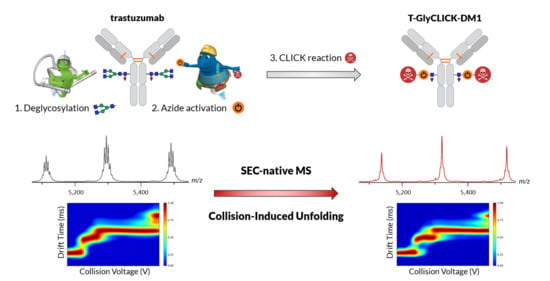
State-of-the-Art Native Mass Spectrometry and Ion Mobility Methods to Monitor Homogeneous Site-Specific Antibody-Drug Conjugates Synthesis
Abstract
1. Introduction
2. Results and Discussion
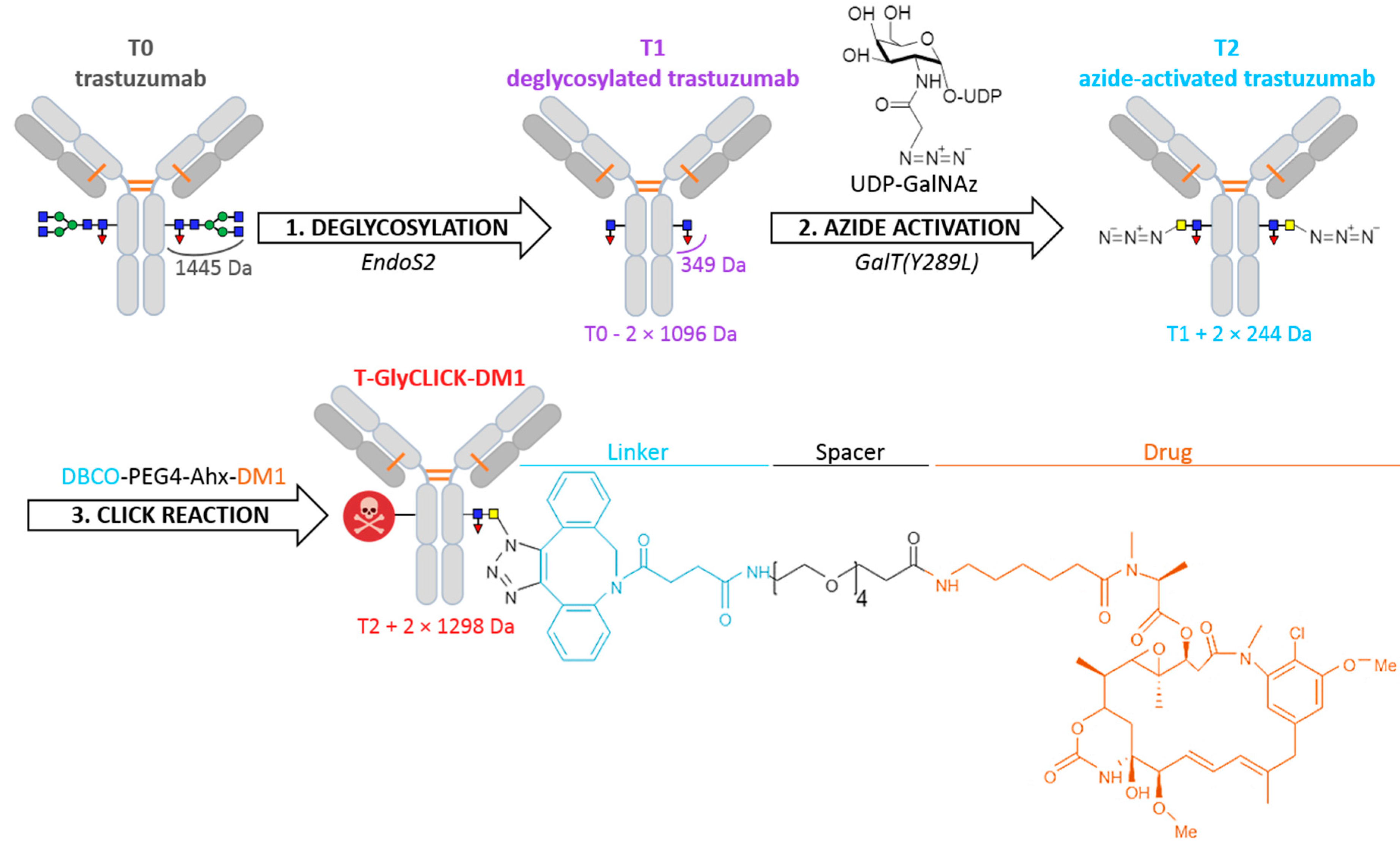
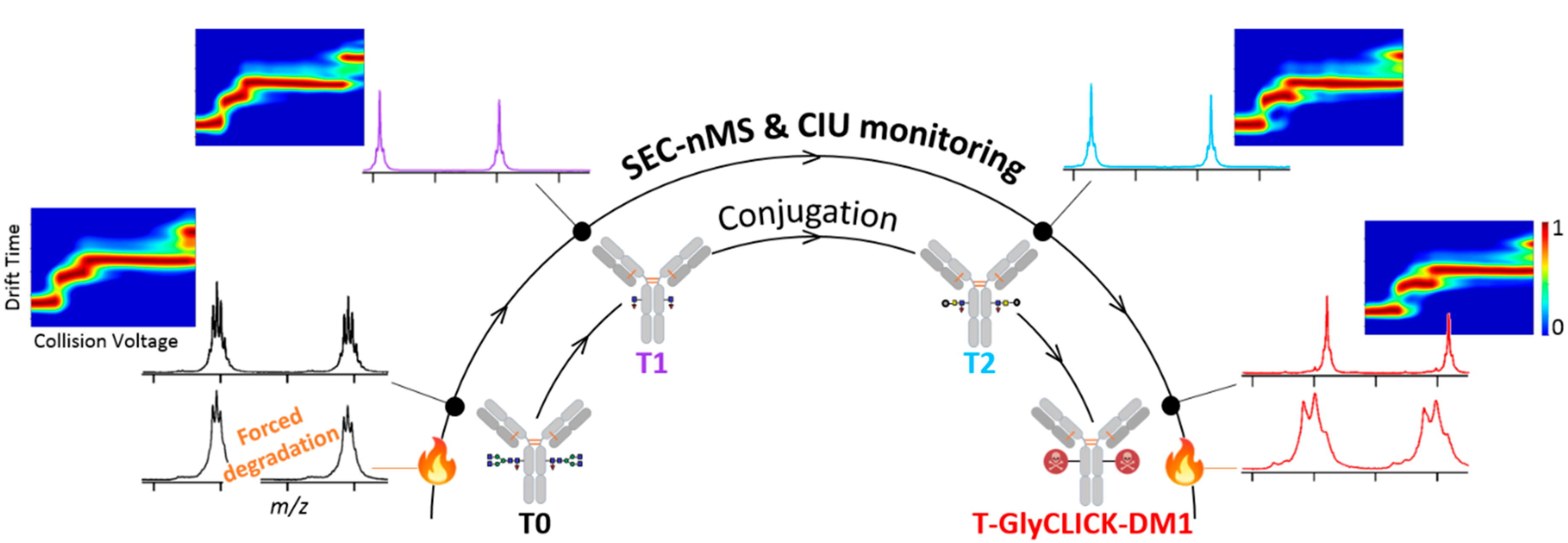
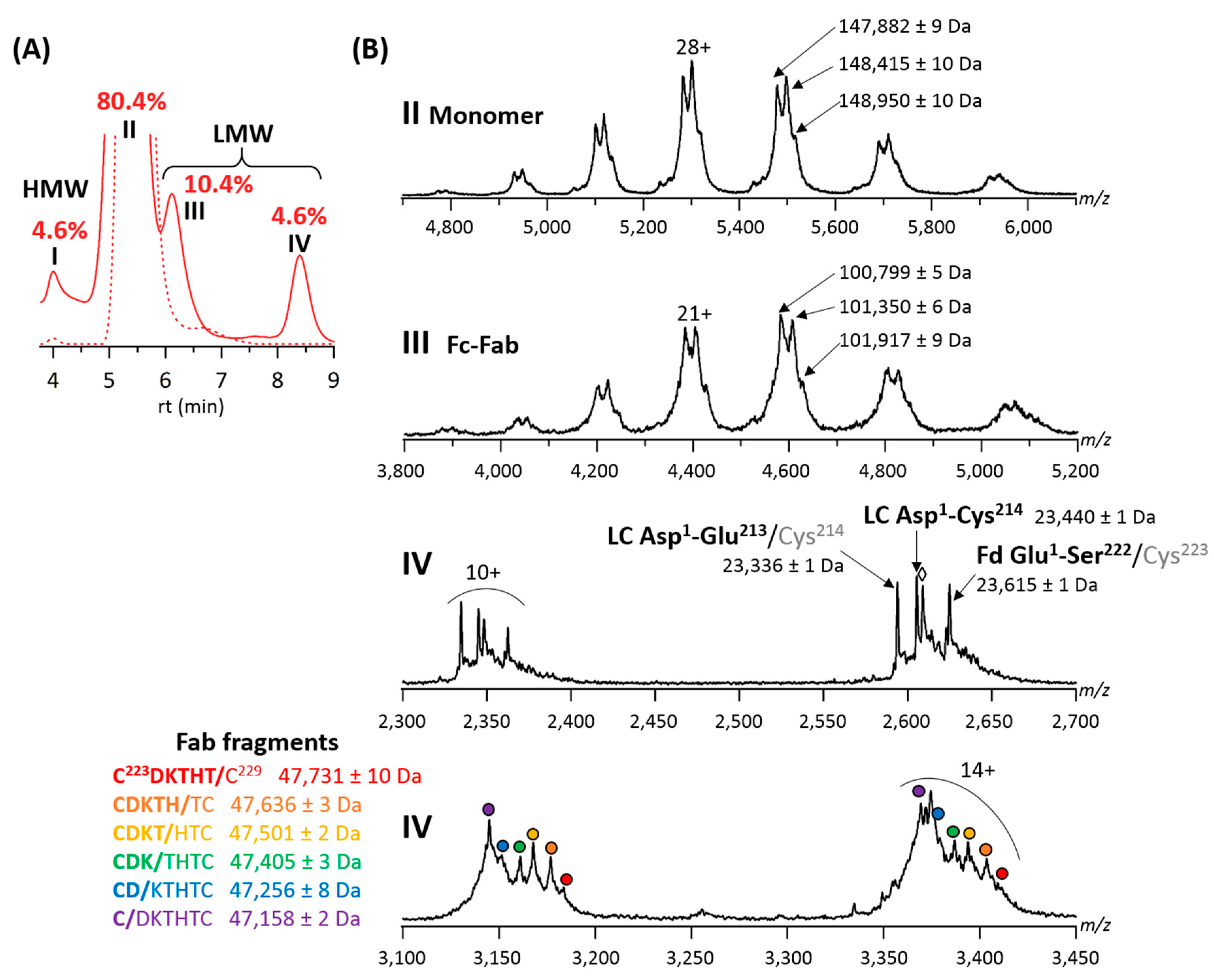
2.1. Online SEC-nMS to Monitor the Conjugation Process
2.2. Forced Degradation Studies
2.3. nIM-MS to Monitor the Conformational Landscape during the Conjugation Process
3. Materials and Methods
3.1. Sample Preparation
3.2. Manual Buffer Exchange
3.3. Online SEC-nMS
3.4. nIM-MS and CIU Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Joubert, N.; Beck, A.; Dumontet, C.; Denevault-Sabourin, C. Antibody–Drug Conjugates: The Last Decade. Pharmaceuticals 2020, 13, 245. [Google Scholar] [CrossRef] [PubMed]
- Vankemmelbeke, M.; Durrant, L. Third-generation antibody drug conjugates for cancer therapy—A balancing act. Ther. Deliv. 2016, 7, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef]
- Busse, A.; Lüftner, D. What Does the Pipeline Promise about Upcoming Biosimilar Antibodies in Oncology? Breast Care 2019, 14, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Lyon, R.P.; Bovee, T.D.; Doronina, S.O.; Burke, P.J.; Hunter, J.H.; Neff-LaFord, H.D.; Jonas, M.; Anderson, M.E.; Setter, J.R.; Senter, P.D. Reducing hydrophobicity of homogeneous antibody-drug conjugates improves pharmacokinetics and therapeutic index. Nat. Biotechnol. 2015, 33, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Donaghy, H. Effects of antibody, drug and linker on the preclinical and clinical toxicities of antibody-drug conjugates. mAbs 2016, 8, 659–671. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, C.F.; Turcott, E.; Westendorf, L.; Webster, J.B.; Alley, S.C.; Kim, K.; Andreyka, J.; Stone, I.; Hamblett, K.J.; Francisco, J.A.; et al. Engineered antibody-drug conjugates with defined sites and stoichiometries of drug attachment. Protein Eng. Des. Sel. 2006, 19, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Junutula, J.R.; Raab, H.; Clark, S.; Bhakta, S.; Leipold, D.D.; Weir, S.; Chen, Y.; Simpson, M.; Tsai, S.P.; Dennis, M.S.; et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008, 26, 925–932. [Google Scholar] [CrossRef]
- Panowski, S.; Bhakta, S.; Raab, H.; Polakis, P.; Junutula, J.R. Site-specific antibody drug conjugates for cancer therapy. mAbs 2013, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Kung Sutherland, M.S.; Walter, R.B.; Jeffrey, S.C.; Burke, P.J.; Yu, C.; Kostner, H.; Stone, I.; Ryan, M.C.; Sussman, D.; Lyon, R.P.; et al. SGN-CD33A: A novel CD33-targeting antibody–drug conjugate using a pyrrolobenzodiazepine dimer is active in models of drug-resistant AML. Blood 2013, 122, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- D’Atri, V.; Pell, R.; Clarke, A.; Guillarme, D.; Fekete, S. Is hydrophobic interaction chromatography the most suitable technique to characterize site-specific antibody-drug conjugates? J. Chromatogr. A 2019, 1586, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Strop, P.; Liu, S.-H.; Dorywalska, M.; Delaria, K.; Dushin Russell, G.; Tran, T.-T.; Ho, W.-H.; Farias, S.; Casas Meritxell, G.; Abdiche, Y.; et al. Location Matters: Site of Conjugation Modulates Stability and Pharmacokinetics of Antibody Drug Conjugates. Chem. Biol. 2013, 20, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Dennler, P.; Chiotellis, A.; Fischer, E.; Brégeon, D.; Belmant, C.; Gauthier, L.; Lhospice, F.; Romagne, F.; Schibli, R. Transglutaminase-Based Chemo-Enzymatic Conjugation Approach Yields Homogeneous Antibody–Drug Conjugates. Bioconj. Chem. 2014, 25, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Farias, S.E.; Strop, P.; Delaria, K.; Galindo Casas, M.; Dorywalska, M.; Shelton, D.L.; Pons, J.; Rajpal, A. Mass Spectrometric Characterization of Transglutaminase Based Site-Specific Antibody–Drug Conjugates. Bioconj. Chem. 2014, 25, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Axup, J.Y.; Bajjuri, K.M.; Ritland, M.; Hutchins, B.M.; Kim, C.H.; Kazane, S.A.; Halder, R.; Forsyth, J.S.; Santidrian, A.F.; Stafin, K.; et al. Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc. Natl. Acad. Sci. USA 2012, 109, 16101–16106. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.L.; Giddens, J.P.; Iavarone, A.T.; Godula, K.; Wang, L.-X.; Bertozzi, C.R. Chemoenzymatic Fc Glycosylation via Engineered Aldehyde Tags. Bioconj. Chem. 2014, 25, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Kolodych, S.; Koniev, O.; Baatarkhuu, Z.; Bonnefoy, J.-Y.; Debaene, F.; Cianférani, S.; Van Dorsselaer, A.; Wagner, A. CBTF: New Amine-to-Thiol Coupling Reagent for Preparation of Antibody Conjugates with Increased Plasma Stability. Bioconj. Chem. 2015, 26, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Koniev, O.; Kolodych, S.; Baatarkhuu, Z.; Stojko, J.; Eberova, J.; Bonnefoy, J.-Y.; Cianférani, S.; Van Dorsselaer, A.; Wagner, A. MAPN: First-in-Class Reagent for Kinetically Resolved Thiol-to-Thiol Conjugation. Bioconj. Chem. 2015, 26, 1863–1867. [Google Scholar] [CrossRef]
- van Geel, R.; Wijdeven, M.A.; Heesbeen, R.; Verkade, J.M.; Wasiel, A.A.; van Berkel, S.S.; van Delft, F.L. Chemoenzymatic Conjugation of Toxic Payloads to the Globally Conserved N-Glycan of Native mAbs Provides Homogeneous and Highly Efficacious Antibody-Drug Conjugates. Bioconj. Chem. 2015, 26, 2233–2242. [Google Scholar] [CrossRef]
- Qasba, P.K. Glycans of Antibodies as a Specific Site for Drug Conjugation Using Glycosyltransferases. Bioconj. Chem. 2015, 26, 2170–2175. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Terral, G.; Debaene, F.; Wagner-Rousset, E.; Marcoux, J.; Janin-Bussat, M.-C.; Colas, O.; Dorsselaer, A.V.; Cianférani, S. Cutting-edge mass spectrometry methods for the multi-level structural characterization of antibody-drug conjugates. Expert Rev. Proteom. 2016, 13, 157–183. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; D’Atri, V.; Ehkirch, A.; Fekete, S.; Hernandez-Alba, O.; Gahoual, R.; Leize-Wagner, E.; François, Y.; Guillarme, D.; Cianférani, S. Cutting-edge multi-level analytical and structural characterization of antibody-drug conjugates: Present and future. Expert Rev. Proteom. 2019, 16, 337–362. [Google Scholar] [CrossRef]
- Chen, T.; Chen, Y.; Stella, C.; Medley, C.D.; Gruenhagen, J.A.; Zhang, K. Antibody-drug conjugate characterization by chromatographic and electrophoretic techniques. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2016, 1032, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Valliere-Douglass, J.F.; McFee, W.A.; Salas-Solano, O. Native Intact Mass Determination of Antibodies Conjugated with Monomethyl Auristatin E and F at Interchain Cysteine Residues. Anal. Chem. 2012, 84, 2843–2849. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yin, S.; Wu, Y.; Ouyang, J. Development of a Native Nanoelectrospray Mass Spectrometry Method for Determination of the Drug-to-Antibody Ratio of Antibody–Drug Conjugates. Anal. Chem. 2013, 85, 1699–1704. [Google Scholar] [CrossRef] [PubMed]
- Hengel, S.M.; Sanderson, R.; Valliere-Douglass, J.; Nicholas, N.; Leiske, C.; Alley, S.C. Measurement of in Vivo Drug Load Distribution of Cysteine-Linked Antibody–Drug Conjugates Using Microscale Liquid Chromatography Mass Spectrometry. Anal. Chem. 2014, 86, 3420–3425. [Google Scholar] [CrossRef] [PubMed]
- Ehkirch, A.; Hernandez-Alba, O.; Colas, O.; Beck, A.; Guillarme, D.; Cianferani, S. Hyphenation of size exclusion chromatography to native ion mobility mass spectrometry for the analytical characterization of therapeutic antibodies and related products. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1086, 176–183. [Google Scholar] [CrossRef]
- Friese, O.V.; Smith, J.N.; Brown, P.W.; Rouse, J.C. Practical approaches for overcoming challenges in heightened characterization of antibody-drug conjugates with new methodologies and ultrahigh-resolution mass spectrometry. mAbs 2018, 10, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.; Pack, L.; Hunter, J.H.; Valliere-Douglass, J.F. Native size-exclusion chromatography-mass spectrometry: Suitability for antibody–drug conjugate drug-to-antibody ratio quantitation across a range of chemotypes and drug-loading levels. mAbs 2019, 12, 1682895. [Google Scholar] [CrossRef]
- Botzanowski, T.; Erb, S.; Hernandez-Alba, O.; Ehkirch, A.; Colas, O.; Wagner-Rousset, E.; Rabuka, D.; Beck, A.; Drake, P.M.; Cianferani, S. Insights from native mass spectrometry approaches for top- and middle- level characterization of site-specific antibody-drug conjugates. mAbs 2017, 9, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Debaene, F.; Bœuf, A.; Wagner-Rousset, E.; Colas, O.; Ayoub, D.; Corvaïa, N.; Van Dorsselaer, A.; Beck, A.; Cianférani, S. Innovative Native MS Methodologies for Antibody Drug Conjugate Characterization: High Resolution Native MS and IM-MS for Average DAR and DAR Distribution Assessment. Anal. Chem. 2014, 86, 10674–10683. [Google Scholar] [CrossRef] [PubMed]
- Marcoux, J.; Champion, T.; Colas, O.; Wagner-Rousset, E.; Corvaia, N.; Van Dorsselaer, A.; Beck, A.; Cianferani, S. Native mass spectrometry and ion mobility characterization of trastuzumab emtansine, a lysine-linked antibody drug conjugate. Protein Sci. 2015, 24, 1210–1223. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Lippens, J.L.; Netirojjanakul, C.; Campuzano, I.D.G.; Ruotolo, B.T. Quantitative collision-induced unfolding differentiates model antibody-drug conjugates. Protein Sci. 2019, 28, 598–608. [Google Scholar] [CrossRef] [PubMed]
- Dixit, S.M.; Polasky, D.A.; Ruotolo, B.T. Collision induced unfolding of isolated proteins in the gas phase: Past, present, and future. Curr. Opin. Chem. Biol. 2018, 42, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Haberger, M.; Leiss, M.; Heidenreich, A.-K.; Pester, O.; Hafenmair, G.; Hook, M.; Bonnington, L.; Wegele, H.; Haindl, M.; Reusch, D.; et al. Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. mAbs 2015, 8, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Nowak, C.; Cheung, J.K.; Dellatore, S.M.; Katiyar, A.; Bhat, R.; Sun, J.; Ponniah, G.; Neill, A.; Mason, B.; Beck, A.; et al. Forced degradation of recombinant monoclonal antibodies: A practical guide. mAbs 2017, 9, 1217–1230. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yu, S.-F.; Yee, S.; Kaur, S.; Xu, K. Characterization of in vivo biotransformations for trastuzumab emtansine by high-resolution accurate-mass mass spectrometry. mAbs 2018, 10, 960–967. [Google Scholar] [CrossRef]
- Halley, J.; Chou, Y.R.; Cicchino, C.; Huang, M.; Sharma, V.; Tan, N.C.; Thakkar, S.; Zhou, L.L.; Al-Azzam, W.; Cornen, S.; et al. An Industry Perspective on Forced Degradation Studies of Biopharmaceuticals: Survey Outcome and Recommendations. J. Pharm. Sci. 2020, 109, 6–21. [Google Scholar] [CrossRef]
- Chaudhuri, R.; Cheng, Y.; Middaugh, C.R.; Volkin, D.B. High-Throughput Biophysical Analysis of Protein Therapeutics to Examine Interrelationships Between Aggregate Formation and Conformational Stability. AAPS J. 2013, 16, 48–64. [Google Scholar] [CrossRef]
- Moritz, B.; Stracke, J.O. Assessment of disulfide and hinge modifications in monoclonal antibodies. Electrophoresis 2017, 38, 769–785. [Google Scholar] [CrossRef]
- Vlasak, J.; Ionescu, R. Fragmentation of monoclonal antibodies. mAbs 2011, 3, 253–263. [Google Scholar] [CrossRef]
- Ross, P.L.; Wolfe, J.L. Physical and Chemical Stability of Antibody Drug Conjugates: Current Status. J. Pharm. Sci. 2016, 105, 391–397. [Google Scholar] [CrossRef]
- Adem, Y.T. Physical Stability Studies of Antibody-Drug Conjugates (ADCs) Under Stressed Conditions. In Antibody-Drug Conjugates. Methods in Molecular Biology; Tumey, L.N., Ed.; Springer Protocols: New York, NY, USA, 2020; Volume 2078, pp. 301–311. [Google Scholar]
- Wakankar, A.A.; Feeney, M.B.; Rivera, J.; Chen, Y.; Kim, M.; Sharma, V.K.; Wang, Y.J. Physicochemical Stability of the Antibody−Drug Conjugate Trastuzumab-DM1: Changes due to Modification and Conjugation Processes. Bioconj. Chem. 2010, 21, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Beckley, N.S.; Lazzareschi, K.P.; Chih, H.-W.; Sharma, V.K.; Flores, H.L. Investigation into Temperature-Induced Aggregation of an Antibody Drug Conjugate. Bioconj. Chem. 2013, 24, 1674–1683. [Google Scholar] [CrossRef]
- Yates, Z.; Gunasekaran, K.; Zhou, H.; Hu, Z.; Liu, Z.; Ketchem, R.R.; Yan, B. Histidine Residue Mediates Radical-induced Hinge Cleavage of Human IgG1. J. Biol. Chem. 2010, 285, 18662–18671. [Google Scholar] [CrossRef]
- Yan, B.; Boyd, D. Breaking the Light and Heavy Chain Linkage of Human Immunoglobulin G1 (IgG1) by Radical Reactions. J. Biol. Chem. 2011, 286, 24674–24684. [Google Scholar] [CrossRef]
- Cohen, S.L.; Price, C.; Vlasak, J. β-Elimination and Peptide Bond Hydrolysis: Two Distinct Mechanisms of Human IgG1 Hinge Fragmentation upon Storage. J. Am. Chem. Soc. 2007, 129, 6976–6977. [Google Scholar] [CrossRef]
- Mohamed, H.E.; Mohamed, A.A.; Al-Ghobashy, M.A.; Fathalla, F.A.; Abbas, S.S. Stability assessment of antibody-drug conjugate Trastuzumab emtansine in comparison to parent monoclonal antibody using orthogonal testing protocol. J. Pharm. Biomed. Anal. 2018, 150, 268–277. [Google Scholar] [CrossRef]
- Botzanowski, T.; Hernandez-Alba, O.; Malissard, M.; Wagner-Rousset, E.; Deslignière, E.; Colas, O.; Haeuw, J.-F.; Beck, A.; Cianférani, S. Middle level IM-MS and CIU experiments for improved therapeutic immunoglobulin subclass fingerprinting. Anal. Chem. 2020, 92, 8827–8835. [Google Scholar] [CrossRef]
- Tian, Y.; Han, L.; Buckner, A.C.; Ruotolo, B.T. Collision Induced Unfolding of Intact Antibodies: Rapid Characterization of Disulfide Bonding Patterns, Glycosylation, and Structures. Anal. Chem. 2015, 87, 11509–11515. [Google Scholar] [CrossRef]
- Upton, R.; Migas, L.G.; Pacholarz, K.J.; Beniston, R.G.; Estdale, S.; Firth, D.; Barran, P.E. Hybrid mass spectrometry methods reveal lot-to-lot differences and delineate the effects of glycosylation on the tertiary structure of Herceptin®. Chem. Sci. 2019, 10, 2811–2820. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, B.T.; Benesch, J.L.P.; Sandercock, A.M.; Hyung, S.-J.; Robinson, C.V. Ion mobility–mass spectrometry analysis of large protein complexes. Nat. Protoc. 2008, 3, 1139–1152. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.F.; Hall, Z.; Giles, K.; Hoyes, J.; Robinson, C.V.; Ruotolo, B.T. Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Anal. Chem. 2010, 82, 9557–9565. [Google Scholar] [CrossRef] [PubMed]
- Polasky, D.A.; Dixit, S.M.; Fantin, S.M.; Ruotolo, B.T. CIUSuite 2: Next-Generation Software for the Analysis of Gas-Phase Protein Unfolding Data. Anal. Chem. 2019, 91, 3147–3155. [Google Scholar] [CrossRef] [PubMed]
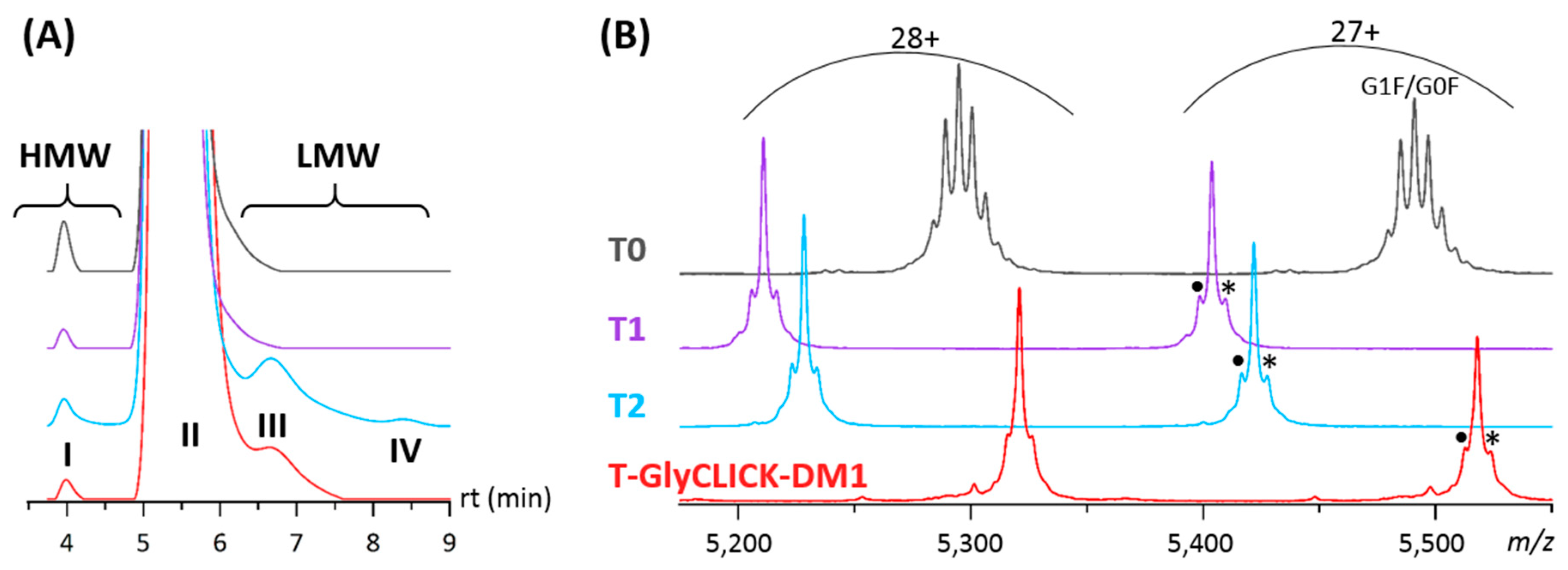
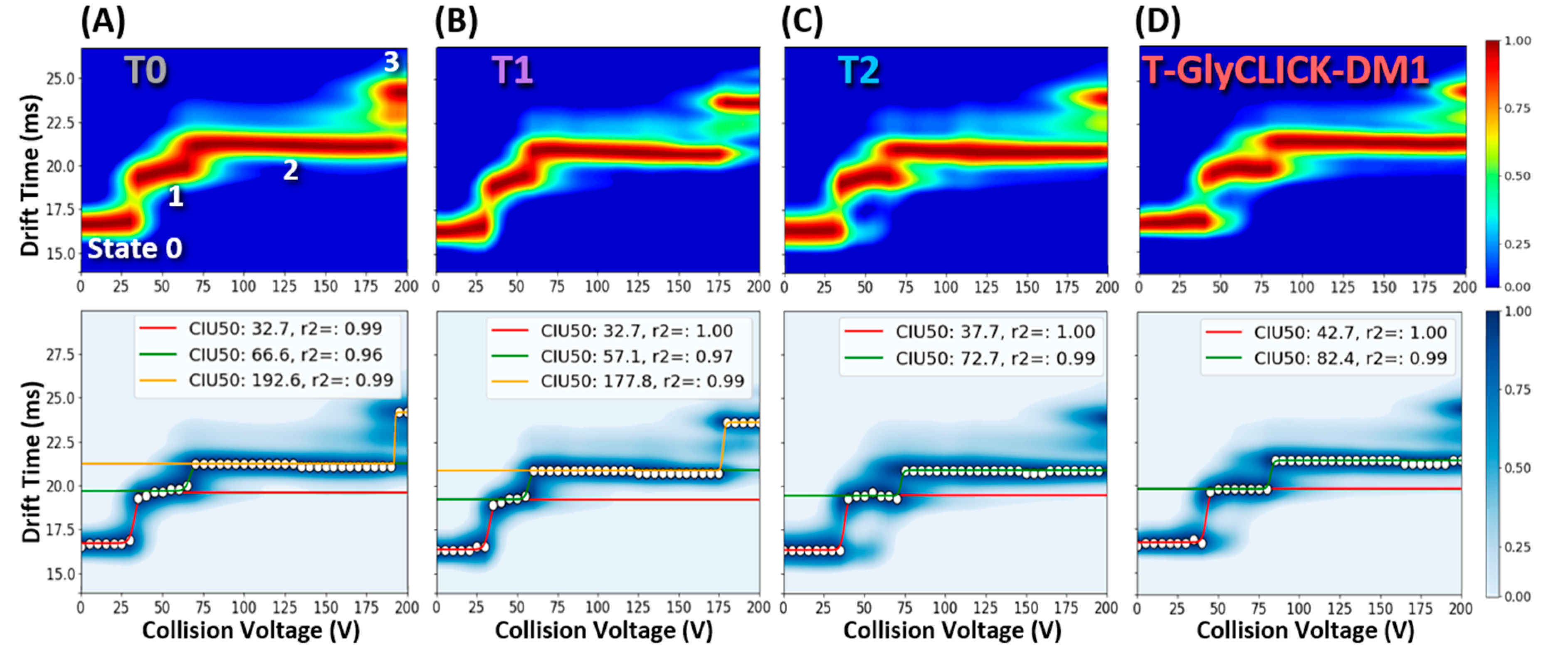
| T0 | T1 | T2 | T-GlyCLICK-DM1 | |||||
|---|---|---|---|---|---|---|---|---|
| Main Product | 99.6% | 99.7% | 96.6% | 98.5% | ||||
| G0F/G0 147,930 ± 4 Da | 145,875 ± 1 Da | 146,372 ± 2 Da | 148,957 ± 1 Da | |||||
| (G0F)2 148,067 ± 4 Da | ||||||||
| G1F/G0F 148,228 ± 2 Da | ||||||||
| (G1F)2 148,387 ± 2 Da | ||||||||
| G2F/G1F 148,548 ± 1 Da | ||||||||
| HMW Dimers | 0.4% | 0.3% | 0.5% | 0.2% | ||||
| 296,828 ± 25 Da | 291,719 ± 25 Da | 292,911 ± 23 Da | ND * | |||||
| LMWS | - | - | 2.7% | Fc-Fab | 99,319 ± 6 Da | 1.3% | Fc-Fab | 101,910 ± 8 Da |
| 0.2% | LC | 23,473 ± 2 Da | <0.1% | LC | 23,474 ± 3 Da | |||
| Fd | 23,618 ± 4 Da | Fd | 23,615 ± 3 Da | |||||
| Fab | 47,129 ± 9 Da | Fab | 47,091 ± 8 Da | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deslignière, E.; Ehkirch, A.; Duivelshof, B.L.; Toftevall, H.; Sjögren, J.; Guillarme, D.; D’Atri, V.; Beck, A.; Hernandez-Alba, O.; Cianférani, S. State-of-the-Art Native Mass Spectrometry and Ion Mobility Methods to Monitor Homogeneous Site-Specific Antibody-Drug Conjugates Synthesis. Pharmaceuticals 2021, 14, 498. https://doi.org/10.3390/ph14060498
Deslignière E, Ehkirch A, Duivelshof BL, Toftevall H, Sjögren J, Guillarme D, D’Atri V, Beck A, Hernandez-Alba O, Cianférani S. State-of-the-Art Native Mass Spectrometry and Ion Mobility Methods to Monitor Homogeneous Site-Specific Antibody-Drug Conjugates Synthesis. Pharmaceuticals. 2021; 14(6):498. https://doi.org/10.3390/ph14060498
Chicago/Turabian StyleDeslignière, Evolène, Anthony Ehkirch, Bastiaan L. Duivelshof, Hanna Toftevall, Jonathan Sjögren, Davy Guillarme, Valentina D’Atri, Alain Beck, Oscar Hernandez-Alba, and Sarah Cianférani. 2021. "State-of-the-Art Native Mass Spectrometry and Ion Mobility Methods to Monitor Homogeneous Site-Specific Antibody-Drug Conjugates Synthesis" Pharmaceuticals 14, no. 6: 498. https://doi.org/10.3390/ph14060498
APA StyleDeslignière, E., Ehkirch, A., Duivelshof, B. L., Toftevall, H., Sjögren, J., Guillarme, D., D’Atri, V., Beck, A., Hernandez-Alba, O., & Cianférani, S. (2021). State-of-the-Art Native Mass Spectrometry and Ion Mobility Methods to Monitor Homogeneous Site-Specific Antibody-Drug Conjugates Synthesis. Pharmaceuticals, 14(6), 498. https://doi.org/10.3390/ph14060498