RNA Phage VLP-Based Vaccine Platforms
Abstract
1. Introduction
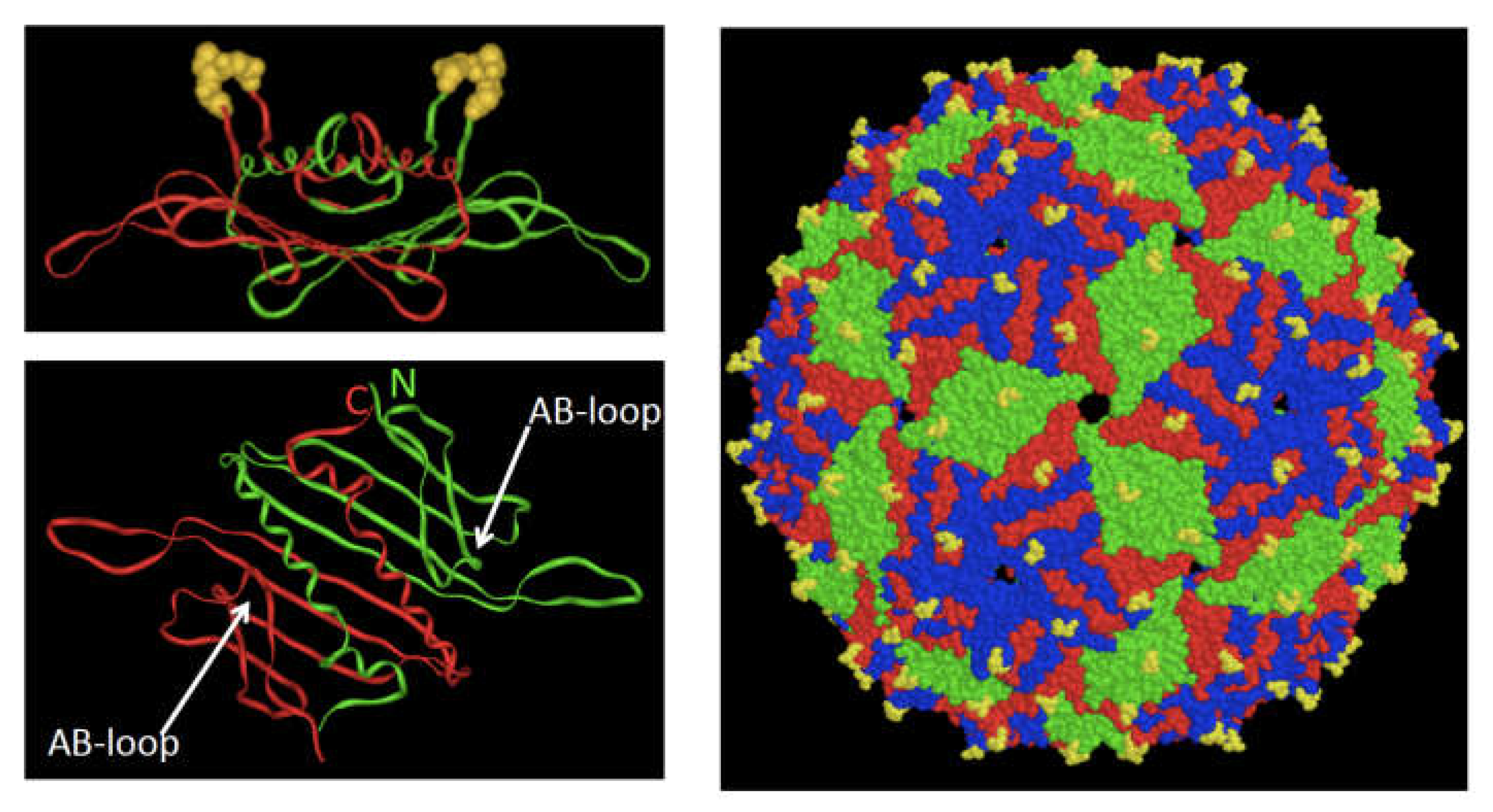
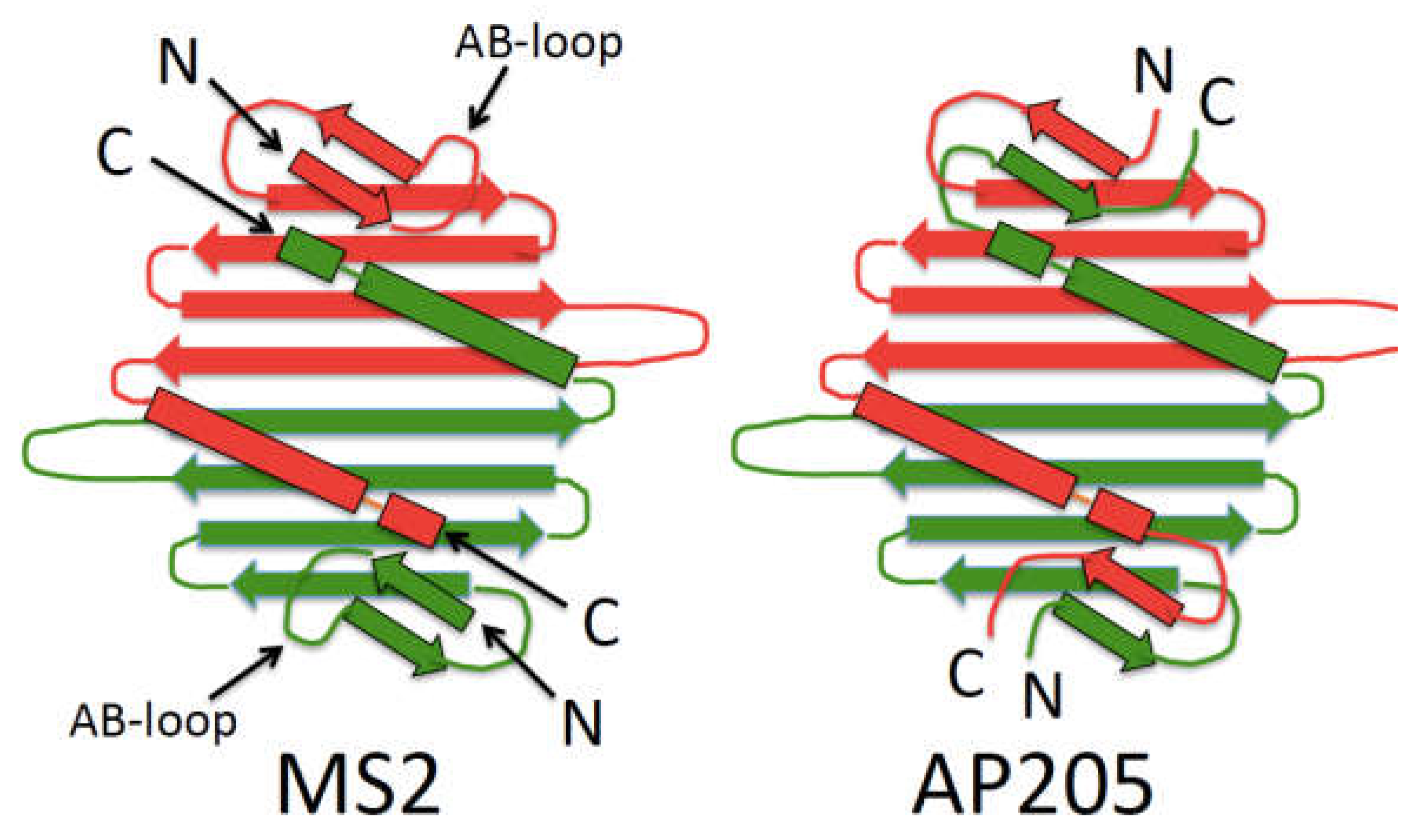
2. The Bacteriophage MS2 VLP as Display Platform
3. Engineered Display of a Known Epitope: An MS2 VLP-Based Universal HPV Vaccine
4. Vaccines by Affinity-Selection
5. Affinity-Selection from Random-Sequence Libraries
6. Antigen-Fragment Libraries
7. PP7 VLPs
8. Genetic Display on Qß VLPs
9. Display on Qß VLPs by Chemical Conjugation
10. AP205 Offers New Display Modes
11. Even More RNA Phages
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fiers, W.; Contreras, R.; Duerinck, F.; Haegeman, G.; Iserentant, D.; Merregaert, J.; Jou, W.M.; Molemans, F.; Raeymaekers, A.; Berghe, A.V.D.; et al. Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene. Nat. Cell Biol. 1976, 260, 500–507. [Google Scholar] [CrossRef]
- Pumpens, P. Single-Stranded RNA Phages: From Moleuclar Biology to Nanotechnology; CRC Press: Boca Raton, FL, USA, 2020; p. 616. [Google Scholar]
- Hyman, P.; Trubi, G.; Abedon, S.T. Virus-Like Particle: Evolving Meanings in Different Disciplines. PHAGE Ther. Appl. Res. 2020, 2, 11–15. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Rohrer, U.H.; Kündig, T.M.; Burki, K.; Hengartner, H.; Zinkernagel, R.M. The influence of antigen organization on B cell responsiveness. Science 1993, 262, 1448–1451. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Zinkernagel, R.M. The influence of virus structure on antibody responses and virus serotype formation. Immunol. Today 1996, 17, 553–558. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Zinkernagel, R.M.; Oxenius, A. Immune responses in the absence of costimulation: Viruses know the trick. J. Immunol. 1998, 161, 5791–5794. [Google Scholar] [PubMed]
- Golmohammadi, R.; Fridborg, K.; Bundule, M.; Valegard, K.; Liljas, L. The crystal structure of bacteriophage Q beta at 3.5 A resolution. Structure 1996, 4, 543–554. [Google Scholar] [CrossRef]
- Golmohammadi, R.; Valegård, K.; Fridborg, K.; Liljas, L. The Refined Structure of Bacteriophage MS2 at 2·8 Å Resolution. J. Mol. Biol. 1993, 234, 620–639. [Google Scholar] [CrossRef] [PubMed]
- Mastico, R.A.; Talbot, S.J.; Stockley, P.G. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993, 74, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S. Subunit Fusion Confers Tolerance to Peptide Insertions in a Virus Coat Protein. Arch. Biochem. Biophys. 1997, 347, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S.; Manifold-Wheeler, B.; Medford, A.; Jordan, S.K.; do Carmo Caldeira, J.; Chackerian, B. Immunogenic Display of Diverse Peptides on Virus-like Particles of RNA Phage MS2. J. Mol. Biol. 2008, 380, 252–263. [Google Scholar] [CrossRef]
- Chackerian, B.; Caldeira, J.D.C.; Peabody, J.; Peabody, D.S. Peptide Epitope Identification by Affinity Selection on Bacteriophage MS2 Virus-Like Particles. J. Mol. Biol. 2011, 409, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S.; Lim, F. Complementation of RNA Binding Site Mutations in MS2 Coat Protein Heterodimers. Nucleic Acids Res. 1996, 24, 2352–2359. [Google Scholar] [CrossRef]
- Peabody, D.S.; Chakerian, A. Asymmetric Contributions to RNA Binding by the Thr45Residues of the MS2 Coat Protein Dimer. J. Biol. Chem. 1999, 274, 25403–25410. [Google Scholar] [CrossRef][Green Version]
- Romanowski, B.; Colares de Borba, P.; Naud, P.S.; Roteli-Mattins, C.M.; De Carvalho, N.S.; Teixeira, J.C.; Aoki, F.; Ramjattan, B.; Shier, R.M.; Socmani, R.; et al. Sustained efficacy and immunogencity of the numan papillomavirus (HPV)-16/18 ASo4-adjuvanted vaccine" analysis of a randomised placebo-controlled tiral up to 6.4 years. Lancet 2009, 374, 1975–1985. [Google Scholar]
- Tumban, E.; Peabody, J.; Peabody, D.S.; Chackerian, B. A Pan-HPV Vaccine Based on Bacteriophage PP7 VLPs Displaying Broadly Cross-Neutralizing Epitopes from the HPV Minor Capsid Protein, L2. PLoS ONE 2011, 6, e23310. [Google Scholar] [CrossRef]
- Tumban, E.; Peabody, J.; Peabody, D.S.; Chackerian, B. A universal virus-like particle-based vaccine for human papillomavirus: Longevity of protection and role of endogenous and exogenous adjuvants. Vaccine 2013, 31, 4647–4654. [Google Scholar] [CrossRef][Green Version]
- Tumban, E.; Peabody, J.; Tyler, M.; Peabody, D.S.; Chackerian, B. VLPs Displaying a Single L2 Epitope Induce Broadly Cross-Neutralizing Antibodies against Human Papillomavirus. PLoS ONE 2012, 7, e49751. [Google Scholar] [CrossRef] [PubMed]
- Twarock, R.; Stockley, P.G. RNA-Mediated Virus Assembly: Mechanisms and Consequences for Viral Evolution and Therapy. Annu. Rev. Biophys. 2019, 48, 495–514. [Google Scholar] [CrossRef]
- Peabody, D.S. Role of the coat Protein-RNA interaction in the life cycle of bacteriophage MS. Mol. Genet. Genom. 1997, 254, 358–364. [Google Scholar] [CrossRef]
- Ord, R.L.; Caldeira, J.C.; Rodriguez, M.; Noe, A.; Chackerian, B.; Peabody, D.S.; Gutiérrez, G.; Lobo, C.A. A malaria vaccine candidate based on an epitope of the Plasmodium falciparum RH5 protein. Malar. J. 2014, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Frietze, K.M.; Pascale, J.M.; Moreno, B.; Chackerian, B.; Peabody, D.S. Ppathogen-specific deep sequence-coupled biopanning: A method for surverying human antibody responses. PLoS ONE 2017, 12, e0171511. [Google Scholar] [CrossRef]
- Kunkel, T. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 1985, 82, 488–492. [Google Scholar] [CrossRef]
- Collar, A.L.; Linville, A.C.; Core, S.B.; Wheeler, C.M.; Geisler, W.M.; Peabody, D.S.; Chackerian, B.; Frietze, K.M. Antibodies to Variable Domain 4 Linear Epitopes of the Chlamydia trachomatis Major Outer Membrane Protein Are Not Associated with Chlamydia Resolution or Reinfection in Women. mSphere 2020, 5, e00654-20. [Google Scholar] [CrossRef]
- Caldeira, J.D.C.; Medford, A.; Kines, R.C.; Lino, C.A.; Schiller, J.T.; Chackerian, B.; Peabody, D.S. Immunogenic display of diverse peptides, including a broadly cross-type neutralizing human papillomavirus L2 epitope, on virus-like particles of the RNA bacteriophage PP7. Vaccine 2010, 28, 4384–4393. [Google Scholar] [CrossRef]
- Zhao, L.; Kopylov, M.; Potter, C.S.; Carragher, B.; Finn, M.G. Engineering the PP7 Virus Capsid as a Peptide Display Platform. ACS Nano 2019, 13, 4443–4454. [Google Scholar] [CrossRef]
- Cielens, I.; Jackevica, L.; Strods, A.; Kazaks, A.; Ose, V.; Bogans, J.; Pumpens, P.; Renhofa, R. Mosaic RNA Phage VLPs Carrying Domain III of the West Nile Virus E Protein. Mol. Biotechnol. 2014, 56, 459–469. [Google Scholar] [CrossRef]
- Kozlovska, T.M.; Cielens, I.; Vasiljeva, I.; Strelnikova, A.; Kazaks, A.; Dislers, A.; Dreilina, D.; Ose, V.; Gusars, I.; Pumpens, P. RNA Phage Qβ Coat Protein as a Carrier for Foreign Epitopes. Intervirology 1996, 39, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Pumpens, P.; Renhofa, R.; Dishlers, A.; Kozlovska, T.; Ose, V.; Pushko, P.; Tars, K.; Grens, E.; Bachmann, M.F. The True Story and Advantages of RNA Phage Capsids as Nanotools. Intervirology 2016, 59, 74–110. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, K.; Kozlovska, T.; Cielens, I.; Strelnikova, A.; Kazaks, A.; Ose, V.; Pumpens, P. Mosaic Qß coats as a new presentation model. FEBS Lett. 1998, 431, 7–11. [Google Scholar] [CrossRef]
- Brown, S.B.; Fiedler, J.D.; Finn, M.G. Assembly of Hybrid Bacteriophage Qß Virus-like Particles. Biochemistry 2009, 48, 11155–11157. [Google Scholar] [CrossRef] [PubMed]
- Skamel, C.; Aller, S.G.; Waffo, A.B. In Vitro Evolution and Affinity-Maturation with Coliphage Qß Display. PLoS ONE 2014, 9, e113069. [Google Scholar] [CrossRef]
- Singleton, R.L.; Sanders, C.A.; Jones, K.; Thorington, B.; Egbo, T.; Coats, M.T.; Waffo, A.B. Function of the RNA Coliphage Qß Proteins in Medical In Vitro Evolution. Methods Protoc. 2018, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Chackerian, B.; Briglio, L.; Albert, P.S.; Lowy, D.R.; Schiller, J.T. Induction of Autoantibodies to CCR5 in Macaques and Subsequent Effects upon Challenge with an R5-Tropic Simian/Human Immunodeficiency Virus. J. Virol. 2004, 78, 4037–4047. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chackerian, B.; Lenz, P.; Lowy, D.R.; Schiller, J.T. Determinants of Autoantibody Induction by Conjugated Papillomavirus Virus-Like Particles. J. Immunol. 2002, 169, 6120–6126. [Google Scholar] [CrossRef] [PubMed]
- Chackerian, B.; Lowy, D.R.; Schiller, J.T. Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proc. Natl. Acad. Sci. USA 1999, 96, 2373–2378. [Google Scholar] [CrossRef] [PubMed]
- Chackerian, B.; Lowy, D.R.; Schiller, J.T. Conjugation of a self-antigen to papillomavirus-like particles allows for efficient induction of protective autoantibodies. J. Clin. Investig. 2001, 108, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Dyer, M.R. Therapeutic vaccination for chronic diseases: A new class of drugs in sight. Nat. Rev. Drug Discov. 2004, 3, 81–88. [Google Scholar] [CrossRef]
- Crossey, E.; Amar, M.J.; Sampson, M.; Peabody, J.; Schiller, J.T.; Chackerian, B.; Remaley, A.T. A cholesterol-lowering VLP vaccine that targets PCSK9. Vaccine 2015, 33, 5747–5755. [Google Scholar] [CrossRef]
- Chackerian, B.; Remaley, A. Vaccine strategies for lowering LDL by immunization against proprotein convertase subtilisin/kexin type. Curr. Opin. Lipidol. 2016, 27, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Chowdhury, S.; McKay, C.S.; Baniel, C.; Wright, W.S.; Bentley, P.; Kaczanowska, K.; Gildersleeve, J.C.; Finn, M.G.; Benmohamed, L.; et al. Significant Impact of Immunogen Designn on the Diversity of Antibodies Generated by Carbohydrate-Based Anticancer Vaccine. ACS Chem. Biol. 2015, 10, 2364–2372. [Google Scholar] [CrossRef]
- Yin, Z.; Comellas-Aragones, M.; Chowdhury, S.; Bentley, P.; Kaczanowska, K.; Benmohamed, L.; Gildersleeve, J.C.; Finn, M.G.; Huang, X. Boosting immunity to small tumor-associated carbohydrates with bacteriophage Qß capsids. ACS Chem. Biol. 2013, 8, 1253–1262. [Google Scholar] [CrossRef]
- Yin, Z.; Dulaney, S.; McKay, C.; Baniel, C.; Kaczanowska, K.; Ramadan, S.; Finn, M.G.; Huang, X. Chemical Synthesis of GM2 Glycans, Bioconjugation with Bacteriophage Qβ, and the Induction of Anticancer Antibodies. Chem.Bio.Chem. 2015, 17, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Cornuz, J.; Zwahlen, S.; Jungi, W.F.; Osterwalder, J.; Klingler, K.; van Mell, G.; Bangala, Y.; Guessous, I.; Muller, P.; Willers, J.; et al. A vaccine against nicotine for smoking cessarion: A raondomized controlled trial. PLoS ONE 2008, 3, e2547. [Google Scholar] [CrossRef] [PubMed]
- Maurer, P.; Bachmann, M.F. Therapeutic vaccines for nicotine dependence. Curr. Opin. Mol. Ther. 2006, 8, 11–16. [Google Scholar] [PubMed]
- Maurer, P.; Bachmann, M.B. Vaccination against nicotine: An emerging therapy for tabacco dependence. Expert Opin. Investig. Drugs 2007, 16, 1775–1783. [Google Scholar] [CrossRef]
- Tissot, A.C.; Renhofa, R.; Schmitz, N.; Cielens, I.; Meijerink, E.; Ose, V.; Jennings, G.T.; Saudan, P.; Pumpens, P.; Bachmann, M.F. Versatile Virus-Like Particle Carrier for Epitope Based Vaccines. PLoS ONE 2010, 5, e9809. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.; Brian, I.J.; Ishizuka, A.; Bachmann, M.F.; Draper, S.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, srep19234. [Google Scholar] [CrossRef] [PubMed]
- Janitzek, C.M.; Matondo, S.; Thrane, S.; Nielsen, M.A.; Kavishe, R.; Mwakalinga, S.B.; Theander, T.G.; Salanti, A.; Sander, A.F. Bacterial superglue generates a full-length circumsporozoite protein virus-like particle vaccine capable of inducing high and durable antibody responses. Malar. J. 2016, 15, 1–9. [Google Scholar] [CrossRef]
- Shishovs, M.; Rūmnieks, J.; Diebolder, C.; Jaudzems, K.; Andreas, L.B.; Stanek, J.; Kazaks, A.; Kotelovica, S.; Akopjana, I.; Pintacuda, G.; et al. Structure of AP205 Coat Protein Reveals Circular Permutation in ssRNA Bacteriophages. J. Mol. Biol. 2016, 428, 4267–4279. [Google Scholar] [CrossRef]
- Xuelan, L.; Chang, X.; Rothen, D.; Dervoni, M.B.; Krenger, P.; TRoongta, S.; Wright, E.; Vogel, M.B.; Tars, K.; Mohsen, M.O.; et al. AP205 VLPs Based on Dimerized Capsid Proteins Accommodate RBM Domain of SARS-CoV-2 and Serve as an Attractive Vaccine Candidate. Vaccines 2021, 9, 403. [Google Scholar]
- Bruun, T.U.J.; Andersson, A.-M.C.; Draper, S.; Howarth, M. Engineering a Rugged Nanoscaffold To Enhance Plug-and-Display Vaccination. ACS Nano 2018, 12, 8855–8866. [Google Scholar] [CrossRef] [PubMed]
- Fougeroux, C.; Goksøyr, L.; Idorn, M.; Soroka, V.; Myeni, S.K.; Dagil, R.; Janitzek, C.M.; Søgaard, M.; Aves, K.L.; Horsted, E.W.; et al. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 2021, 12, 324. [Google Scholar] [CrossRef]
- Janitzek, C.M.; Peabody, J.; Thrane, S.; Carlsen, P.H.R.; Theander, T.G.; Salanti, A.; Chackerian, B.; Nielsen, M.A.; Sander, A.F. A proof-of-concept study for the design of a VLP-based combinatorial HPV and placental malaria vaccine. Sci. Rep. 2019, 9, 5260. [Google Scholar] [CrossRef] [PubMed]
- Thrane, S.; Aves, K.-L.; Uddbäck, I.E.M.; Janitzek, C.M.; Han, J.; Yang, Y.R.; Ward, A.B.; Theander, T.G.; Nielsen, M.A.; Salanti, A.; et al. A Vaccine Displaying a Trimeric Influenza-A HA Stem Protein on Capsid-Like Particles Elicits Potent and Long-Lasting Protection in Mice. Vaccines 2020, 8, 389. [Google Scholar] [CrossRef]
- Thrane, S.; Janitzek, C.M.; Matondo, S.; Resende, M.; Gustavsson, T.; De Jongh, W.A.; Clemmensen, S.; Roeffen, W.; Van De Vegte-Bolmer, M.; Van Gemert, G.J.; et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J. Nanobiotechnol. 2016, 14, 1–16. [Google Scholar] [CrossRef]
- Govasli, M.L.; Diaz, Y.; Puntervoll, P. Virus-like particle-display of the enterotoxigenic Escherichia coli heat-stable toxoid STh-A14T elicits neutralizing antibodies in mice. Vaccines 2019, 37, 6405–6414. [Google Scholar] [CrossRef]
- Yenkoidiok-Douti, L.; Williams, A.E.; Canepa, G.; Molina-Cruz, A.; Barillas-Mury, C. Engineering a Virus-Like Particle as an Antigenic Platform for a Pfs47-Targeted Malaria Transmission-Blocking Vaccine. Sci. Rep. 2019, 9, 16833–16839. [Google Scholar] [CrossRef]
- Liekniņa, I.; Kalniņš, G.; Akopjana, I.; Bogans, J.; Šišovs, M.; Jansons, J.; Rūmnieks, J.; Tārs, K. Production and characterization of novel ssRNA bacteriophage virus-like particles from metagenomic sequencing data. J. Nanobiotechnol. 2019, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Rūmnieks, J.; Liekniņa, I.; Kalniņš, G.; Šišovs, M.; Akopjana, I.; Bogans, J.; Tārs, K. Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes. Sci. Adv. 2020, 6, eabc0023. [Google Scholar] [CrossRef] [PubMed]
- Liekniņa, I.; Černova, D.; Rūmnieks, J.; Tārs, K. Novel ssRNA phage VLP platform for displaying foreign epitopes by genetic fusion. Vaccines 2020, 38, 6019–6026. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peabody, D.S.; Peabody, J.; Bradfute, S.B.; Chackerian, B. RNA Phage VLP-Based Vaccine Platforms. Pharmaceuticals 2021, 14, 764. https://doi.org/10.3390/ph14080764
Peabody DS, Peabody J, Bradfute SB, Chackerian B. RNA Phage VLP-Based Vaccine Platforms. Pharmaceuticals. 2021; 14(8):764. https://doi.org/10.3390/ph14080764
Chicago/Turabian StylePeabody, David S., Julianne Peabody, Steven B. Bradfute, and Bryce Chackerian. 2021. "RNA Phage VLP-Based Vaccine Platforms" Pharmaceuticals 14, no. 8: 764. https://doi.org/10.3390/ph14080764
APA StylePeabody, D. S., Peabody, J., Bradfute, S. B., & Chackerian, B. (2021). RNA Phage VLP-Based Vaccine Platforms. Pharmaceuticals, 14(8), 764. https://doi.org/10.3390/ph14080764





