been reported that patients may still experience bleeding complications within therapeutic INRs, and even at sub-therapeutic INRs [4,5]. Age, hypertension, and concomitant aspirin use—in addition to high INR—are known as patient-related risk factors for complications [3]; however, it needs to be explained further, and genetic factors can be an answer. According to Pourgholi et al., CYP2C9 and NQO1 variants may affect bleeding complications [6]. Our previous research also identified several gene polymorphisms (e.g., APOB and GATA4), which can affect warfarin bleedings [7,8]. However, compared to pharmacogenomic studies for warfarin dose [9], previous studies have rarely assessed the genetic effects on bleeding complications during anticoagulation therapy.
The renin–angiotensin system (RAS) consists of four major components: renin (REN), angiotensinogen (AGT), angiotensin I-converting enzyme (ACE), and angiotensin receptors (AGTR1 and AGTR2). The RAS is known to play an important role in the regulation of electrolyte balance, vasoconstriction, vascular remodeling, and fibrinolysis [10,11]. Previous studies have demonstrated clinically significant associations between RAS polymorphisms and several cardiovascular diseases (e.g., hypertension, coronary heart disease, and stroke) [12]. According to a recent meta-analysis, it has been demonstrated that ACE I/D polymorphism is associated with intracranial hemorrhage [13]. According to Shiotani et al., AGT A-20C was associated with gastrointestinal bleeding [14]. As the RAS is highly involved in cardiovascular functions, including fibrinolysis, RAS polymorphisms may affect bleeding complications.
Machine learning is a subdomain of artificial intelligence, which makes machines mimic human intelligence [15]. With powerful and fast development, machine learning has been applied to several fields, including medicine [16]. In terms of cardiology, machine learning can help clinicians by interpreting high-dimensional data (e.g., biomedical and clinical big data, multi-omics data, and images) [17] and predicting outcomes (e.g., coronary artery disease and heart failure) [18]. Several studies also have developed machine learning algorithms to predict warfarin dose [19,20,21]. However, previous studies have rarely investigated bleeding complications by applying machine learning algorithms.
Therefore, we aimed to investigate the effects of genetic variants and haplotypes of RAS-related genes on bleeding complications among mechanical heart valve patients maintaining therapeutic INRs, and we used supervised machine learning to build predictive models for bleeding occurrence.
2. Results
From among the 229 patients enrolled, 142 were included in the analysis. Of the 87 who were excluded, 28 did not reach stable INR, 4 had bleeding complications at supra-therapeutic INRs, and 55 reported minimal bleeding complications not verified by health professionals. Among the 55 patients who were excluded from this study because of the lack of professional verification, 8 had bleeding complications before achieving a stable INR, and 32, 6, and 9 patients had bleeding complications at therapeutic, supra-, and sub-therapeutic INRs, respectively.
As shown in Table 1, the median age and male proportion of included patients was 60 y and 36.6%, respectively. The mean follow-up period was 14.3 ± 6.4 y. Among patients included in the analysis, 21 patients reported bleedings while maintaining therapeutic INRs (11 with minor and 10 with minimal bleeding complications). Atrial fibrillation was the only significant factor for bleeding complications among demographic characteristics (p = 0.045).

Table 1.
Patient characteristics of study patients.
As shown in Table 2, statistically significant associations between genotypes and bleeding complications were found for rs5050 of AGT, rs4341 and rs4353 of ACE, and rs2640543 of AGTR1. In the case of rs5050, 13 out of 45 patients (28.9%) with the G allele had bleeding complications, whereas 8 out of 97 patients (8.2%) with the TT genotype had bleeding complications (p = 0.001). For rs4341 and rs4353, patients with the wild allele were at a higher risk of bleeding, compared with those who had the variant homozygotes (19.1% vs. 6.3%, p = 0.041; 19.2% vs. 2.6%, p = 0.014, respectively). C allele carriers of rs1800764 experienced more bleeding complications than those with the TT genotype with marginal significance (18.8% vs. 6.5%, p = 0.055). For rs2640543, patients with the A allele had higher bleeding risks, compared with those with the GG genotype (23.4% vs. 10.5%, p = 0.042).

Table 2.
Genetic factors associated with bleeding complications at therapeutic INR.
Since all of the ACE polymorphisms analyzed in the study were in a moderate linkage disequilibrium (LD) (r2 range: 0.63–0.79; D’ range: 0.82–0.98; Figure S1), and each SNP showed similar association with bleeding complications, we constructed haplotypes of the ACE gene. Five haplotypes were detected at a frequency more than 1%: H1 (CGA, 41.8%), H2 (TCG, 47.0%), H3 (TCA, 4.7%), H4 (CCG, 4.1%), and H5 (TGA, 1.4%). The most frequent haplotype was H2, containing variant alleles at every locus, and patients with H2/H2 experienced fewer bleeding complications than the others (3.0% vs. 18.3%, p = 0.028).
After adjusting for related covariates, patients with H2/H2 were revealed to have an approximately 0.12-fold higher risk of bleeding complications than the others (Table 3). Patients with G allele of rs5050 and A allele of rs2640543 had 5.0- and 3.2-fold higher rates of bleeding complications at therapeutic INRs, compared with those with the TT and GG genotypes, respectively. In constructed models, the attributable risks related to H2, rs5050 and rs2640543 were 88.0%, 80.2%, and 68.5%, respectively. The area under the receiver-operating curve (AUROC) value (mean, 95% confidence interval (CI)) was 0.771 (0.656–0.886) (Figure 1) and the Hosmer–Lemeshow test showed that the fitness of model was satisfactory (χ2=3.162, 5 degrees of freedom, p = 0.675). The AUROC values (mean, 95% CI) across 10 random iterations using five-fold cross-validated multivariable logistic regression, elastic net, random forest (RF), support vector machine (SVM)–linear kernel, and SVM–radial kernel models were 0.732 (0.694–0.771), 0.741 (0.612–0.870), 0.723 (0.589–0.857), 0.673 (0.517–0.828), and 0.680 (0.528–0.832), respectively.

Table 3.
Multivariable analysis to identify predictors for bleeding complications at therapeutic INR.
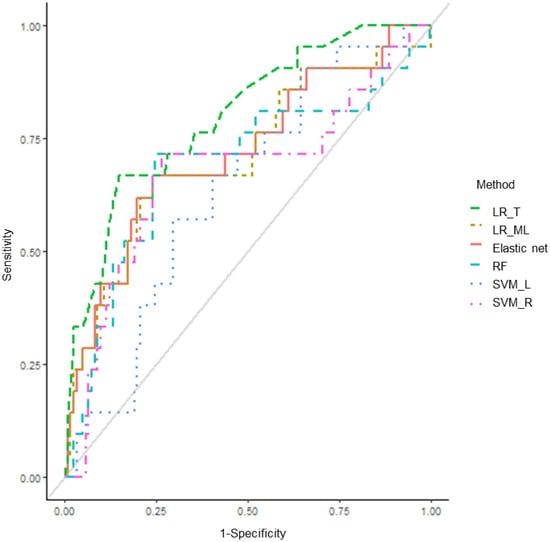
Figure 1.
Area under the receiver operating characteristic curve (AUROC) for bleeding complications at therapeutic INRs. LR_T: traditional logistic regression; LR_ML: machine learning-based logistic regression; RF: random forest; SVM_L: support vector machine–linear kernel; SVM_R: support vector machine–radial kernel.
We calculated the number needed to genotype (NNG) for preventing one patient with a high-risk allele or haplotype from experiencing a higher incidence of bleeding complications using each model, and the values were 8, 14, and 19 for H2, rs5050, and rs2640543, respectively. In weighted risk score (WRS) analysis, patients with bleeding complications showed a significantly higher WRS than those without (3.6 ± 1.2 vs. 2.3 ± 1.3, p < 0.001). As shown in Table 4, the incidence of bleeding complication displayed increasing patterns, according to the quartile of WRS. The highest quartile group (≥75th percentile) of WRS had 12.0 (95% CI 3.1–46.6, p < 0.001) times increased risk of bleeding, compared to the 25–75th percentile group.

Table 4.
Weighted risk score analysis of patients with bleeding complications.
3. Discussion
The main finding of the present study suggested that the ACE haplotype, H2 (TCG), carrying three SNPs (rs1800764, rs4341, and rs4353) in addition to rs5050 of AGT and rs2640543 of AGTR1 were associated with bleeding complications at therapeutic INRs among patients who received warfarin therapy after mechanical heart valve replacement surgery. Patients with H2/H2 experienced a 0.12-fold higher risk of bleeding complications than the others. G allele carriers of rs5050 and A allele carriers of rs2640543 were 5.0 and 3.2 times more likely to experience bleeding complications compared with TT genotype and GG genotype carriers, respectively. In the five-fold cross-validated multivariable logistic regression, elastic net, RF, and SVM models, the mean AUROC values ranged between 0.67 and 0.74.
The RAS has essential roles in the cardiovascular system. Among its many functions, the RAS—particularly angiotensin II—is involved in vascular pathophysiology, including vascular cells growth/apoptosis, vascular smooth muscle cell differentiation/proliferation, and extracellular matrix remodeling [11]. The RAS is also known to be involved in the balance of coagulation and fibrinolysis by two processes: (1) tissue plasminogen activator (t-PA) production by bradykinin, which is metabolized by ACE, and (2) plasminogen activator inhibitor-1 (PAI-1) induction by angiotensin II [10].
The ACE gene, located on human chromosome 17, consists of 26 exons and 25 introns and is considered highly polymorphic. Among genetic polymorphisms of ACE, I/D polymorphism has been most extensively studied in several cardiovascular diseases [22,23]. Rs4341, an intronic variant of ACE, is known to be in perfect LD with ACE I/D polymorphism in Caucasian and Asian populations, and this is commonly used as an alternative method of determining ACE I/D polymorphism; GG genotype carriers of rs4341 are considered to be DD genotype carriers of the ACE I/D polymorphism [24]. In a study with healthy subjects [25], ACE I/D polymorphism was related to serum ACE concentrations, accounting for 47% of the total variance of serum ACE concentrations. A recent meta-analysis including 39 case–control studies showed that ACE I/D polymorphism was significantly associated with intracranial hemorrhage, especially among Asians [13]. Although some studies have described the DD genotype as a potent thrombophilic factor [26,27], several studies have reported that D allele carriers of ACE have increased risk of hemorrhagic stroke [28] and blood loss after hip surgery [29], which is also consistent with our results.
Rs1800764 and rs4353, included in the associated haplotype with bleeding complications in our study, were located in the upstream region and intron 19 of ACE, respectively. Chung et al. showed that rs1800764 had a significant relation with young-onset hypertension and rs4353 was significantly associated with ACE activity [30]. Furthermore, the haplotype containing the A allele of rs4353 was reportedly related to increased serum concentrations of ACE and increased hypertension risk [31]. eQTL analysis performed on GTEx also supported our results [32]; both rs1800764 and rs4353 were recorded as significant expression quantitative trait loci with ACE transcript (p = 3.0 × 10−21 and 9.8 × 10−17, respectively), showing higher expression with wild-type alleles in fibroblast tissues. In consideration of the above findings, our results might be explained by the increased serum concentrations of ACE.
Angiotensinogen, encoded by AGT, is the only precursor of the RAS and is sequentially cleaved by renin and ACE [33]. Gould et al. showed that plasma AGT concentrations were closely related to the Km of renin, indicating that plasma AGT concentrations might affect the angiotensin II level accordingly [34]. Located in the promoter region of the AGT gene, rs5050 is reported to be essential for the transcription of AGT [35], and the variant allele of rs5050 has been reported to increase the promoter activity of AGT [35,36]. Ishigami et al. also revealed that this variant was associated with high AGT plasma concentrations [37]. In studies with patients taking low-dose aspirin, rs5050 was reportedly associated with bleeding [14,38]. Our study also revealed that this variant was significantly associated with increased bleeding risk.
AGTR1, expressed in all different organs, is the principal receptor that mediates major actions of angiotensin II [39]. Although there have been no studies that examine the bleeding risk of rs2640543, several studies have demonstrated the functional effect of rs2640543, which is linked with cardiovascular disease [40,41]. Su et al. reported that, in a study with Chinese patients, the haplotype-containing wild allele of rs2640543 was associated with decreased systolic blood pressure reduction in response to benazepril, an ACE inhibitor [40]. eQTL results also showed that the wild allele of rs2640543 also showed higher expression in both fibroblast and aorta artery (p = 2.5 × 10−12 and p = 3.2 × 10−5, respectively) [32]. Accordingly, the increased RAS activity seems to affect the bleeding.
In our study, VKORC1 and CYP2C9 polymorphisms, the well-known genetic factors for warfarin dose prediction [1], were not significantly associated with bleeding complications. As our study patients had already achieved therapeutic INRs after dosing adjustments, VKORC1 and CYP2C9 polymorphisms were expected not to affect bleeding risk.
This study applied several machine-learning-based methodologies based on the significant factors in the univariate analyses to predict bleeding complications. The AUROC values from the five-fold cross-validated multivariable logistic regression, elastic net, and RF revealed the favorable performance of these models (higher than 0.7). The elastic net is a penalized linear regression model that combines the penalties of the lasso and ridge methods [41]. RF is an ensemble method, which increases the diversity by using a random subset of available features at each node and provides a more accurate prediction than a single decision tree [42,43,44]. In the case of SVM models, the AUROC values were around 0.67. In this study, SVMs were implemented using linear and radial basis function kernels. Linear kernel SVMs have a single tuning parameter, C, which is the cost parameter of the error term, whereas radial kernel SVMs have an additional hyperparameter, sigma, which determines the width for Gaussian distribution [44,45].
Since this study dealt with patients with INRs between 2 and 3, only minimal or minor bleeding events were observed. Although it is obvious that major hemorrhages are of importance, minor bleedings are also clinically important, because they serve as warnings for subsequent major bleedings and may increase the number of visits to clinics, resulting in additional medical costs.
To evaluate the potential clinical value of genotyping SNPs, we calculated the WRS based on our logistic regression models. The highest quartile group of WRS had 12-fold significantly higher bleeding complications than the 25–75th percentile group, implying the possibility of discriminating the high-risk group of bleeding complications in patients on stable warfarin therapy.
The limitations of our study are its retrospective study design and small sample size. In addition, we did not consider the social and clinical histories (e.g., alcohol use and bleeding/stroke history), which could affect bleeding, due to the insufficient data. However, to our knowledge, this is the first study to investigate and predict the warfarin-induced bleeding complications with RAS-related genetic variants using machine learning algorithms.
4. Materials and Methods
4.1. Study Patients and Data Collection
This is a retrospective analysis of prospectively collected blood samples. The details for study patients have already been described in previous papers [7,8]. Study patients were recruited from the previous study cohort, entitled the Ewha-Severance Treatment (EAST) Group of Warfarin. Briefly, 229 patients were included who received mechanical heart valve replacement and were treated with warfarin between January 1982 and December 2009 at the Severance Cardiovascular Hospital of Yonsei University College of Medicine. Among the EAST cohort, patients with a stable INR, which was defined as at least three consecutive INR values in the therapeutic range (2–3) at the outpatient clinic, were recruited for the study. Patients whose bleedings occurred at supra- or sub-therapeutic INRs or were not confirmed by doctors were excluded.
Patients were routinely followed up at the outpatient clinic until warfarin discontinuation, loss to follow up, death, or the end of the study, whichever came first. Blood samples were collected at the outpatient visit. By reviewing patients’ medical records between January 1982 and August 2017, the following data were collected: age, sex, weight, height, body mass index, position and type of valve prosthesis, comorbidities, co-medications, follow-up time, INR values, and bleeding occurrence. Each bleeding event was confirmed by a doctor at the hospital, and the INR was checked at the time of the event. Bleeding complications were assessed using the Platelet Inhibition and Patient Outcomes (PLATO) classification (i.e., major fetal/life-threatening, major other, minor, and minimal) [46].
The Institutional Review Board of the Yonsei University Medical Center approved this study (approval number: 4-2009-0283). This study followed the ethical standards of the Institutional Review Board of the Yonsei University Medical Center and Helsinki declaration principles. Written informed consent was taken from all patients.
4.2. Genotyping Methods
The SNPs of RAS-associated genes were selected by reviewing the following sources: (1) Haploreg 4.1 for minor allele frequencies and LD patterns of SNPs in Asian populations [47]; (2) the PharmGKB database for clinical annotations [48]; and (3) previous studies [30,49,50]. Finally, a total of 16 SNPs were investigated: four SNPs of AGT (rs7079, rs699, rs5050, and rs11122576), two SNPs of REN (rs2368564 and rs12750834), three SNPs of ACE (rs1800764, rs4341, and rs4353), four SNPs of AGTR1 (rs275651, rs2640543, rs5186, and rs5182), and one SNP of AGTR2 (rs1403543), along with the two well-known SNPs for warfarin stable doses (VKORC1 rs9934438 and CYP2C9 rs1057910).
We extracted genomic DNA from patients’ blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). All samples were genotyped using the TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, USA) based on a real-time PCR system or SNaPShot multiplex kits (Applied Biosystems, Foster City, CA, USA) based on a single-base primer extension assay.
4.3. Statistical Analysis and Machine Learning Methods
We calculated the LD (r2 and D’) for each SNP pair in a gene by Haploview 4.2 [51] and performed haplotype analysis using Plink [52]. To compare the patients with and without bleeding complications, the chi-square test and independent t-test were used for categorical and continuous variables, respectively. To determine independent risk factors related to bleeding complications, we performed multivariable logistic regression analysis with backward elimination using variables whose p-value was less than 0.05 in univariate analysis, in addition to clinical confounders (age and sex). We obtained odds ratios (ORs) and adjusted odds ratios (AORs) by logistic analyses and calculated attributable risk (%) by the formula of ((AOR − 1)/AOR) × 100. The model was tested by Hosmer–Lemeshow statistics and AUROC analysis.
To predict bleeding complications, we utilized machine learning algorithms, including five-fold cross-validated multivariable logistic regression, elastic net, RF, and a SVM. In each algorithm, we used 10 repeat iterations. To evaluate model performance, we used the AUROC with a 95% CI.
The NNG, which represents the number of patients for preventing one additional bleeding complication, was calculated by the following equations [7]:
where Riskno genotyping was defined as the risk of higher incidence of bleeding complications without genotyping. To assess the cumulative effect of clinical factors and multiple SNPs on bleeding complications, the WRS was created based on variables that were included in each model in this study. The point assigned to each variable was determined by the variable’s beta coefficient from the logistic regression model in the current study, and the WRS was the sum of the points for variables that patients had.
Relative risk reduction (RRR) = (AOR − 1)/AOR;
Absolute risk reduction (ARR) = RRR × Riskno genotyping;
NNG = 1/ARR,
A p-value of <0.05 was considered statistically significant. All analyses were performed with SPSS 20.0 (IBM, Armonk, NY, USA) and R package caret.
5. Conclusions
This study demonstrated that RAS-related polymorphisms, including the H2 haplotype of ACE, rs5050 of AGT, and rs2640543 of AGTR1 could affect bleeding complications during warfarin treatment for patients with mechanical heart valves. Our results could be used to develop individually tailored intervention strategies to prevent warfarin-induced bleeding.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14080824/s1, Figure S1. Linkage disequilibrium patterns and relative position of SNPs among study patients. Present SNPs were in AGT ((a) and (e)), REN ((b) and (f)), ACE ((c) and (g)) and AGTR1 ((d) and (h)), respectively. Values in squares are the pairwise calculation of D’ ((a)–(d)) or r2 ((e)–(h)). Empty squares indicate D’ = 1.0 or r2 = 1.0.
Author Contributions
B.-C.C. and H.-S.G. were responsible for the study concept and design. J.-H.K. and J.Y. participated in data collection and data analysis. J.-H.K., J.Y. and H.-S.G. contributed to the manuscript writing and discussion. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education [grant numberNRF-2017R1D1A1B03034033]. This research was also supported by a grant from the Ministry of Food and Drug Safety of Korea [grant number 21153MFDS431].
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of the Yonsei University Medical Center (approval number: 4-2009-0283).
Informed Consent Statement
Written informed consent was obtained from all patients involved in the study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available in the Mendeley Data repository, http://dx.doi.org/10.17632/s5cngbxmy7.1 (accessed on 20 August 2021).
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
| ACE | angiotensin I-converting enzyme |
| AGT | angiotensinogen |
| AGTR | angiotensin receptor |
| AORs | adjusted odds ratios |
| AUROC | area under the receiver-operating curve |
| INRs | international normalized ratios |
| LD | linkage disequilibrium |
| NNG | number needed to genotype |
| ORs | odds ratios |
| PAI-1 | plasminogen activator inhibitor-1 |
| PLATO | Platelet Inhibition and Patient Outcomes |
| RAS | renin–angiotensin system |
| REN | renin |
| RF | random forest |
| SVM | support vector machine |
| SNPs | single nucleotide polymorphisms |
| WRS | weighted risk score |
References
- Elias, D.J.; Topol, E.J. Warfarin pharmacogenomics: A big step forward for individualized medicine: Enlightened dosing of warfarin. Eur. J. Hum. Genet. 2008, 16, 532–534. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.A.; Otto, C.M.; Bonow, R.O.; Carabello, B.A.; Erwin, J.P.; Guyton, R.A.; O’Gara, P.T.; Ruiz, C.E.; Skubas, N.J.; Sorajja, P.; et al. 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: Executive summary: A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014, 129, 2440–2492. [Google Scholar] [CrossRef]
- Marie, I.; Leprince, P.; Menard, J.F.; Tharasse, C.; Levesque, H. Risk factors of vitamin K antagonist overcoagulation. QJM Int. J. Med. 2012, 105, 53–62. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miura, T.; Nishinaka, T.; Terada, T.; Yonezawa, K. Relationship between aging and dosage of warfarin: The current status of warfarin anticoagulant therapy for Japanese outpatients in a department of cardiovascular medicine. J. Cardiol. 2009, 53, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Moffett, B.S.; Ung, M.; Bomgaars, L. Risk factors for elevated INR values during warfarin therapy in hospitalized pediatric patients. Pediatr. Blood Cancer 2012, 58, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Pourgholi, L.; Goodarzynejad, H.; Mandegary, A.; Ziaee, S.; Talasaz, A.H.; Jalali, A.; Boroumand, M. Gene polymorphisms and the risk of warfarin-induced bleeding complications at therapeutic international normalized ratio (INR). Toxicol. Appl. Pharmacol. 2016, 309, 37–43. [Google Scholar] [CrossRef]
- Yee, J.; Kim, W.; Chang, B.C.; Chung, J.E.; Lee, K.E.; Gwak, H.S. APOB gene polymorphisms may affect the risk of minor or minimal bleeding complications in patients on warfarin maintaining therapeutic INR. Eur. J. Hum. Genet. 2019, 27, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.; Kim, W.; Chang, B.C.; Chung, J.E.; Lee, K.E.; Gwak, H.S. Genetic variations in the transcription factors GATA4 and GATA6 and bleeding complications in patients receiving warfarin therapy. Drug Des. Devel. Ther. 2019, 13, 1717–1727. [Google Scholar] [CrossRef]
- Kaye, J.B.; Schultz, L.E.; Steiner, H.E.; Kittles, R.A.; Cavallari, L.H.; Karnes, J.H. Warfarin Pharmacogenomics in Diverse Populations. Pharmacotherapy 2017, 37, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.E. The renin-angiotensin system and fibrinolysis. Am. J. Cardiol. 1997, 79, 12–16. [Google Scholar] [CrossRef]
- Ruiz-Ortega, M.; Lorenzo, O.; Rupérez, M.; Esteban, V.; Suzuki, Y.; Mezzano, S.; Plaza, J.J.; Egido, J. Role of the renin-angiotensin system in vascular diseases: Expanding the field. Hypertension 2001, 38, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.G.; Staessen, J.A. Genetic polymorphisms in the renin-angiotensin system: Relevance for susceptibility to cardiovascular disease. Eur. J. Pharmacol. 2000, 410, 289–302. [Google Scholar] [CrossRef]
- Li, Z.; Wang, S.; Jiao, X.; Wei, G. Genetic association of angiotensin-converting enzyme I/D polymorphism with intracranial hemorrhage: An updated meta-analysis of 39 case-control studies. World Neurosurg. 2019, 134, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Shiotani, A.; Nishi, R.; Yamanaka, Y.; Murao, T.; Matsumoto, H.; Tarumi, K.-I.; Kamada, T.; Sakakibara, T.; Haruma, K. Renin-angiotensin system associated with risk of upper GI mucosal injury induced by low dose aspirin: Renin angiotensin system genes’ polymorphism. Dig. Dis. Sci. 2011, 56, 465–471. [Google Scholar] [CrossRef]
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Pannu, A. Artificial intelligence and its application in different areas. Artif. Intell. 2015, 4, 79–84. [Google Scholar]
- Mathur, P.; Srivastava, S.; Xu, X.; Mehta, J.L. Artificial intelligence, machine learning, and cardiovascular disease. Clin. Med. Insights Cardiol. 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Perillo, T.; De Rosa, E.; Ugga, L.; Petretta, M. Current applications of big data and machine learning in cardiology. J. Geriatr. Cardiol. 2019, 16, 601–607. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, P.; Gao, Z.; Wang, R.; Khalighi, K. Ensemble of machine learning algorithms using the stacked generalization approach to estimate the warfarin dose. PLoS ONE 2018, 13, e0205872. [Google Scholar] [CrossRef] [PubMed]
- Cosgun, E.; Limdi, N.A.; Duarte, C.W. High-dimensional pharmacogenetic prediction of a continuous trait using machine learning techniques with application to warfarin dose prediction in African Americans. Bioinformatics 2011, 27, 1384–1389. [Google Scholar] [CrossRef]
- Hu, Y.H.; Wu, F.; Lo, C.L.; Tai, C.T. Predicting warfarin dosage from clinical data: A supervised learning approach. Artif. Intell. Med. 2012, 56, 27–34. [Google Scholar] [CrossRef]
- Hubert, C.; Houot, A.; Corvol, P.; Soubrier, F. Structure of the angiotensin I-converting enzyme gene. Two alternate promoters correspond to evolutionary steps of a duplicated gene. J. Biol. Chem. 1991, 266, 15377–15383. [Google Scholar] [CrossRef]
- Rigat, B.; Hubert, C.; Corvol, P.; Soubrier, R. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP1) (dipeptidyl carboxypeptidase 1). Nucleic Acids Res. 1992, 20, 1433. [Google Scholar] [CrossRef] [PubMed]
- Glenn, K.L.; Du, Z.-Q.; Eisenmann, J.C.; Rothschild, M.F. An alternative method for genotyping of the ACE I/D polymorphism. Mol. Biol. Rep. 2008, 36, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Investig. 1990, 86, 1343–1346. [Google Scholar] [CrossRef]
- Dilley, A.; Saidi, P.; Evatt, B.; Austin, H.; Zawadsky, J.; Harwood, D.; Ellingsen, D.; Barnhart, E.; Phillips, D.; Hooper, W.C.; et al. Deletion polymorphism in the angiotensin-converting enzyme gene as a thrombophilic risk factor after hip arthroplasty. Thromb. Haemost. 1998, 80, 869–873. [Google Scholar] [CrossRef]
- Jackson, A.; Brown, K.; Langdown, J.; Luddington, R.; Baglin, T. Effect of the angiotensin-converting enzyme gene deletion polymorphism on the risk of venous thromboembolism. Br. J. Haematol. 2000, 111, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Roy, S.; Sharma, V.; Kaul, S.; Jyothy, A.; Munshi, A. Association of ACE gene I/D polymorphism and ACE levels with hemorrhagic stroke: Comparison with ischemic stroke. Neurol. Sci. 2015, 36, 137–142. [Google Scholar] [CrossRef]
- Pola, E.; Gaetani, E.; Pola, R.; Papaleo, P.; Flex, A.; Aloi, F.; De Santis, V.; Santoliquido, A.; Pola, P. Angiotensin-converting enzyme gene polymorphism may influence blood loss in a geriatric population undergoing total hip arthroplasty. J. Am. Geriatr. Soc. 2002, 50, 2025–2028. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-M.; Wang, R.-Y.; Fann, C.S.J.; Chen, J.-W.; Jong, Y.-S.; Jou, Y.-S.; Yang, H.-C.; Kang, C.-S.; Chen, C.-C.; Chang, H.-C.; et al. Fine-mapping angiotensin-converting enzyme gene: Separate QTLs identified for hypertension and for ACE activity. PLoS ONE 2013, 8, e56119. [Google Scholar] [CrossRef][Green Version]
- Martínez-Rodríguez, N.; Posadas-Romero, C.; Molina, T.V.; Vallejo, M.; Del-Valle-Mondragón, L.; Rámirez-Bello, J.; Valladares, A.; Cruz-López, M.; Vargas-Alarcón, G. Single nucleotide polymorphisms of the angiotensin-converting enzyme (ACE) gene are associated with essential hypertension and increased ACE enzyme levels in Mexican individuals. PLoS ONE 2013, 8, e65700. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Cassis, L.A.; Kooi, C.W.V.; Daugherty, A. Structure and functions of angiotensinogen. Hypertens. Res. 2016, 39, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.B.; Green, D. Kinetics of the human renin and human substrate reaction. Cardiovasc. Res. 1971, 5, 86–89. [Google Scholar] [CrossRef]
- Yanai, K.; Nibu, Y.; Murakami, K.; Fukamizu, A. A cis-acting DNA element located between TATA box and transcription initiation site is critical in response to regulatory sequences in human angiotensinogen gene. J. Biol. Chem. 1996, 271, 15981–15986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Y.; Zhou, J.; Narayanan, C.S.; Cui, Y.; Kumar, A. Role of C/A polymorphism at -20 on the expression of human angiotensinogen gene. Hypertension 1999, 33, 108–115. [Google Scholar] [CrossRef]
- Ishigami, T.; Tamura, K.; Fujita, T.; Kobayashi, I.; Hibi, K.; Kihara, M.; Toya, Y.; Ochiai, H.; Umemura, S. Angiotensinogen gene polymorphism near transcription start site and blood pressure: Role of a T-to-C transition at intron I. Hypertension 1999, 34, 430–434. [Google Scholar] [CrossRef]
- Negovan, A.; Voidăzan, S.; Pantea, M.; Moldovan, V.; Bățagă, S.; Cozlea, L.; Mocan, S.; Banescu, C. AGT A-20C (rs5050) gene polymorphism and ulcer occurrence in patients treated with low-dose aspirin: A case-control study. Rev. Romana Med. Lab. 2015, 23, 179–187. [Google Scholar] [CrossRef]
- Helin, K.; Stoll, M.; Meffert, S.; Stroth, U.; Unger, T. The role of angiotensin receptors in cardiovascular diseases. Revista Română Medicină Laborator 1997, 29, 23–29. [Google Scholar] [CrossRef]
- Su, X.; Lee, L.; Li, X.; Lv, J.; Hu, Y.; Zhan, S.; Cao, W.; Mei, L.; Tang, Y.-M.; Wang, D.; et al. Association between angiotensinogen, angiotensin II receptor genes, and blood pressure response to an angiotensin-converting enzyme inhibitor. Circulation 2007, 115, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.; Olshen, R. Classification and Regression Trees; Chapman and Hall: London, UK, 1984. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Mehran, R.; Rao, S.V.; Bhatt, D.L.; Gibson, C.M.; Caixeta, A.; Eikelboom, J.; Kaul, S.; Wiviott, S.D.; Menon, V.; Nikolsky, E.; et al. Standardized bleeding definitions for cardiovascular clinical trials: A consensus report from the Bleeding Academic Research Consortium. Circulation 2011, 123, 2736–2747. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo, M.; McDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Altman, R.B.; Klein, T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Philippe, J.; Germain, S.; Mathieu, F.; Jeunemaître, X.; Corvol, P.; Pinet, F. Functionality of two new polymorphisms in the human renin gene enhancer region. J. Hypertens. 2002, 20, 2391–2398. [Google Scholar] [CrossRef] [PubMed]
- Griessenauer, C.J.; Tubbs, R.S.; Foreman, P.M.; Chua, M.H.; Vyas, N.A.; Lipsky, R.H.; Lin, M.; Iyer, R.; Haridas, R.; Walters, B.C.; et al. Association of renin-angiotensin system genetic polymorphisms and aneurysmal subarachnoid hemorrhage J. Neurosurg. 2018, 128, 86–93. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).