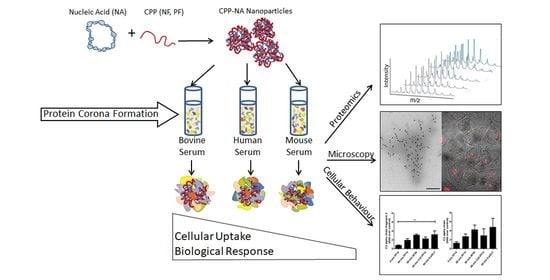
Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona
Abstract
:1. Introduction
2. Results and Discussion
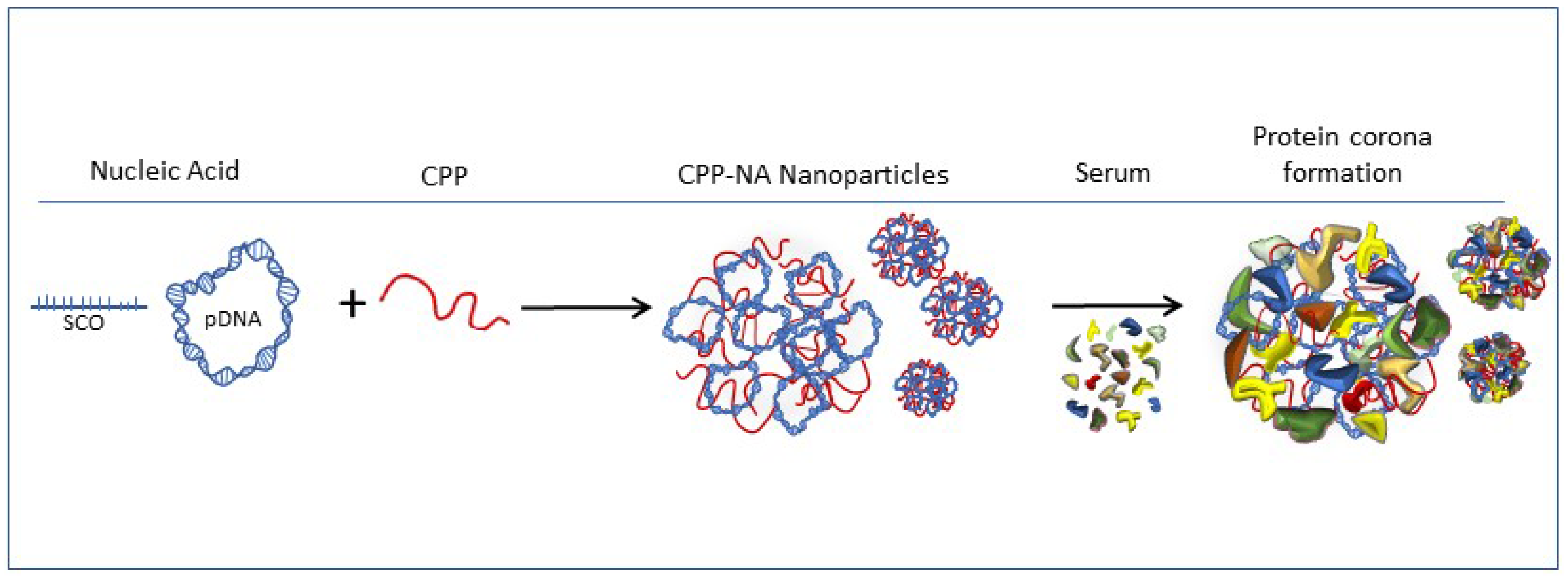
2.1. Formation of Protein Corona on CPP-pDNA Nanoparticles Depends on Used Serum
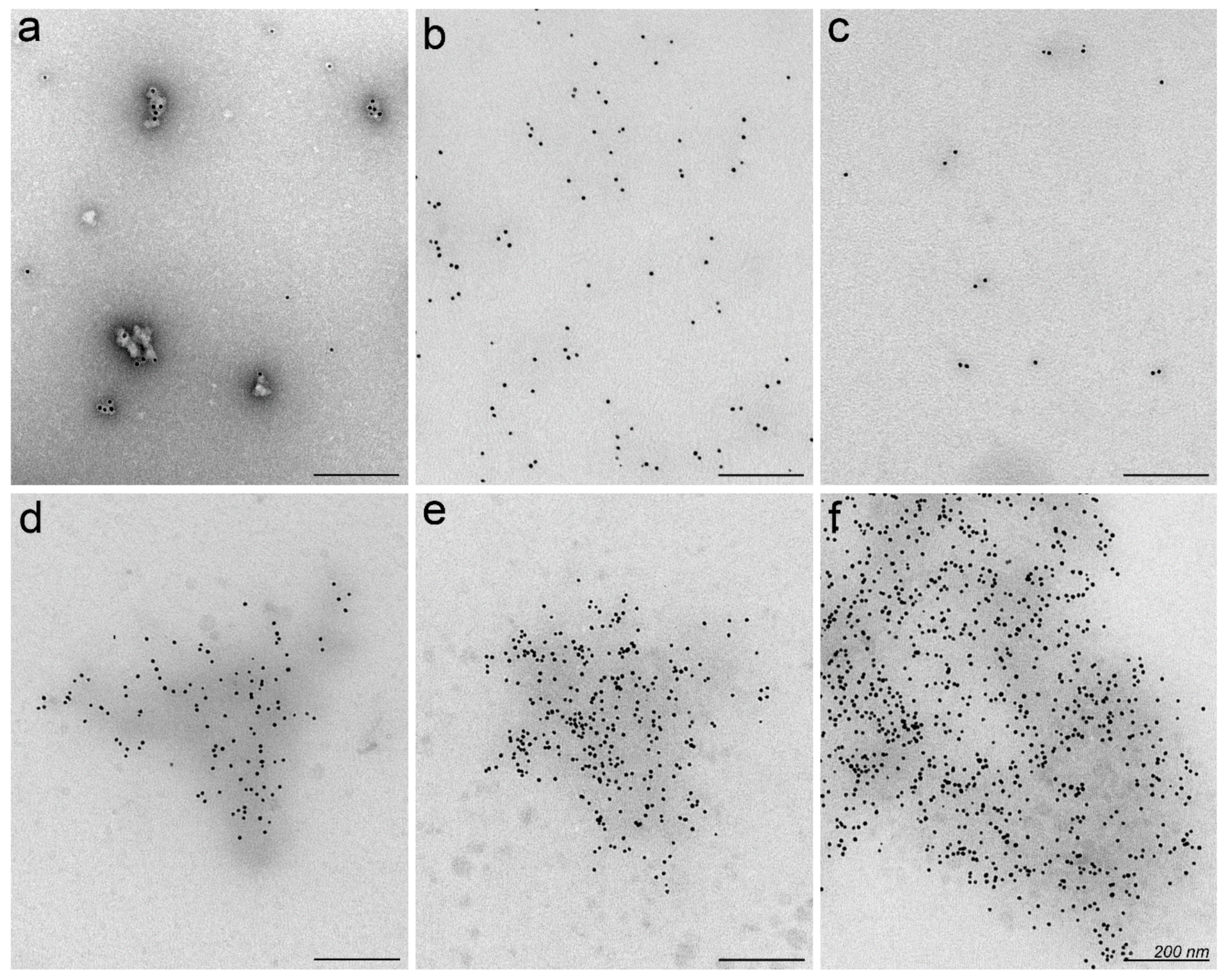
2.2. Nanoparticles May Aggregate in Serum Containing Environment
2.3. Proteins Involved in Transport and Haemostasis Are Major Constituents of Protein Corona
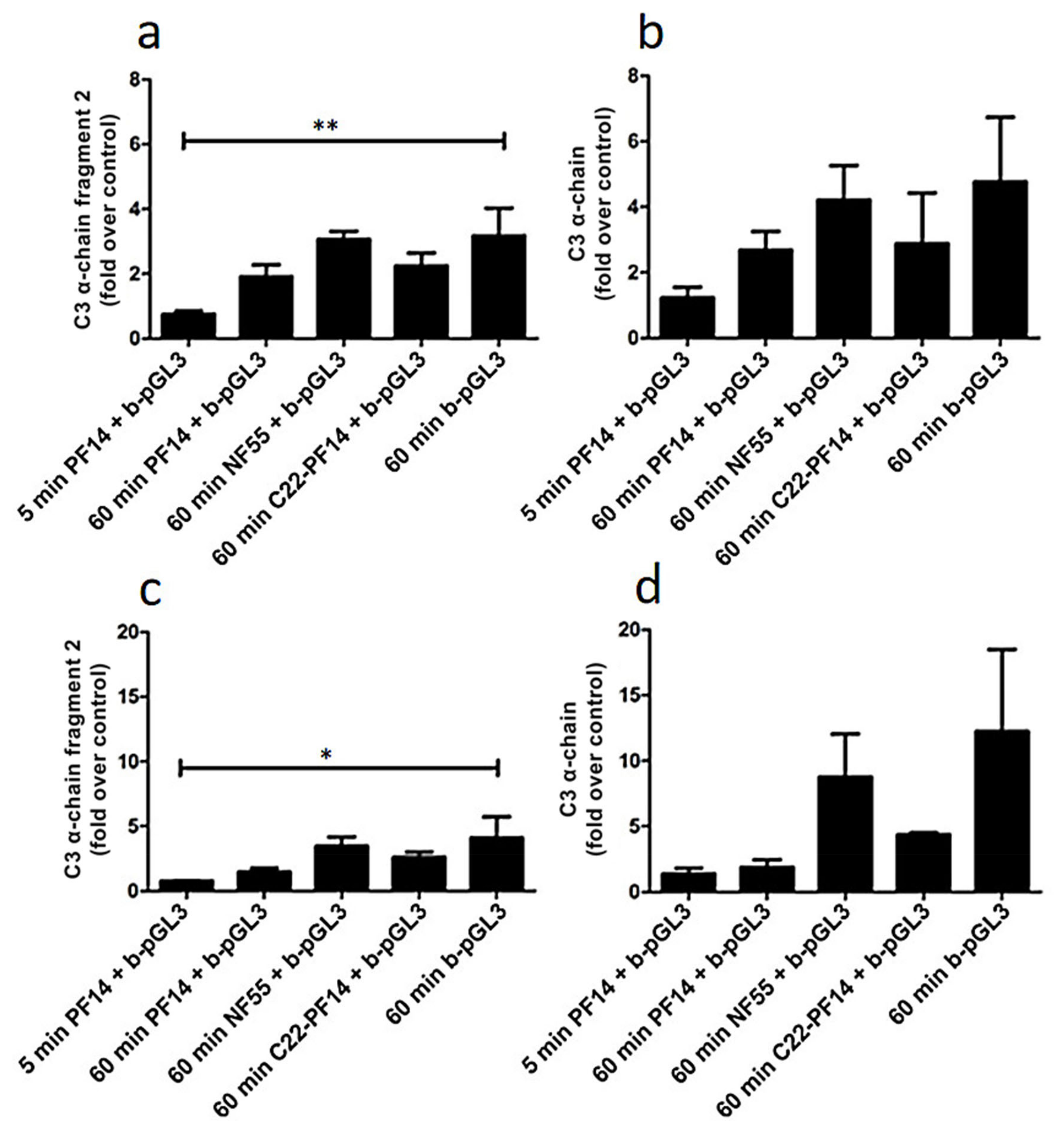
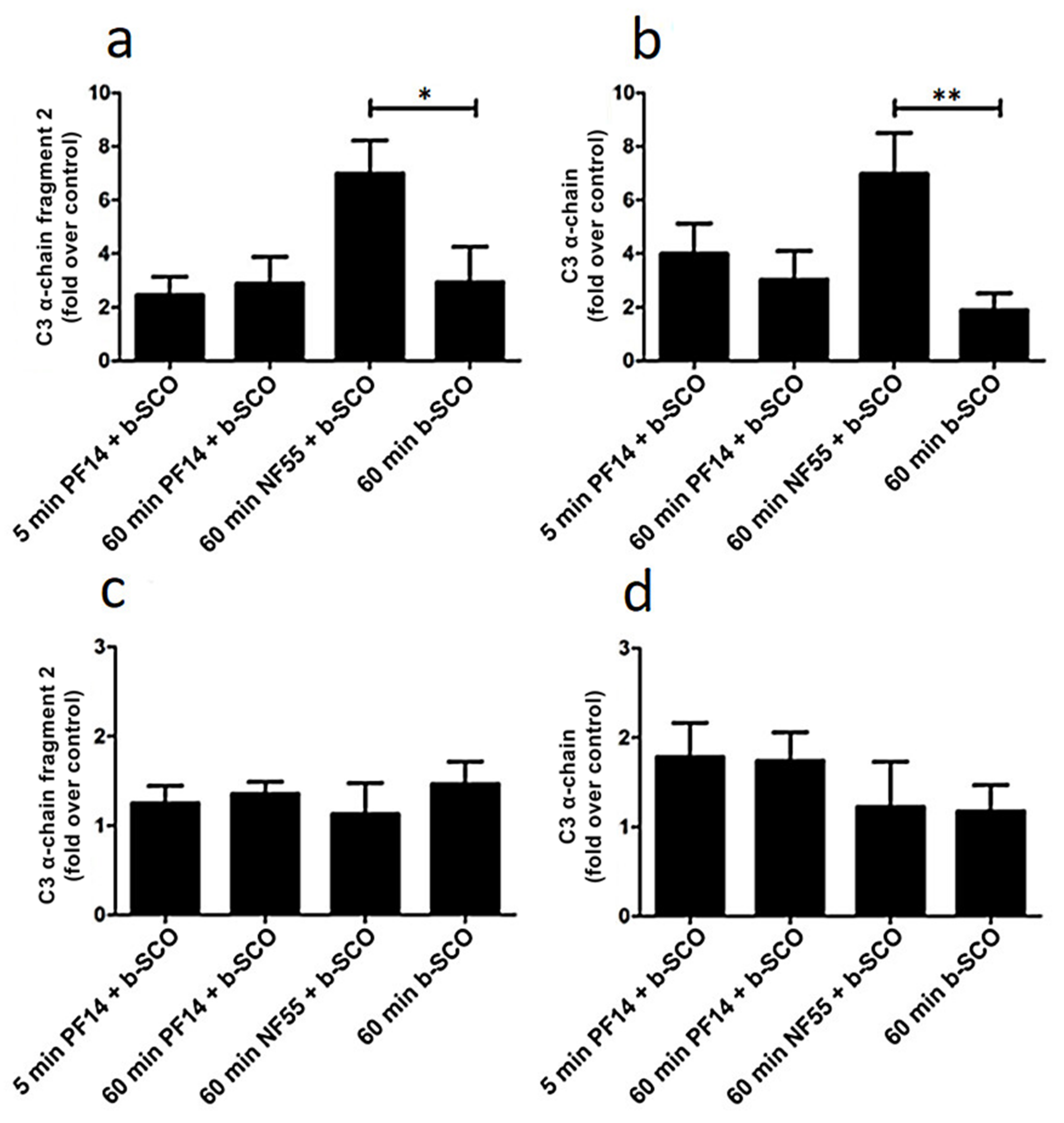
2.4. Adsorption of Complement Component 3 Depends on Nanoparticle Characteristics
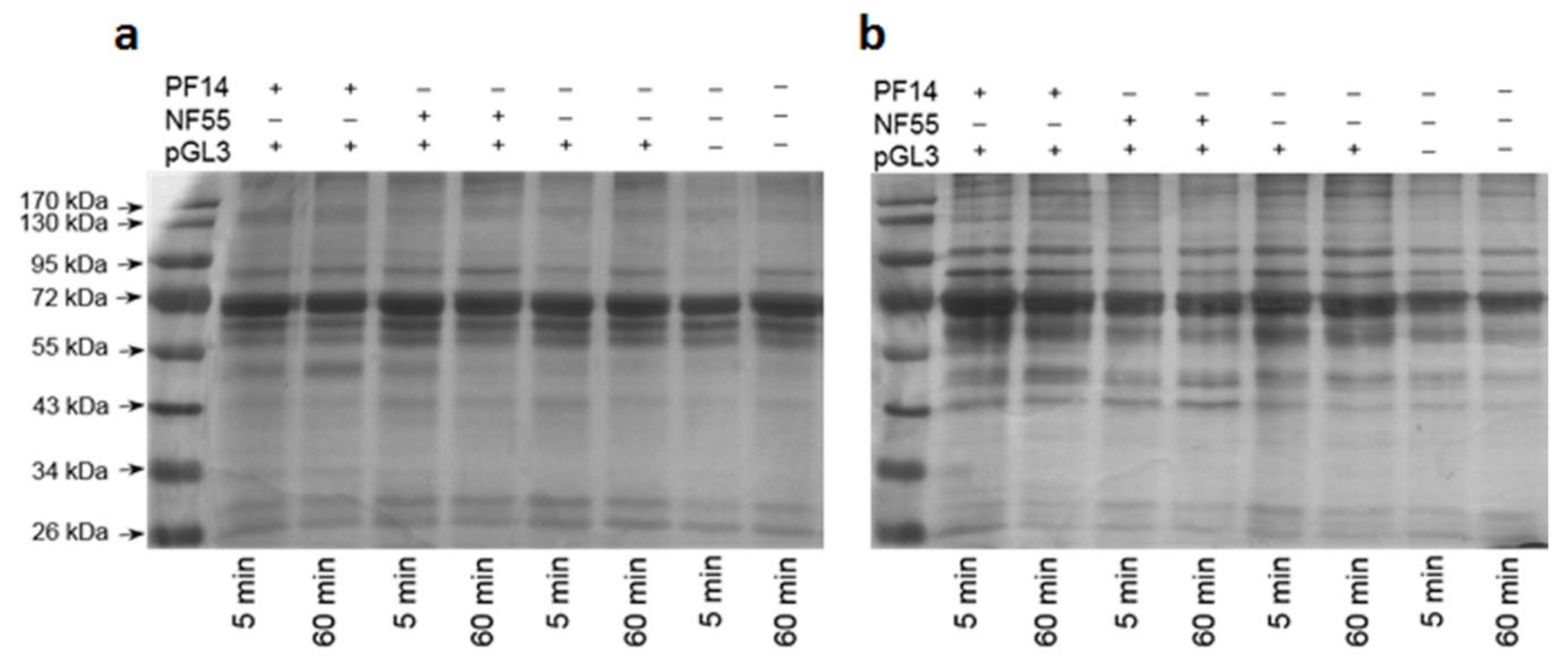
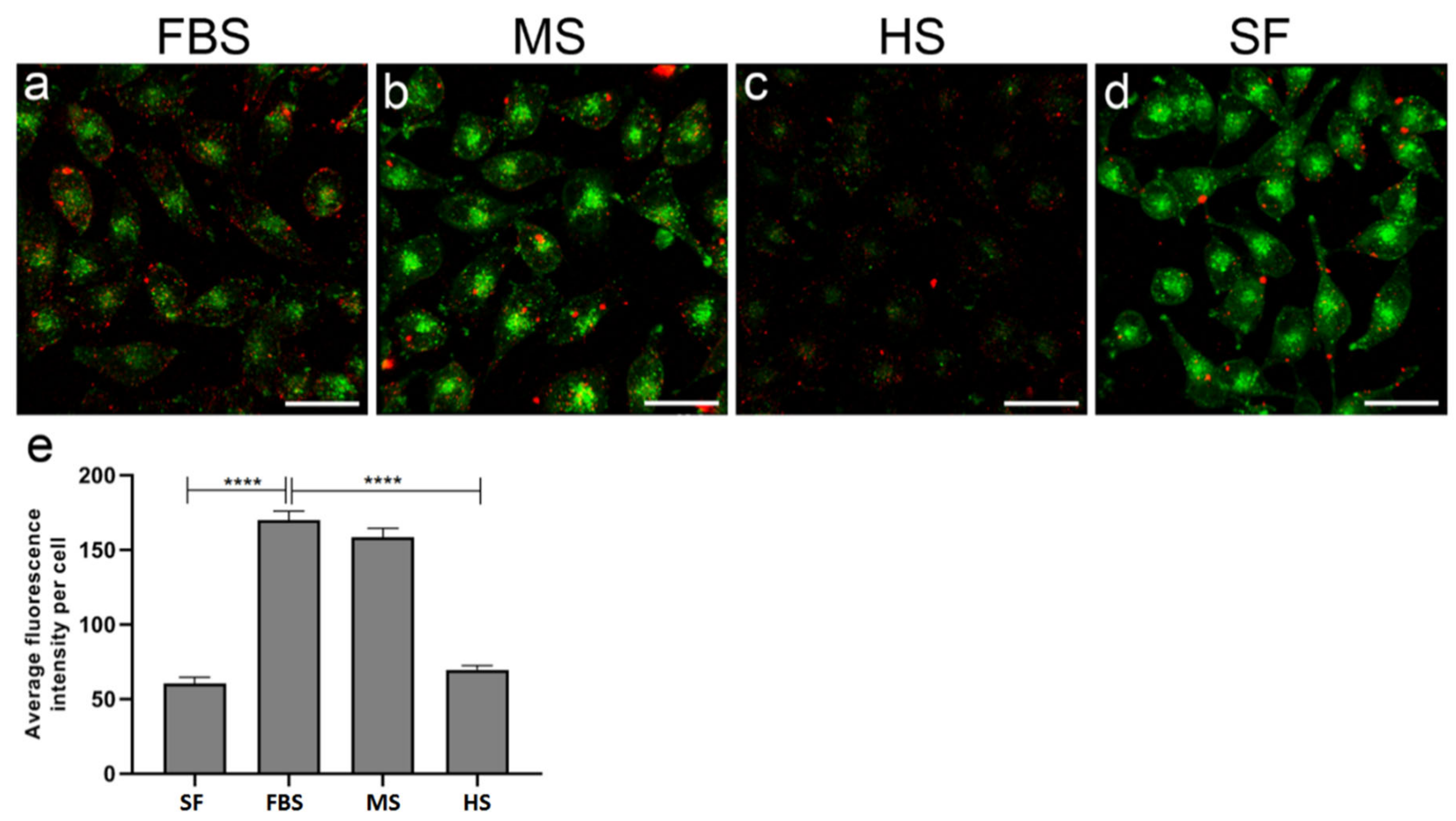
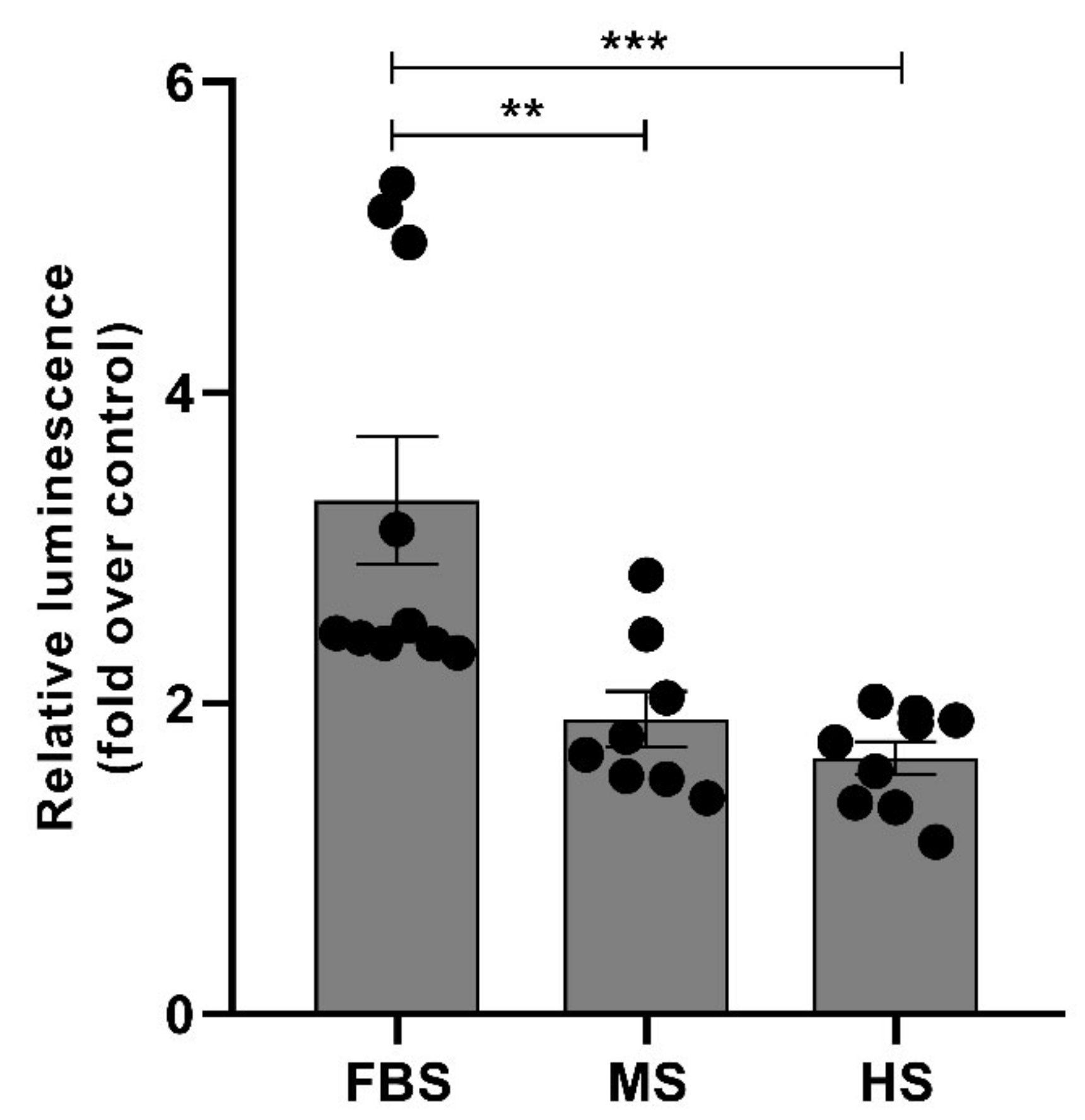
2.5. Protein Corona on CPP-SCO Nanoparticles Controls the Efficiency of Internalisation and Splicing Switching by SCO in Cells
3. Materials and Methods
3.1. Plasma and Serum Collection
3.2. Labelling of pDNA with Biotin
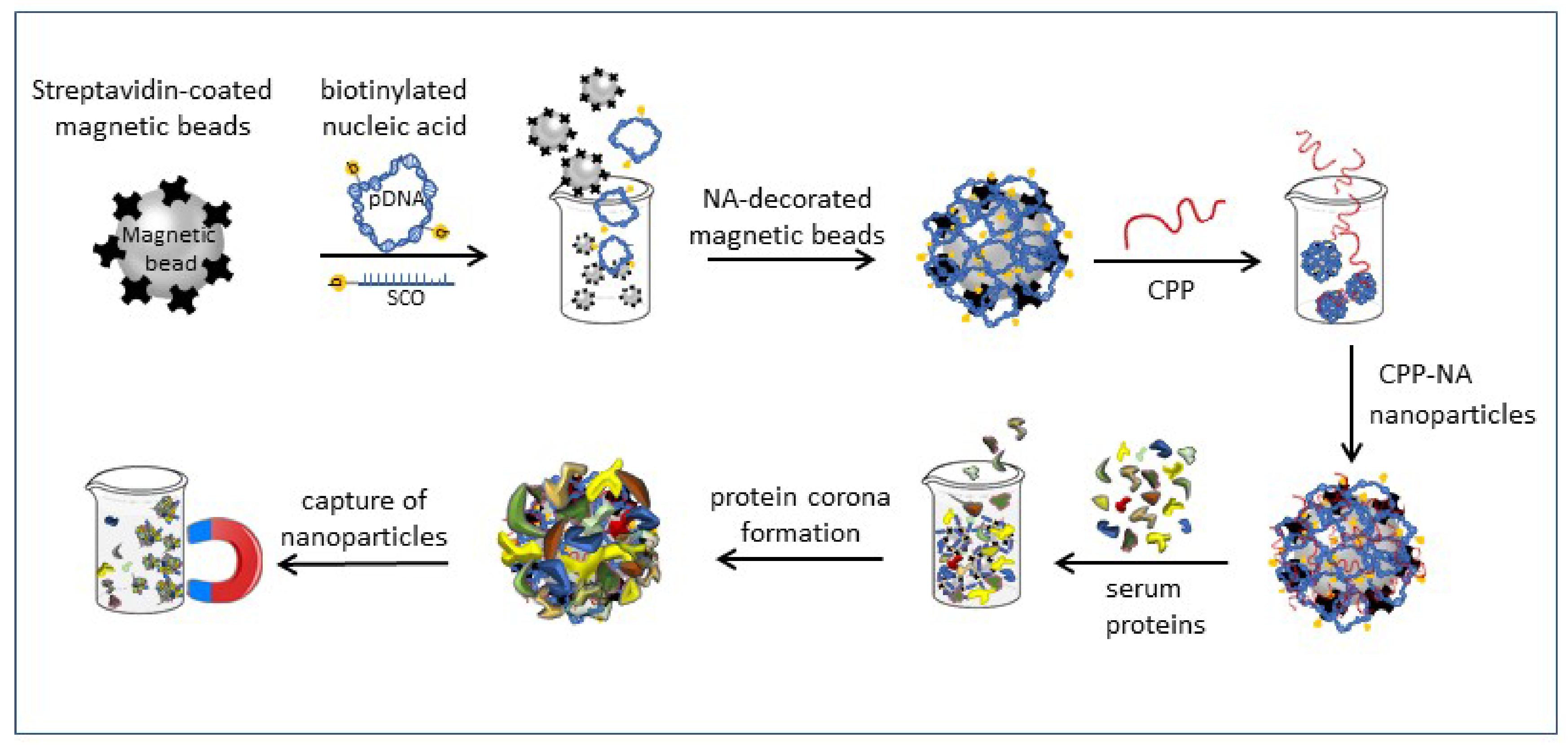
3.3. Nanoparticle Formation and Pull-Down
3.4. Dynamic Light Scattering (DLS) and Zeta Potential Measurements
3.5. Protein Separation and Silver Staining
3.6. Western Blot
3.7. Mass-Spectrometry
3.8. Electron Microscopy of Nanoparticles
3.9. Translocation of PF14-SCO Nanoparticles into Cells in Different Sera Containing Media and Splicing Correction by SCO
3.10. Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| C3 | component 3 of complement system |
| C22-PF14 | PepFect 14 analogue with a 22 carbon-fatty acid (behenyl) |
| CPP | cell-penetrating peptide |
| MS | mass-spectrometry |
| NF55 | NickFect 55 peptide |
| pDNA | plasmid DNA |
| PF14 | PepFect 14 peptide |
| SCO | splice-correcting oligonucleotide |
References
- Riley, R.S.; June, C.H.; Langer, R.; Mitchell, M.J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019, 18, 175–196. [Google Scholar] [CrossRef] [PubMed]
- Fenton, O.S.; Olafson, K.N.; Pillai, P.S.; Mitchell, M.J.; Langer, R. Advances in Biomaterials for Drug Delivery. Adv. Mater. 2018, 30, e1705328. [Google Scholar] [CrossRef]
- Jones, M.R.; Seeman, N.C.; Mirkin, C.A. Nanomaterials. Programmable materials and the nature of the DNA bond. Science 2015, 347, 1260901. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Copolovici, D.M.; Langel, K.; Eriste, E.; Langel, U. Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef] [PubMed]
- Peraro, L.; Kritzer, J.A. Emerging Methods and Design Principles for Cell-Penetrant Peptides. Angew. Chem. Int. Ed. Engl. 2018, 57, 11868–11881. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Margus, H.; Padari, K.; Pooga, M. Insights into cell entry and intracellular trafficking of peptide and protein drugs provided by electron microscopy. Adv. Drug Deliv. Rev. 2013, 65, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Urgard, E.; Lorents, A.; Klaas, M.; Padari, K.; Viil, J.; Runnel, T.; Langel, K.; Kingo, K.; Tkaczyk, E.; Langel, U.; et al. Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J. Control. Release 2016, 235, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Ukidve, A.; Krishnan, V.; Mitragotri, S. Effect of physicochemical and surface properties on in vivo fate of drug nanocarriers. Adv. Drug Deliv Rev. 2019, 143, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Francia, V.; Schiffelers, R.M.; Cullis, P.R.; Witzigmann, D. The Biomolecular Corona of Lipid Nanoparticles for Gene Therapy. Bioconjug. Chem. 2020, 31, 2046–2059. [Google Scholar] [CrossRef] [PubMed]
- Paunovska, K.; Sago, C.D.; Monaco, C.M.; Hudson, W.H.; Castro, M.G.; Rudoltz, T.G.; Kalathoor, S.; Vanover, D.A.; Santangelo, P.J.; Ahmed, R.; et al. A Direct Comparison of in Vitro and in Vivo Nucleic Acid Delivery Mediated by Hundreds of Nanoparticles Reveals a Weak Correlation. Nano Lett. 2018, 18, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Malcolm, D.W.; Varghese, J.J.; Sorrells, J.E.; Ovitt, C.E.; Benoit, D.S.W. The Effects of Biological Fluids on Colloidal Stability and siRNA Delivery of a pH-Responsive Micellar Nanoparticle Delivery System. ACS Nano 2018, 12, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Saher, O.; Lehto, T.; Gissberg, O.; Gupta, D.; Gustafsson, O.; Andaloussi, S.E.; Darbre, T.; Lundin, K.E.; Smith, C.I.E.; Zain, R. Sugar and Polymer Excipients Enhance Uptake and Splice-Switching Activity of Peptide-Dendrimer/Lipid/Oligonucleotide Formulations. Pharmaceutics 2019, 11, 666. [Google Scholar] [CrossRef] [PubMed]
- Lundin, K.E.; Gissberg, O.; Smith, C.I.E.; Zain, R. Chemical Development of Therapeutic Oligonucleotides. Methods Mol. Biol. 2019, 2036, 3–16. [Google Scholar] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, P.; Hall, J.B.; McLeland, C.B.; Dobrovolskaia, M.A.; McNeil, S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009, 61, 428–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggard, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.H.; Lee, B.J. Protein corona: A new approach for nanomedicine design. Int. J. Nanomed. 2017, 12, 3137–3151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbo, C.; Molinaro, R.; Parodi, A.; Toledano Furman, N.E.; Salvatore, F.; Tasciotti, E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caracciolo, G.; Cardarelli, F.; Pozzi, D.; Salomone, F.; Maccari, G.; Bardi, G.; Capriotti, A.L.; Cavaliere, C.; Papi, M.; Lagana, A. Selective targeting capability acquired with a protein corona adsorbed on the surface of 1,2-dioleoyl-3-trimethylammonium propane/DNA nanoparticles. ACS Appl Mater. Interfaces 2013, 5, 13171–13179. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Querbes, W.; De, S.; Qin, J.; Frank-Kamenetsky, M.; Jayaprakash, K.N.; Jayaraman, M.; Rajeev, K.G.; Cantley, W.L.; Dorkin, J.R.; et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010, 18, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Love, K.T.; Dorkin, J.R.; Sirirungruang, S.; Zhang, Y.; Chen, D.; Bogorad, R.L.; Yin, H.; Chen, Y.; Vegas, A.J.; et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. USA 2014, 111, 3955–3960. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Kuipers, F.; Havekes, L.M.; Havinga, R.; Dontje, B.; Poelstra, K.; Scherphof, G.L.; Kamps, J.A. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem. Biophys. Res. Commun. 2005, 328, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef] [PubMed]
- Valeur, E.; Jimonet, P. New Modalities, Technologies, and Partnerships in Probe and Lead Generation: Enabling a Mode-of-Action Centric Paradigm. J. Med. Chem. 2018, 61, 9004–9029. [Google Scholar] [CrossRef]
- Porosk, L.; Arukuusk, P.; Pohako, K.; Kurrikoff, K.; Kiisholts, K.; Padari, K.; Pooga, M.; Langel, U. Enhancement of siRNA transfection by the optimization of fatty acid length and histidine content in the CPP. Biomater. Sci. 2019, 7, 4363–4374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cagliani, R.; Gatto, F.; Bardi, G. Protein Adsorption: A Feasible Method for Nanoparticle Functionalization? Materials 2019, 12, 1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvati, A.; Pitek, A.S.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.R.; Kelly, P.M.; Aberg, C.; Mahon, E.; Dawson, K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francia, V.; Montizaan, D.; Salvati, A. Interactions at the cell membrane and pathways of internalization of nano-sized materials for nanomedicine. Beilstein J. Nanotechnol. 2020, 11, 338–353. [Google Scholar] [CrossRef] [PubMed]
- Saha, K.; Kim, S.T.; Yan, B.; Miranda, O.R.; Alfonso, F.S.; Shlosman, D.; Rotello, V.M. Surface functionality of nanoparticles determines cellular uptake mechanisms in mammalian cells. Small 2013, 9, 300–305. [Google Scholar] [CrossRef]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned—Challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessner, A.; Lieske, A.; Paulke, B.; Muller, R. Influence of surface charge density on protein adsorption on polymeric nanoparticles: Analysis by two-dimensional electrophoresis. Eur. J. Pharm. Biopharm. 2002, 54, 165–170. [Google Scholar] [CrossRef]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, M.; Abdelmonem, A.M.; Behzadi, S.; Clement, J.H.; Dutz, S.; Ejtehadi, M.R.; Hartmann, R.; Kantner, K.; Linne, U.; Maffre, P.; et al. Temperature: The "ignored" factor at the NanoBio interface. ACS Nano 2013, 7, 6555–6562. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Lohse, S.E.; Murphy, C.J.; Fathizadeh, A.; Montazeri, A.; Suslick, K.S. Variation of protein corona composition of gold nanoparticles following plasmonic heating. Nano Lett 2014, 14, 6–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barran-Berdon, A.L.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Lagana, A. Time evolution of nanoparticle-protein corona in human plasma: Relevance for targeted drug delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Augustsson, C.; Lilja, M.; Lundkvist, K.; Dahlback, B.; Linse, S.; Cedervall, T. The nanoparticle protein corona formed in human blood or human blood fractions. PLoS ONE 2017, 12, e0175871. [Google Scholar] [CrossRef] [Green Version]
- Schottler, S.; Klein, K.; Landfester, K.; Mailander, V. Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale 2016, 8, 5526–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirshafiee, V.; Kim, R.; Mahmoudi, M.; Kraft, M.L. The importance of selecting a proper biological milieu for protein corona analysis in vitro: Human plasma versus human serum. Int. J. Biochem. Cell Biol. 2016, 75, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Colapicchioni, V.; Tilio, M.; Digiacomo, L.; Gambini, V.; Palchetti, S.; Marchini, C.; Pozzi, D.; Occhipinti, S.; Amici, A.; Caracciolo, G. Personalized liposome-protein corona in the blood of breast, gastric and pancreatic cancer patients. Int. J. Biochem. Cell Biol. 2016, 75, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Tabatabaei, M.; Farokhzad, O.C.; Mahmoudi, M. Personalized protein corona on nanoparticles and its clinical implications. Biomater. Sci. 2017, 5, 378–387. [Google Scholar] [CrossRef]
- Xiao, W.; Xiong, J.; Zhang, S.; Xiong, Y.; Zhang, H.; Gao, H. Influence of ligands property and particle size of gold nanoparticles on the protein adsorption and corresponding targeting ability. Int. J. Pharm 2018, 538, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alvarez, R.; Hadjidemetriou, M.; Sanchez-Iglesias, A.; Liz-Marzan, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [Green Version]
- Arvizo, R.R.; Miranda, O.R.; Moyano, D.F.; Walden, C.A.; Giri, K.; Bhattacharya, R.; Robertson, J.D.; Rotello, V.M.; Reid, J.M.; Mukherjee, P. Modulating pharmacokinetics, tumor uptake and biodistribution by engineered nanoparticles. PLoS ONE 2011, 6, e24374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Gao, H. The impact of protein corona on the behavior and targeting capability of nanoparticle-based delivery system. Int. J. Pharm 2018, 552, 328–339. [Google Scholar] [CrossRef]
- Van Asbeck, A.H.; Beyerle, A.; McNeill, H.; Bovee-Geurts, P.H.; Lindberg, S.; Verdurmen, W.P.; Hallbrink, M.; Langel, U.; Heidenreich, O.; Brock, R. Molecular parameters of siRNA--cell penetrating peptide nanocomplexes for efficient cellular delivery. ACS Nano 2013, 7, 3797–3807. [Google Scholar] [CrossRef]
- Margus, H.; Arukuusk, P.; Langel, Ü.; Pooga, M. Characteristics of Cell-Penetrating Peptide/Nucleic Acid Nanoparticles. Mol. Pharm. 2016, 13, 172–179. [Google Scholar] [CrossRef]
- Arukuusk, P.; Pärnaste, L.; Margus, H.; Eriksson, N.K.; Vasconcelos, L.; Padari, K.; Pooga, M.; Langel, Ü. Differential endosomal pathways for radically modified peptide vectors. Bioconjug. Chem. 2013, 24, 1721–1732. [Google Scholar] [CrossRef]
- Deshayes, S.; Konate, K.; Dussot, M.; Chavey, B.; Vaissiere, A.; Van, T.N.N.; Aldrian, G.; Padari, K.; Pooga, M.; Vives, E.; et al. Deciphering the internalization mechanism of WRAP:siRNA nanoparticles. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183252. [Google Scholar] [CrossRef]
- Ezzat, K.; Helmfors, H.; Tudoran, O.; Juks, C.; Lindberg, S.; Padari, K.; El-Andaloussi, S.; Pooga, M.; Langel, U. Scavenger receptor-mediated uptake of cell-penetrating peptide nanocomplexes with oligonucleotides. FASEB J. 2012, 26, 1172–1180. [Google Scholar] [CrossRef]
- Bolhassani, A.; Kardani, K.; Vahabpour, R.; Habibzadeh, N.; Aghasadeghi, M.R.; Sadat, S.M.; Agi, E. Prime/boost immunization with HIV-1 MPER-V3 fusion construct enhances humoral and cellular immune responses. Immunol. Lett. 2015, 168, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Konate, K.; Lindberg, M.F.; Vaissiere, A.; Jourdan, C.; Aldrian, G.; Margeat, E.; Deshayes, S.; Boisguerin, P. Optimisation of vectorisation property: A comparative study for a secondary amphipathic peptide. Int. J. Pharm. 2016, 509, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Kurrikoff, K.; Veiman, K.L.; Kunnapuu, K.; Peets, E.M.; Lehto, T.; Parnaste, L.; Arukuusk, P.; Langel, U. Effective in vivo gene delivery with reduced toxicity, achieved by charge and fatty acid -modified cell penetrating peptide. Sci. Rep. 2017, 7, 17056. [Google Scholar] [CrossRef]
- Lehto, T.; Vasconcelos, L.; Margus, H.; Figueroa, R.; Pooga, M.; Hällbrink, M.; Langel, Ü. Saturated Fatty Acid Analogues of Cell-Penetrating Peptide PepFect14: Role of Fatty Acid Modification in Complexation and Delivery of Splice-Correcting Oligonucleotides. Bioconjug. Chem. 2017, 28, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Freimann, K.; Arukuusk, P.; Kurrikoff, K.; Parnaste, L.; Raid, R.; Piirsoo, A.; Pooga, M.; Langel, U. Formulation of Stable and Homogeneous Cell-Penetrating Peptide NF55 Nanoparticles for Efficient Gene Delivery In Vivo. Mol. Ther. Nucleic Acids 2018, 10, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Bombelli, F.B.; Dawson, K.A. Physical-chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Aberg, C.; Salvati, A.; Dawson, K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Gaus, H.J.; Gupta, R.; Chappell, A.E.; Ostergaard, M.E.; Swayze, E.E.; Seth, P.P. Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay. Nucleic Acids Res. 2018, 47, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Oskolkov, N.; Arukuusk, P.; Copolovici, D.M.; Lindberg, S.; Margus, H.; Padari, K.; Pooga, M.; Langel, Ü. NickFects, Phosphorylated Derivatives of Transportan 10 for Cellular Delivery of Oligonucleotides. Int. J. Peptide Res. Ther. 2011, 17, 147–157. [Google Scholar] [CrossRef]
- Breitner, E.K.; Hussain, S.M.; Comfort, K.K. The role of biological fluid and dynamic flow in the behavior and cellular interactions of gold nanoparticles. J. Nanobiotechnol. 2015, 13, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sork, H.; Nordin, J.Z.; Turunen, J.J.; Wiklander, O.P.; Bestas, B.; Zaghloul, E.M.; Margus, H.; Padari, K.; Duru, A.D.; Corso, G.; et al. Lipid-based Transfection Reagents Exhibit Cryo-induced Increase in Transfection Efficiency. Mol. Ther. Nucleic Acids 2016, 5, e290. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Tong, H.; Garewal, M.; Ren, G. Optimized negative-staining electron microscopy for lipoprotein studies. Biochim. Biophys. Acta 2013, 1830, 2150–2159. [Google Scholar] [CrossRef] [Green Version]
- Chollet, P.; Favrot, M.C.; Hurbin, A.; Coll, J.L. Side-effects of a systemic injection of linear polyethylenimine-DNA complexes. J. Gene Med. 2002, 4, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Madhunapantula, S.V.; Robertson, G.P. Toxicological considerations when creating nanoparticle-based drugs and drug delivery systems. Expert Opin. Drug. Metab. Toxicol. 2012, 8, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Farokhzad, O.C.; Mahmoudi, M. Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona. Trends Biotechnol. 2017, 35, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Grenier, P.; Mahmoudi, M.; Lima, E.M.; Appel, E.A.; Dormont, F.; Lim, J.M.; Karnik, R.; Langer, R.; Farokhzad, O.C. Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun. 2017, 8, 777. [Google Scholar] [CrossRef]
- Kosuge, M.; Takeuchi, T.; Nakase, I.; Jones, A.T.; Futaki, S. Cellular internalization and distribution of arginine-rich peptides as a function of extracellular peptide concentration, serum, and plasma membrane associated proteoglycans. Bioconjug. Chem. 2008, 19, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Chan, W.C. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, K.; Furumoto, K.; Nagayama, S.; Minato, K.; Higaki, K.; Kai, T.; Kimura, T. Pre-coating with serum albumin reduces receptor-mediated hepatic disposition of polystyrene nanosphere: Implications for rational design of nanoparticles. J. Control. Release 2004, 100, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.T.; Kuhlmann, M.; Hvam, M.L.; Howard, K.A. Albumin-based drug delivery: Harnessing nature to cure disease. Mol. Cell Ther. 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntosh, D.P.; Tan, X.Y.; Oh, P.; Schnitzer, J.E. Targeting endothelium and its dynamic caveolae for tissue-specific transcytosis in vivo: A pathway to overcome cell barriers to drug and gene delivery. Proc. Natl. Acad. Sci. USA 2002, 99, 1996–2001. [Google Scholar] [CrossRef] [Green Version]
- Maas, C.; Oschatz, C.; Renne, T. The plasma contact system 2.0. Semin. Thromb. Hemost. 2011, 37, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Contact pathway of coagulation and inflammation. Thromb. J. 2015, 13, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagayama, S.; Ogawara, K.; Minato, K.; Fukuoka, Y.; Takakura, Y.; Hashida, M.; Higaki, K.; Kimura, T. Fetuin mediates hepatic uptake of negatively charged nanoparticles via scavenger receptor. Int. J. Pharm. 2007, 329, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, C.S.; Yu, R.Z.; Gaus, H.J.; Seth, P.P.; Swayze, E.E.; Bennett, F.C.; Geary, R.S.; Henry, S.P.; Wang, Y. Pharmacokinetic and Pharmacodynamic Investigations of ION-353382, a Model Antisense Oligonucleotide: Using Alpha-2-Macroglobulin and Murinoglobulin Double-Knockout Mice. Nucleic Acid Ther. 2016, 26, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Charbgoo, F.; Nejabat, M.; Abnous, K.; Soltani, F.; Taghdisi, S.M.; Alibolandi, M.; Thomas Shier, W.; Steele, T.W.J.; Ramezani, M. Gold nanoparticle should understand protein corona for being a clinical nanomaterial. J. Control. Release 2018, 272, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, A.; Mohtashami, M.; Sheijani, S.S.; Aliakbari, K. Chitosan-genipin nanohydrogel as a vehicle for sustained delivery of alpha-1 antitrypsin. Res. Pharm. Sci. 2015, 10, 523–534. [Google Scholar] [PubMed]
- Fujioka, Y.; Taniguchi, T.; Ishikawa, Y.; Yokoyama, M. Significance of acidic sugar chains of apolipoprotein B-100 in cellular metabolism of low-density lipoproteins. J. Lab. Clin. Med. 2000, 136, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Freimann, K.; Arukuusk, P.; Kurrikoff, K.; Vasconcelos, L.D.F.; Veiman, K.L.; Uusna, J.; Margus, H.; Garcia-Sosa, A.T.; Pooga, M.; Langel, U. Optimization of in vivo DNA delivery with NickFect peptide vectors. J. Control. Release 2016, 241, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Griffin, J.I.; Inturi, S.; Brenneman, B.; Banda, N.K.; Holers, V.M.; Moghimi, S.M.; Simberg, D. In Vitro and In Vivo Differences in Murine Third Complement Component (C3) Opsonization and Macrophage/Leukocyte Responses to Antibody-Functionalized Iron Oxide Nanoworms. Front. Immunol. 2017, 8, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, V.P.; Gifford, G.B.; Chen, F.; Benasutti, H.; Wang, G.; Groman, E.V.; Scheinman, R.; Saba, L.; Moghimi, S.M.; Simberg, D. Immunoglobulin deposition on biomolecule corona determines complement opsonization efficiency of preclinical and clinical nanoparticles. Nat. Nanotechnol. 2019, 14, 260–268. [Google Scholar] [CrossRef]
- Szebeni, J.; Storm, G. Complement activation as a bioequivalence issue relevant to the development of generic liposomes and other nanoparticulate drugs. Biochem. Biophys. Res. Commun. 2015, 468, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.T.; van der Vlies, A.J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K.; Swartz, M.A.; Hubbell, J.A. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164. [Google Scholar] [CrossRef]
- Kardani, K.; Milani, A.; Sameneh, H.S.; Bolhassani, A. Cell penetrating peptides: The potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019, 16, 1227–1258. [Google Scholar] [CrossRef] [PubMed]
- Katragadda, M.; Magotti, P.; Sfyroera, G.; Lambris, J.D. Hydrophobic effect and hydrogen bonds account for the improved activity of a complement inhibitor, compstatin. J. Med. Chem. 2006, 49, 4616–4622. [Google Scholar] [CrossRef]
- Lindman, S.; Lynch, I.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity. Nano Lett. 2007, 7, 914–920. [Google Scholar] [CrossRef]
- Juks, C.; Padari, K.; Margus, H.; Kriiska, A.; Etverk, I.; Arukuusk, P.; Koppel, K.; Ezzat, K.; Langel, Ü.; Pooga, M. The role of endocytosis in the uptake and intracellular trafficking of PepFect14-nucleic acid nanocomplexes via class A scavenger receptors. Biochim. Biophys. Acta 2015, 1848, 3205–3216. [Google Scholar] [CrossRef] [Green Version]
- Valeur, E.; Knerr, L.; Olwegard-Halvarsson, M.; Lemurell, M. Targeted delivery for regenerative medicines: An untapped opportunity for drug conjugates. Drug Discov. Today 2017, 22, 841–847. [Google Scholar] [CrossRef]
- Juks, C.; Lorents, A.; Arukuusk, P.; Langel, U.; Pooga, M. Cell-penetrating peptides recruit type A scavenger receptors to the plasma membrane for cellular delivery of nucleic acids. FASEB J. 2017, 31, 975–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, J.; Klein, P.M.; Al Danaf, N.; Nordin, J.Z.; Reinhard, S.; Loy, D.M.; Hohn, M.; El Andaloussi, S.; Lamb, D.C.; Wagner, E.; et al. Supramolecular Assembly of Aminoethylene-Lipopeptide PMO Conjugates into RNA Splice-Switching Nanomicelles. Adv. Funct. Mater. 2019, 29, 1906432. [Google Scholar] [CrossRef]
- Tamberg, N.; Tahk, S.; Koit, S.; Kristjuhan, K.; Kasvandik, S.; Kristjuhan, A.; Ilves, I. Keap1-MCM3 interaction is a potential coordinator of molecular machineries of antioxidant response and genomic DNA replication in metazoa. Sci. Rep. 2018, 8, 12136. [Google Scholar] [CrossRef] [PubMed]
- Padari, K.; Säälik, P.; Hansen, M.; Koppel, K.; Raid, R.; Langel, Ü.; Pooga, M. Cell transduction pathways of transportans. Bioconjug. Chem. 2005, 16, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Palchetti, S.; Digiacomo, L.; Chiozzi, R.Z.Z.; Capriotti, A.L.; Amenitsch, H.; Tentori, P.M.; Palmieri, V.; Papi, M.; Cardarelli, F.; et al. Human Biomolecular Corona of Liposomal Doxorubicin: The Overlooked Factor in Anticancer Drug Delivery. ACS Appl Mater. Interfaces 2018, 10, 22951–22962. [Google Scholar] [CrossRef]
- Kang, S.H.; Cho, M.J.; Kole, R. Up-regulation of luciferase gene expression with antisense oligonucleotides: Implications and applications in functional assay development. Biochemistry 1998, 37, 6235–6239. [Google Scholar] [CrossRef]
- van den Brand, D.; Gorris, M.A.J.; van Asbeck, A.H.; Palmen, E.; Ebisch, I.; Dolstra, H.; Hallbrink, M.; Massuger, L.; Brock, R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019, 141, 180–190. [Google Scholar] [CrossRef]
- Carreras-Badosa, G.; Maslovskaja, J.; Periyasamy, K.; Urgard, E.; Padari, K.; Vaher, H.; Tserel, L.; Gestin, M.; Kisand, K.; Arukuusk, P.; et al. NickFect type of cell-penetrating peptides present enhanced efficiency for microRNA-146a delivery into dendritic cells and during skin inflammation. Biomaterials 2020, 262, 120316. [Google Scholar] [CrossRef]
| Name | Sequence | MW (kDa) | Nucleobases | Strands |
|---|---|---|---|---|
| Cell-penetrating peptides | ||||
| PF14 | Stearyl-AGYLLGKLLOOLAAAALOOLL | 2.406 | ||
| C22-PF14 | Behenyl-AGYLLGKLLOOLAAAALOOLL | 2.462 | ||
| NF55 | Stearyl-AGYLLGO * INLKALAALAKAIL | 2.378 | ||
| Nucleic acids | ||||
| SCO | CCU CUU ACC UCA GUU ACA # | 6.098 | 18 | single-stranded |
| SCO-Cy5 | Cy5-CCU CUU ACC UCA GUU ACA # | 6.760 | 18 | single-stranded |
| b-SCO | biotin-CCU CUU ACC UCA GUU ACA # | 6.485 | 18 | single-stranded |
| pDNA (pGL3) | ~3055 | ~9400 | double-stranded | |
| Nanoparticle/Complex, Molar or Charge Ratio | Size ± SD (nm) | PdI (Polydispersity Index) | Zeta Potential ± SD (mV) |
|---|---|---|---|
| PF14 + SCO, MR5 | 91.7 ± 4.1 | 0.242 | −20.4 ± 0.4 |
| PF14 + pGL3, CR2 | 135.7 ± 5.6 | 0.271 | 18.3 ± 1.8 |
| C22-14 + SCO, MR5 | 94.3 ± 1.2 | 0.258 | −23.3 ± 3.6 |
| C22-14 + pGL3, CR2 | 208.5 ± 9.1 | 0.600 | 21.7 ± 1.6 |
| NF55 + SCO, MR5 | 102.1 ± 3.0 | 0.512 | −19.9 ± 3.4 |
| NF55 + pGL3, CR2 | 128.8 ± 2.8 | 0.303 | 16.1 ± 1.2 |
| Protein Group | Beads | + b-pGL3 | + PF14 + b-pGL3 |
|---|---|---|---|
| Serum albumin | 25.5 ± 0.5 | 67.5 ± 1.5 | 71.5 ± 1.5 |
| α-2-HS-glycoprotein | 7.0 ± 0.0 | 17.0 ± 0.0 | 19.5 ± 0.5 |
| Apolipoprotein A-II | 3.5 ± 0.5 | 9.5 ± 0.5 | 10.0 ± 0.0 |
| α-1-antiproteinase | 7.0 ± 0.0 | 27.0 ± 1.0 | 27.5 ± 0.5 |
| Apolipoprotein A-I | 6.5 ± 0.5 | 28.0 ± 0.0 | 35.0 ± 0.0 *** |
| Serotransferrin | 5.5 ± 0.5 | 52.5 ± 7.5 | 61.5 ± 2.5 |
| Haemoglobin subunit α | 4.0 ± 0.0 | 9.0 ± 0.0 | 10.0 ± 0.0 ** |
| Haemoglobin foetal subunit β | 4.0 ± 0.0 | 15.0 ± 0.0 | 16.5 ± 0.5 |
| α-2-macroglobulin | 4.5 ± 0.5 | 59.0 ± 7.0 | 69.5 ± 0.5 |
| Fetuin-B | 0.0 ± 0.0 | 12.0 ± 0.0 | 13.0 ± 0.0 ** |
| Inter-alpha-trypsin inhibitor heavy chain H2 | 6.0 ± 0.0 | 25.5 ± 0.5 | 34.5 ± 0.5 ** |
| Vitamin D-binding protein | 3.0 ± 1.0 | 25.5 ± 3.5 | 27.5 ± 0.5 |
| Prothrombin | 5.0 ± 1.0 | 13.0 ± 1.0 | 34.0 ± 1.0 * |
| Complement component 3 | 6.5 ± 0.5 | 35.0 ± 4.0 | 62.5 ± 0.5 |
| α-1-acid glycoprotein | 0.0 ± 0.0 | 7.0 ± 1.0 | 10.0 ± 0.0 |
| Protein Group | b-pGL3 | PF14 + b-pGL3 |
|---|---|---|
| Serum albumin | 44.5 ± 0.5 | 91.5 ± 0.5 *** |
| Serotransferrin | 15.5 ± 0.5 | 76.0 ± 0.0 ** |
| α-2-macroglobulin | 20.0 ± 1.0 | 89.5 ± 1.5 * |
| α-1-antitrypsin | 23.0 ± 0.0 | 33.5 ± 1.5 |
| Apolipoprotein A-I | 15.5 ± 0.5 | 30.0 ± 0.0 * |
| Haptoglobin | 16.0 ± 1.0 | 27.5 ± 0.5 |
| Complement component 3 | 16.0 ± 1.0 | 119.5 ± 0.5 ** |
| Ig γ-1 chain C region | 9.0 ± 1.0 | 15.0 ± 0.0 |
| Ig γ-3 chain C region | 6. ± 0.5 | 17.5 ± 0.5 |
| Ig κ-chain C region | 7.0 ± 0.0 | 12.0 ± 0.0 |
| α-1-acid glycoprotein 1 | 5.0 ± 0.0 | 12.0 ± 0.0 *** |
| Ig α-1 chain C region | 7.0 ± 1.0 | 20.0 ± 0.0 * |
| Complement component 4-B | 4.0 ± 0.0 | 78.0 ± 1.0 ** |
| Hemopexin | 0.0 ± 0.0 | 25.0 ± 0.0 *** |
| Apolipoprotein A-II | 5.5 ± 0.5 | 8.0 ± 0.0 |
| Protein Group | 5 min PF14 + b-pGL3 | 60 min PF14 + b-pGL3 | 5 min NF55 + b-pGL3 | 5 min b-pGL3 |
|---|---|---|---|---|
| Serum albumin | 78.7 ± 4.1 | 69.7 ± 0.3 * | 80.7 ± 2.3 | 86.0 ± 1.7 |
| Vitronectin | 35.7 ± 0.3 *** | 36.3 ± 0.9 *** | 32.3 ± 0.7 *** | 21.0 ± 0.0 |
| Apolipoprotein A-I | 46.7 ± 1.5 | 44.7 ± 1.5 | 42.3 ± 2.2 | 45.7 ± 0.7 |
| Apolipoprotein B-100 | 340.0 ± 3.6 *** | 362.3 ± 8.2 *** | 320.3 ± 15.7 *** | 141.7 ± 15.0 |
| α-2-macroglobulin | 93.3 ± 3.5 | 90.3 ± 1.9 | 94.7 ± 2.7 | 94.0 ± 0.6 |
| Complement component 3 | 153.3 ± 2.6 * | 160.3 ± 2.6 ** | 150.0 ± 5.1 | 139.7 ± 1.5 |
| Serotransferrin | 70.3 ± 5.7 | 62.7 ± 1.2 | 73.0 ± 1.7 | 74.7 ± 1.2 |
| Streptavidin | 9.0 ± 0.6 | 8.7 ± 0.3 | 9.0 ± 0.0 | 9.7 ± 0.3 |
| Haemoglobin subunit α | 12.0 ± 0.6 | 11.7 ± 0.3 | 12.0 ± 0.6 | 12.7 ± 0.7 |
| Fibronectin | 172.0 ± 4.7 ** | 177.3 ± 2.4 ** | 139.3 ± 12.6 | 137.7 ± 7.2 |
| Prothrombin | 53.3 ± 1.2 ** | 51.7 ± 1.2 ** | 46.0 ± 0.6 | 44.0 ± 0.6 |
| Fibrinogen γ-chain | 38.3 ± 2.2 | 39.7 ± 1.5* | 34.3 ± 3.2 | 35.3 ± 22 |
| Murinoglobulin-1 | 97.7 ± 3.2 | 96.0 ± 1.5 | 92.0 ± 4.9 | 96.3 ± 0.3 |
| Fibrinogen β-chain | 64.3 ± 2.7 | 64.0 ± 1.5 | 54.7 ± 6.9 | 58.0 ± 2.5 |
| α-1-antitrypsin 1–4 | 22.3 ± 2.3 | 20.0 ± 1.0 | 23.3 ± 0.7 | 25.0 ± 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorents, A.; Maloverjan, M.; Padari, K.; Pooga, M. Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona. Pharmaceuticals 2021, 14, 667. https://doi.org/10.3390/ph14070667
Lorents A, Maloverjan M, Padari K, Pooga M. Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona. Pharmaceuticals. 2021; 14(7):667. https://doi.org/10.3390/ph14070667
Chicago/Turabian StyleLorents, Annely, Maria Maloverjan, Kärt Padari, and Margus Pooga. 2021. "Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona" Pharmaceuticals 14, no. 7: 667. https://doi.org/10.3390/ph14070667
APA StyleLorents, A., Maloverjan, M., Padari, K., & Pooga, M. (2021). Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona. Pharmaceuticals, 14(7), 667. https://doi.org/10.3390/ph14070667