Engineered EVs for Oxidative Stress Protection
Abstract
:1. Introduction
2. Results
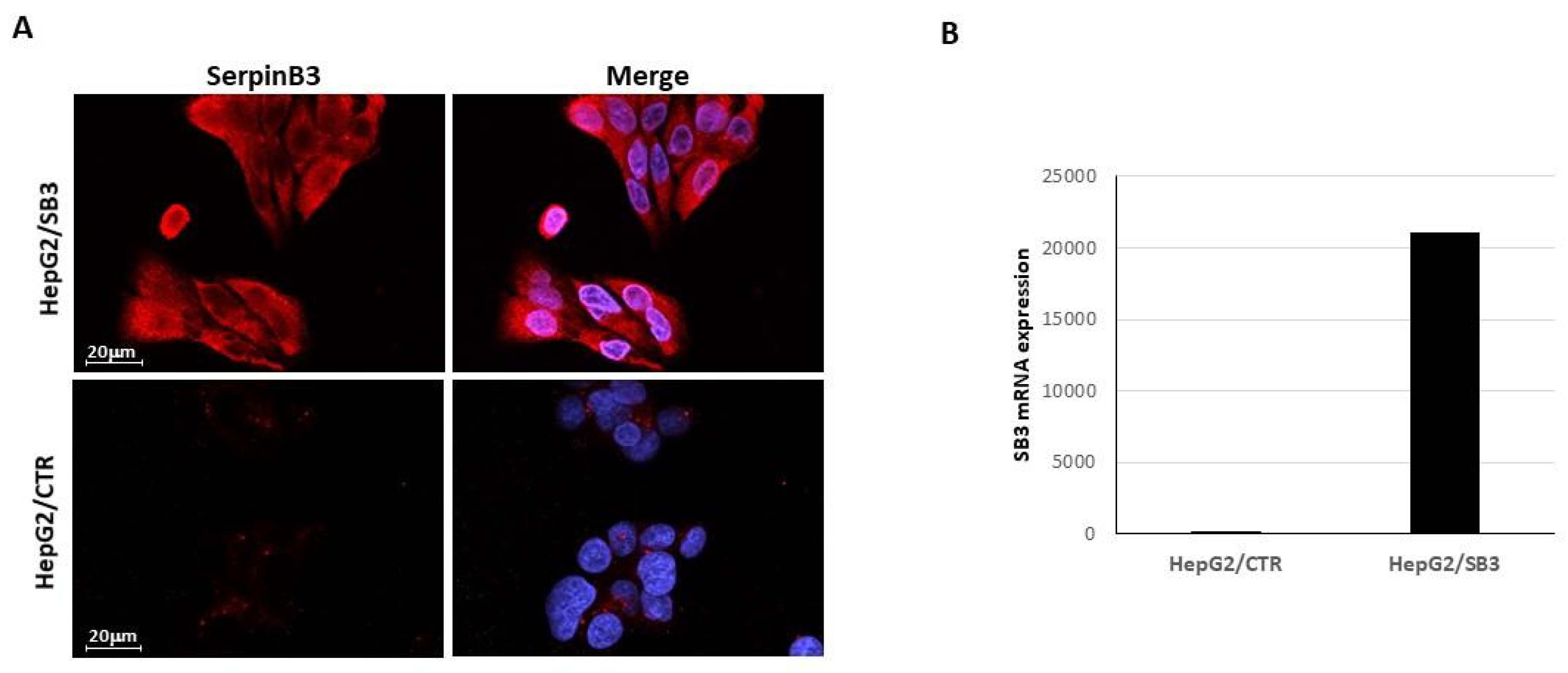
2.1. EV-SerpinB3 Production and Characterization
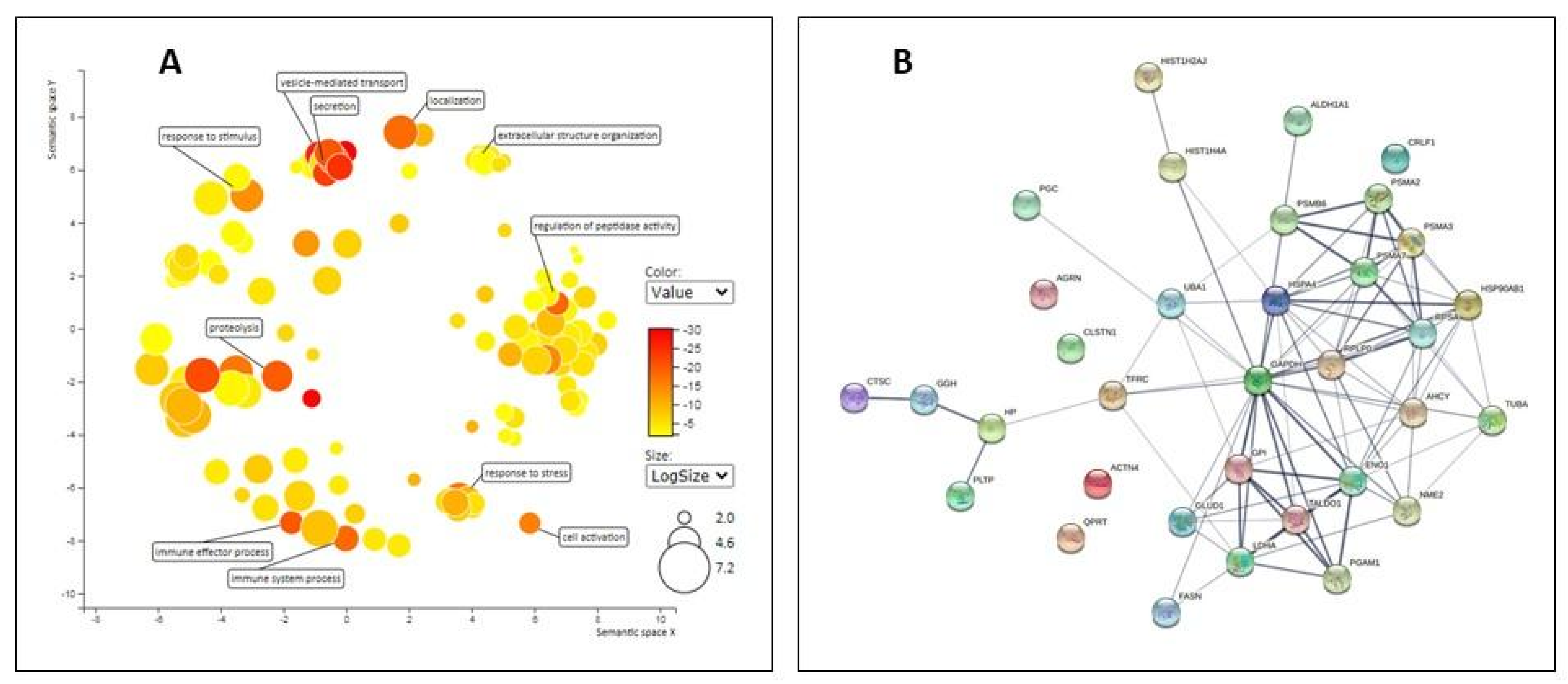
2.2. Proteomic Analysis of EV
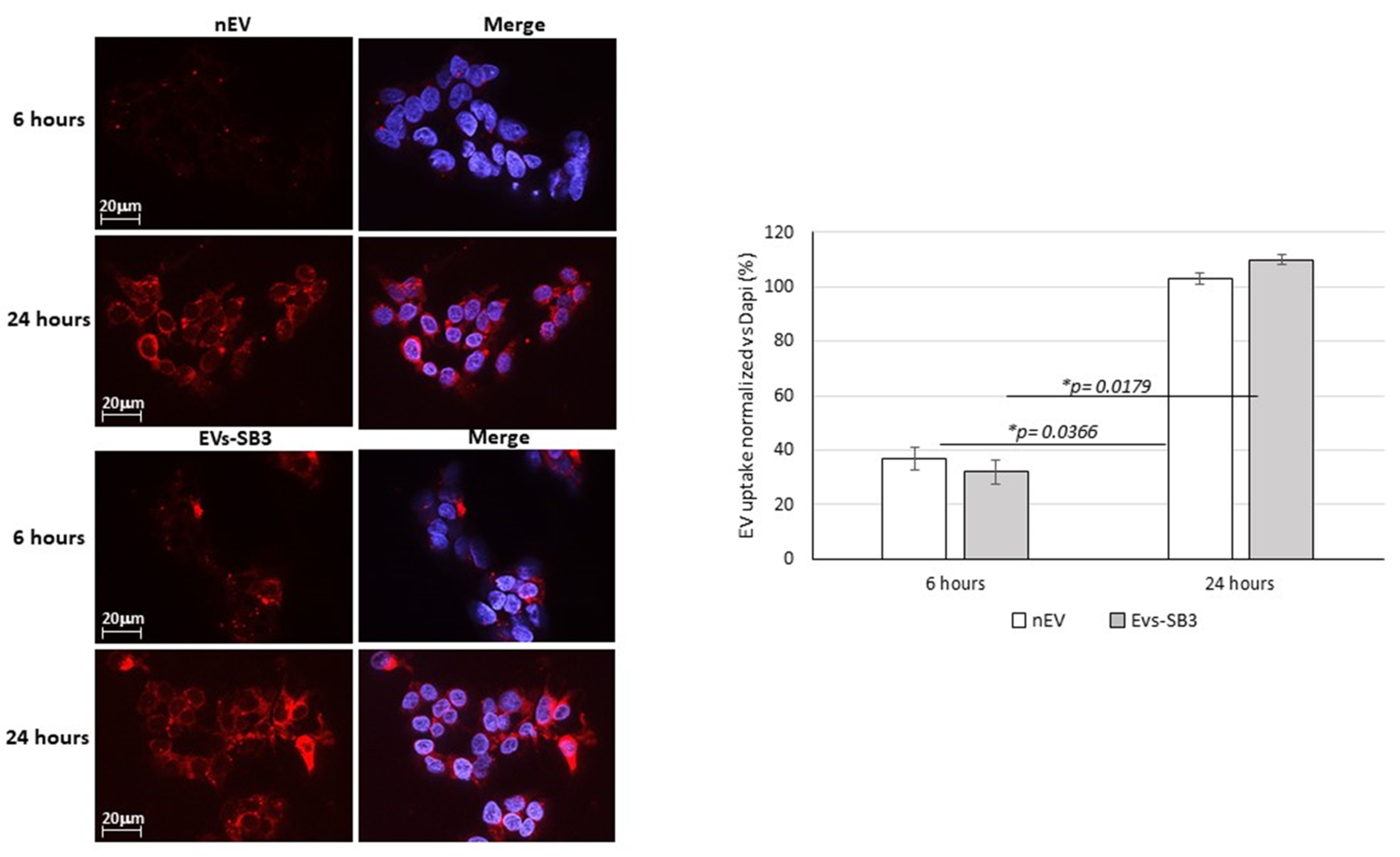
EV Uptake Detection
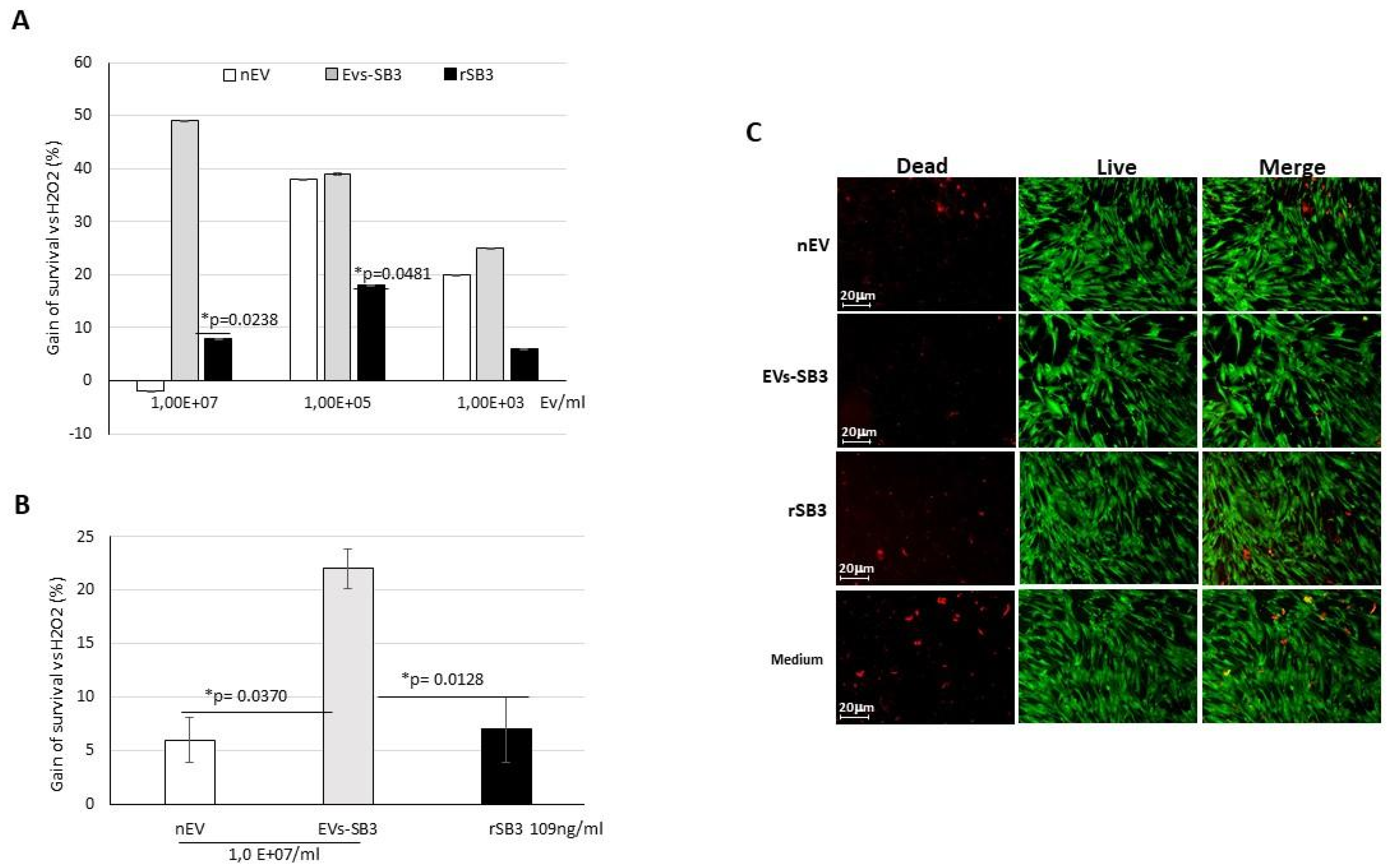
2.3. Biological Activity of EV-SerpinB3 In Vitro
3. Material and Methods
3.1. Characterization of SerpinB3 Expression in Transfected HepG2 Cells
3.1.1. Immunofluorescence
3.1.2. Quantitative Real-Time RT-PCR
3.2. Extracellular Vesicle Production and Characterization
3.2.1. Transmission Electron Microscopy
3.2.2. SerpinB3 Quantification by ELISA
3.3. Proteomic Analysis of EVs
3.4. Western Blot Analysis of Selected EV Proteins
3.5. Biological Activity of EV-SerpinB3 In Vitro
3.5.1. HepG2 Cells
3.5.2. Cardiomyocyte Primary Cells
3.6. EV Labeling and Uptake In Vitro
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andaloussi, S.E.L.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Dis. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Gettins, P.G. Serpin Structure, Mechanism, and Function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, G.A.; Whisstock, J.C.; Bottomley, S.P.; Huntington, J.A.; Kaiserman, D.; Luke, C.J.; Pak, S.C.; Reichhart, J.M.; Bird, P.I. Serpins flex their muscle: I. Putting the clamps, on proteolysis in diverse biological systems. J. Biol. Chem. 2010, 285, 24299–24305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villano, G.; Turato, C.; Quarta, S.; Ruvoletto, M.; Ciscato, F.; Terrin, L.; Semeraro, R.; Paternostro, C.; Parola, M.; Alvaro, D.; et al. Hepatic progenitor cells express SerpinB3. BMC Cell Biol. 2014, 15, 5. [Google Scholar] [CrossRef] [Green Version]
- Vidalino, L.; Doria, A.; Quarta, S.; Zen, M.; Gatta, A.; Pontisso, P. SERPINB3, apoptosis and autoimmunity. Autoimmun. Rev. 2009, 9, 108–112. [Google Scholar] [CrossRef]
- Katagiri, C.; Nakanishi, J.; Kadoya, K.; Hibino, T. Serpin squamous cell carcinoma antigen inhibits UV-induced apoptosis via suppression of c-JUN NH2-terminal kinase. J. Cell Biol. 2006, 172, 983–990. [Google Scholar] [CrossRef] [Green Version]
- Murakami, A.; Suminami, Y.; Hirakawa, H.; Nawata, S.; Numa, F.; Kato, H. Squamous cell carcinoma antigen suppresses radiation-induced cell death. Br. J. Cancer 2001, 84, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Ciscato, F.; Sciacovelli, M.; Villano, G.; Turato, C.; Bernardi, P.; Rasola, A.; Pontisso, P. SERPINB3 protects from oxidative damage by chemotherapeutics through inhibition of mitochondrial respiratory complex I. Oncotarget 2014, 5, 2418–2427. [Google Scholar] [CrossRef]
- Rasola, A.; Sciacovelli, M.; Pantic, B.; Bernardi, P. Signal transduction to the permeability transition pore. FEBS Lett. 2010, 584, 1989–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Witwer, K.V.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkim-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesc. 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Wassell, J. Haptoglobin: Function and polymorphism. Clin. Lab. 2000, 46, 547–552. [Google Scholar] [PubMed]
- Battisti, I.; Ebinezer, L.B.; Lomolino, G.; Masi, A.; Arrigoni, G. Protein profile of commercial soybean milks analyzed by label-free quantitative proteomics. Food Chem. 2021, 352, 129299. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Raudvere, U.; Liis Kolberg, L.; Ivan Kuzmin, I.; Tambet Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turato, C.; Biasiolo, A.; Pengo, P.; Frecer, V.; Quarta, S.; Fasolato, S.; Ruvoletto, M.; Beneduce, L.; Zuin, J.; Fassina, G.; et al. Increased antiprotease activity of the SERPINB3 polymorphc variant SCCA-PD. Exp. Biol. Med. 2011, 236, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.A.; Schiffelers, R.M.; Zarovni, N.; Vago, R. Modulation of Tissue Tropism and Biological Activity of Exosomes and Other Extracellular Vesicles: New Nanotools for Cancer Treatment. Pharmacol. Res. 2016, 111, 487–500. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular Vesicle-Based Therapeutics: Natural versus Engineered Targeting and Trafficking. Exp. Mol. Med. 2019, 51, 32. [Google Scholar] [CrossRef]
- Reshke, R.; Taylor, J.A.; Savard, A.; Guo, H.; Rhym, L.H.; Kowalski, P.S.; Trung, M.T.; Campbell, C.; Little, W.; Anderson, D.G.; et al. Reduction of the Therapeutic Dose of Silencing RNA by Packaging It in Extracellular Vesicles via a Pre-MicroRNA Backbone. Nat. Biomed. Eng. 2020, 4, 52–68. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Christiansen, G.; Gurevich, L.; Moos, T.; Duroux, M. Evaluation of Electroporation-Induced Adverse Effects on Adipose-Derived Stem Cell Exosomes. Cytotechnology 2016, 68, 2125–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active Loading into Extracellular Vesicles Significantly Improves the Cellular Uptake and Photodynamic Effect of Porphyrins. J. Control. Release 2015, 205, 35–44. [Google Scholar] [CrossRef]
- Cannito, S.; Foglia, B.; Villano, G.; Turato, C.; Delgado, T.C.; Morello, E.; Pin, F.; Novo, E.; Napione, L.; Quarta, S.; et al. SerpinB3 differently up-regulates hypoxia inducible factors-1α and -2α in hepatocellularevealing novel potential therapeutic targets. Cancers carcinoma: Mechanisms. Cancers 2019, 11, 1933. [Google Scholar] [CrossRef] [Green Version]
- Estelles, A.; Sperinde, J.; Roulon, T.; Aguilar, B.; Bonner, C.; LePecq, J.B.; Delcayre, A. Exosome Nanovesicles Displaying G Protein-Coupled Receptors for Drug Discovery. Int. J. Nanomed. 2007, 2, 751–760. [Google Scholar]
- Koliha, N.; Wiencek, Y.; Heider, U.; Jüngst, C.; Kladt, N.; Krauthäuser, S.; Johnston, I.C.D.; Bosio, A.; Schauss, A.; Wild, S. A Novel Multiplex Bead-Based Platform Highlights the Diversity of Extracellular Vesicles. J. Extracell. Vesc. 2016, 5, 29975. [Google Scholar] [CrossRef]
- Fasolato, S.; Ruvoletto, M.; Nardo, G.; Rasola, A.; Sciacovelli, M.; Zanus, G.; Turato, C.; Quarta, S.; Terrin, L.; Fadini, G.P.; et al. Low P66shc with High SerpinB3 Levels Favors Necroptosis and Better Survival in Hepatocellular Carcinoma. Biology 2021, 10, 363. [Google Scholar] [CrossRef]
- Tailor, J.; Monteith, G.; Bebawy, M. Ca2+ medicates extracellular vescicle biogenesis thrpugh alternate pathways in malignancy. J. Extracell. Vesc. 2020, 9, 1734326. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, R.M. Maturation of Reticulocytes: Formation of Exosomes as a Mechanism for Shedding Membrane Proteins. Biochem. Cell Biol. 1991, 70, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.; Bayraktar, E.; Helal, E.G.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef] [Green Version]
- Yong, T.; Zhang, X.; Bie, N.; Zhang, H.; Zhang, X.; Li, F.; Hakeem, A.; Hu, J.; Gan, L.; Sanatos, H.A.; et al. Tumor Exosome-Based Nanoparticles Are Efficient Drug Carriers for Chemotherapy. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tsiapalis, D.; O’Driscoll, L. Mesenchymal Stem Cell Derived Extracellular Vesicles for Tissue Engineering and Regenerative Medicine Applications. Cells 2020, 9, 991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-Derived Exosomes for Cell-Free Therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giebel, B.; Kordelas, L.; Börger, V. Clinical Potential of Mesenchymal Stem/Stromal Cell-Derived Extracellular Vesicles. Stem Cell Investig. 2017, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene Name | # Unique Peptides | Fold Change (SB3 vs. CTR) | p Value |
|---|---|---|---|
| HP | 17 | 2.1 | 8.6 × 104 |
| GGH | 4 | −2.2 | 1.9 × 102 |
| ACTN4 | 17 | −2.3 | 3.3 × 105 |
| TUBA1B | 3 | −2.4 | 3.4 × 102 |
| PSMA3 | 2 | −2.6 | 3.9 × 102 |
| ENO1 | 17 | −2.7 | 4.6 × 102 |
| TALDO1 | 2 | −2.8 | 5.0 × 102 |
| LDHA | 8 | −3.1 | 1.8 × 102 |
| NME2 | 8 | −3.2 | 1.2 × 103 |
| CLSTN1 | 13 | −3.2 | 6.5 × 103 |
| HSP90AB1 | 18 | −3.4 | 4.8 × 102 |
| QPRT | 4 | −3.5 | 3.5 × 102 |
| PSMB6 | 3 | −3.7 | 1.3 × 102 |
| PSMA7 | 4 | −3.7 | 4.1 × 102 |
| GAPDH | 14 | −3.7 | 4.9 × 102 |
| GPI | 5 | −3.7 | 0.0 × 100 |
| HIST1H2AJ | 5 | −3.7 | 8.0 × 104 |
| HSPA4 | 8 | −3.8 | 2.8 × 104 |
| ALDH1A1 | 11 | −4.0 | 0.0 × 100 |
| RPSA | 4 | −4.1 | 1.5 × 102 |
| CTSC | 2 | −4.2 | 4.1 × 108 |
| PGAM1 | 2 | −4.2 | 2.9 × 106 |
| AHCY | 9 | −4.3 | 5.0 × 103 |
| UBA1 | 5 | −4.3 | 2.2 × 102 |
| FASN | 44 | −5.0 | 1.6 × 102 |
| PSMA2 | 2 | −5.8 | 1.2 × 102 |
| GLUD1 | 15 | −6.4 | 2.2 × 103 |
| HIST1H4A | 7 | −7.2 | 1.8 × 102 |
| TFRC | 5 | −14.6 | 1.4 × 107 |
| AGRN | 10 | −20.2 | 8.8 × 106 |
| ACLY | 3 | Found only in CTR | --- |
| APEH | 2 | Found only in CTR | --- |
| VCAN | 7 | Found only in CTR | --- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tolomeo, A.M.; Quarta, S.; Biasiolo, A.; Ruvoletto, M.; Pozzobon, M.; De Lazzari, G.; Malvicini, R.; Turato, C.; Arrigoni, G.; Pontisso, P.; et al. Engineered EVs for Oxidative Stress Protection. Pharmaceuticals 2021, 14, 703. https://doi.org/10.3390/ph14080703
Tolomeo AM, Quarta S, Biasiolo A, Ruvoletto M, Pozzobon M, De Lazzari G, Malvicini R, Turato C, Arrigoni G, Pontisso P, et al. Engineered EVs for Oxidative Stress Protection. Pharmaceuticals. 2021; 14(8):703. https://doi.org/10.3390/ph14080703
Chicago/Turabian StyleTolomeo, Anna Maria, Santina Quarta, Alessandra Biasiolo, Mariagrazia Ruvoletto, Michela Pozzobon, Giada De Lazzari, Ricardo Malvicini, Cristian Turato, Giorgio Arrigoni, Patrizia Pontisso, and et al. 2021. "Engineered EVs for Oxidative Stress Protection" Pharmaceuticals 14, no. 8: 703. https://doi.org/10.3390/ph14080703
APA StyleTolomeo, A. M., Quarta, S., Biasiolo, A., Ruvoletto, M., Pozzobon, M., De Lazzari, G., Malvicini, R., Turato, C., Arrigoni, G., Pontisso, P., & Muraca, M. (2021). Engineered EVs for Oxidative Stress Protection. Pharmaceuticals, 14(8), 703. https://doi.org/10.3390/ph14080703








