In Vitro Activity of a Novel Siderophore-Cephalosporin LCB10-0200 (GT-1), and LCB10-0200/Avibactam, against Carbapenem-Resistant Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa Strains at a Tertiary Hospital in Korea
Abstract
1. Introduction
2. Results
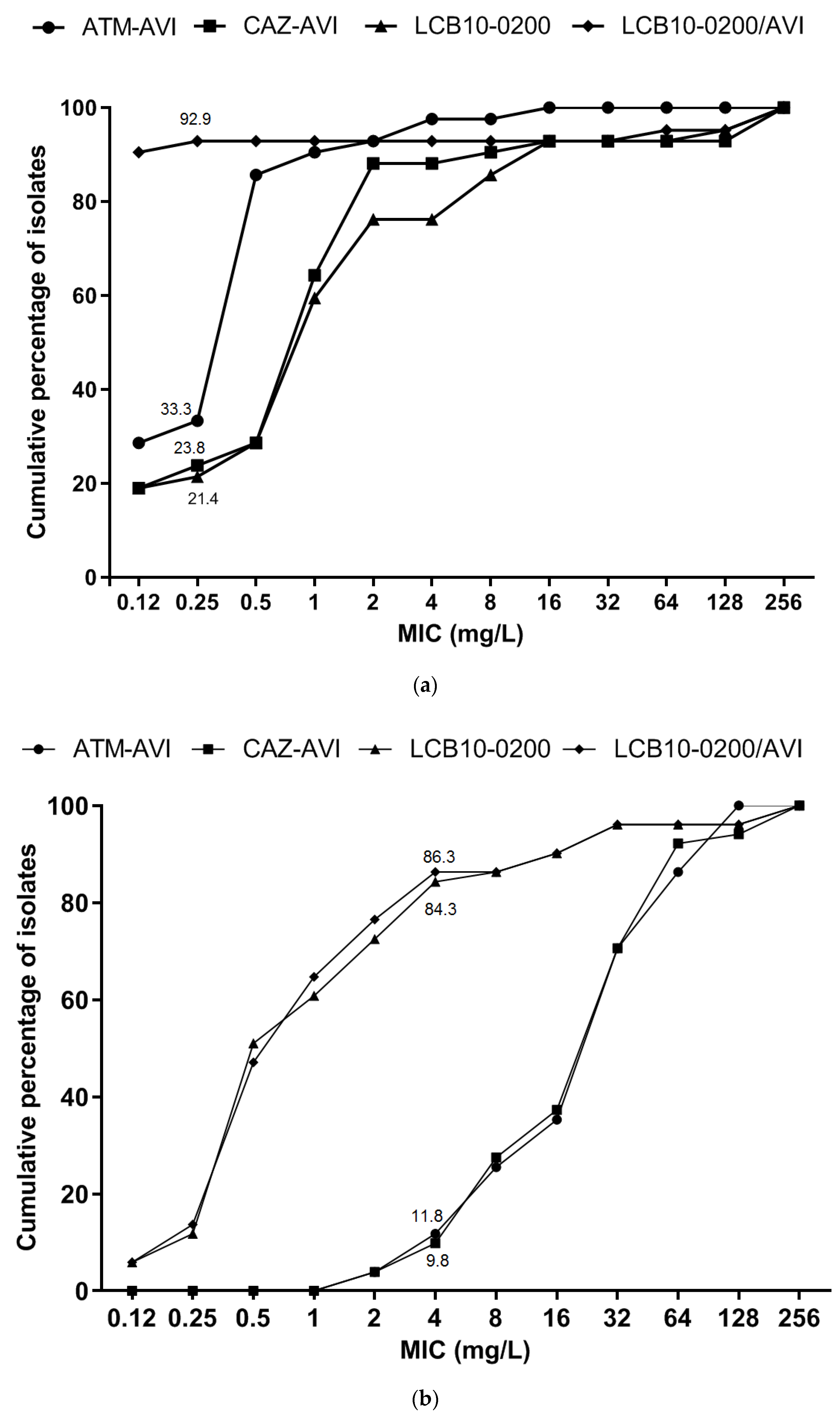
2.1. LCB10-0200, and LCB10-0200/AVI Displayed Potent Activity against Carbapenem-Resistant E. coli and K. pneumoniae Strains
2.2. In Vitro Activity of LCB10-0200, and LCB10-0200/AVI against A. baumannii, and P. aeruginosa Strains
3. Discussion
4. Materials and Methods
4.1. Specimen Collection and Antibiotics
4.2. Susceptibility Tests and MIC Determinations
4.3. DNA Extraction and Whole Genome Sequencing
4.4. Sequence Assembly, Genome Annotation, Multi-Locus Sequence Typing (MLST) and Resistome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Petrosillo, N.; Taglietti, F.; Granata, G. Treatment Options for Colistin Resistant Klebsiella pneumoniae: Present and Future. J. Clin. Med. 2019, 8, 934. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Jánvári, L.; Tóth, Á.; Terhes, G.; Burián, K. Detection of VIM, NDM and OXA-48 producing carbapenem resistant Enterobacterales among clinical isolates in Southern Hungary. Acta Microbiol. Immunol. Hung. 2020, 67, 209–215. [Google Scholar]
- Bush, K. A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents 2015, 46, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Kuroda, M.; Suzuki, S.; Mu, J.-J. Emergence of an Escherichia coli strain co-harbouring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J. Glob. Antimicrob. Resist. 2019, 16, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-Producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aires, C.A.M.; da Conceição-Neto, O.C.; Tavares E Oliveira, T.R.; Dias, C.F.; Montezzi, L.F.; Picão, R.C.; Albano, R.M.; Asensi, M.D.; Carvalho-Assef, A.P.D. Emergence of the Plasmid-Mediated mcr-1 Gene in Clinical KPC-2-Producing Klebsiella pneumoniae Sequence Type 392 in Brazil. Antimicrob. Agents Chemother. 2017, 61, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.P.; Pinto, N.A.; Vu, T.N.; Mai, H.; Pham, A.H.; Lee, H.; Cho, Y.L.; Byun, J.-H.; D’Souza, R.; Yong, D. Resistome Profiles, Plasmid Typing, and Whole-Genome Phylogenetic Tree Analyses of BlaNDM-9 and Mcr-1 Co-Harboring Escherichia coli ST617 from a Patient without a History of Farm Exposure in Korea. Pathogens 2019, 8, 212. [Google Scholar] [CrossRef]
- Oh, S.-H.; Park, H.-S.; Kim, H.-S.; Yun, J.-Y.; Oh, K.; Cho, Y.-L.; Kwak, J.-H. Antimicrobial activities of LCB10-0200, a novel siderophore cephalosporin, against the clinical isolates of Pseudomonas aeruginosa and other pathogens. Int. J. Antimicrob. Agents 2017, 50, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.P.; Pinto, N.A.; Vu, T.N.; Lee, H.; Cho, Y.L.; Byun, J.-H.; D’Souza, R.; Yong, D. In Vitro Activity of a Novel Siderophore-Cephalosporin, GT-1 and Serine-Type β-Lactamase Inhibitor, GT-055, against Escherichia coli, Klebsiella pneumoniae and Acinetobacter spp. Panel Strains. Antibiotics 2020, 9, 267. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, R.; Pinto, N.A.; Hwang, I.; Cho, Y.; Yong, D.; Choi, J.; Lee, K.; Chong, Y. Panel strain of Klebsiella pneumoniae for beta-lactam antibiotic evaluation: Their phenotypic and genotypic characterization. PeerJ 2017, 5, 2896. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, R.; Pinto, N.A.; Phuong, N.L.; Higgins, P.G.; Vu, T.N.; Byun, J.-H.; Cho, Y.L.; Choi, J.R.; Yong, D. Phenotypic and Genotypic Characterization of Acinetobacter spp. Panel Strains: A Cornerstone to Facilitate Antimicrobial Development. Front. Microbiol. 2019, 10, 559. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, S.D.; Mangani, S.; Durand-Reville, T.; Benvenuti, M.; De Luca, F.; Sanyal, G.; Docquier, J.-D. Structural Insight into Potent Broad-Spectrum Inhibition with Reversible Recyclization Mechanism: Avibactam in Complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-Lactamases. Antimicrob. Agents Chemother. 2013, 57, 2496–2505. [Google Scholar] [CrossRef]
- Lahiri, S.D.; Johnstone, M.R.; Ross, P.L.; McLaughlin, R.E.; Olivier, N.B.; Alm, R.A. Avibactam and Class C β-Lactamases: Mechanism of Inhibition, Conservation of the Binding Pocket, and Implications for Resistance. Antimicrob. Agents Chemother. 2014, 58, 5704–5713. [Google Scholar] [CrossRef] [PubMed]
- King, D.T.; King, A.M.; Lal, S.M.; Wright, G.D.; Strynadka, N.C.J. Molecular Mechanism of Avibactam-Mediated β-Lactamase Inhibition. ACS Infect. Dis. 2015, 1, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, J.; Melchers, M.J.; Van Mil, A.C.; Nichols, W.W.; Mouton, J.W. In VitroActivity of Ceftazidime-Avibactam Combination inIn VitroCheckerboard Assays. Antimicrob. Agents Chemother. 2014, 59, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M.; Meunier, D.; Hopkins, K.L.; Doumith, M.; Hill, R.; Pike, R.; Staves, P.; Woodford, N. Activity of ceftazidime/avibactam against problem Enterobacteriaceae and Pseudomonas aeruginosa in the UK, 2015–2016. J. Antimicrob. Chemother. 2018, 73, 648–657. [Google Scholar] [CrossRef]
- Andrei, S.; Valeanu, L.; Chirvasuta, R.; Stefan, M.-G. New FDA approved antibacterial drugs: 2015–2017. Discoveries 2018, 6, e81. [Google Scholar] [CrossRef] [PubMed]
- Ransom, E.; Bhatnagar, A.; Patel, J.B.; Machado, M.-J.; Boyd, S.; Reese, N.; Lutgring, J.D.; Lonsway, D.; Anderson, K.; Brown, A.C.; et al. Validation of Aztreonam-Avibactam Susceptibility Testing Using Digitally Dispensed Custom Panels. J. Clin. Microbiol. 2020, 58, 1–9. [Google Scholar] [CrossRef]
- A Cornely, O.; Cisneros, J.M.; Torre-Cisneros, J.; Rodríguez-Hernández, M.J.; Tallón-Aguilar, L.; Calbo, E.; Horcajada, J.P.; Queckenberg, C.; Zettelmeyer, U.; Arenz, D.; et al. Pharmacokinetics and safety of aztreonam/avibactam for the treatment of complicated intra-abdominal infections in hospitalized adults: Results from the REJUVENATE study. J. Antimicrob. Chemother. 2020, 75, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing Twenty-Eighth Informational Supplement M100-S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 1-562388-39-8. [Google Scholar]
- O’Neill, J. Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2015, 510, 15–16. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2013; CDC: Atlanta, GA, USA, 2013.
- CDC. Bigesst Threats and Data|Antibiotic/Antimicrobial Resistance; CDC: Atlanta, GA, USA, 2019.
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensiv. Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vidal, C.; Sanjuan, G.; Moreno-García, E.; Puerta-Alcalde, P.; Garcia-Pouton, N.; Chumbita, M.; Fernandez-Pittol, M.; Pitart, C.; Inciarte, A.; Bodro, M.; et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: A retrospective cohort study. Clin. Microbiol. Infect. 2021, 27, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Schwartz, I.S.; Kula, B.; Nguyen, M.H. Bacterial Superinfections Among Persons With Coronavirus Disease 2019: A Comprehensive Review of Data From Postmortem Studies. Open Forum Infect. Dis. 2021, 8, ofab065. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Van Duin, D.; Barlow, G.; Nathwani, D. The impact of the COVID-19 pandemic on antimicrobial resistance: A debate. JAC Antimicrob. Resist. 2020, 2, 4–5. [Google Scholar] [CrossRef]
- Årdal, C.; Balasegaram, M.; Laxminarayan, R.; McAdams, D.; Outterson, K.; Rex, J.H.; Sumpradit, N. Antibiotic development—Economic, regulatory and societal challenges. Nat. Rev. Genet. 2019, 18, 267–274. [Google Scholar] [CrossRef]
- Renwick, M.J.; Simpkin, V.; Mossialos, E. Targeting Innovation in Antibiotic Drug Discovery and Development: The Need for a One Health—One Europe—One World Framework; WHO: Geneva, Switzerland, 2016; ISBN 978 92 890 5040 1. [Google Scholar]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Kostyanev, T.; Bonten, M.J.M.; Obrien, S.J.; Steel, H.; Ross, S.T.; Francois, B.; Tacconelli, E.; Winterhalter, M.; Stavenger, R.A.; Karlen, A.; et al. The Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-Private partnerships for the development of new strategies to tackle antibiotic resistance. J. Antimicrob. Chemother. 2016, 71, 290–295. [Google Scholar] [CrossRef]
- Larsen, J.C.; Disbrow, G.L. Project BioShield and the Biomedical Advanced Research Development Authority: A ten year progress report on meeting U.S. preparedness objectives for threat agents. Clin. Infect. Dis. 2017, 64, 1430–1434. [Google Scholar] [CrossRef]
- Piddock, L.J. Gardp The Global Antibiotic Research and Development Partnership (GARDP): A not-For-Profit antibiotic development organisation. Lancet Infect. Dis. 2018, 18, 1304–1305. [Google Scholar] [CrossRef]
- Alm, R.A.; Gallant, K. Innovation in Antimicrobial Resistance: The CARB-X Perspective. ACS Infect. Dis. 2020, 6, 1317–1322. [Google Scholar] [CrossRef]
- Billington, J.K. The ABCs of the US Broad Spectrum Antimicrobials Program: Antibiotics, Biosecurity, and Congress. Health Secur. 2015, 13, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Dhand, R. The Rationale and Evidence for Use of Inhaled Antibiotics to ControlPseudomonas aeruginosaInfection in Non-Cystic Fibrosis Bronchiectasis. J. Aerosol Med. Pulm. Drug Deliv. 2018, 31, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Y.; Van Der Mei, H.C.; Ren, Y.; Busscher, H.J.; Shi, L. Lipid-Based Antimicrobial Delivery-Systems for the Treatment of Bacterial Infections. Front. Chem. 2020, 7, 872. [Google Scholar] [CrossRef] [PubMed]
- Durrant, J.D.; Amaro, R.E. Machine-learning techniques applied to antibacterial drug discovery. Chem. Biol. Drug Des. 2014, 85, 14–21. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; Macnair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef]
- Räisänen, K.; Koivula, I.; Ilmavirta, H.; Puranen, S.; Kallonen, T.; Lyytikäinen, O.; Jalava, J. Emergence of ceftazidime-avibactam-resistant Klebsiella pneumoniae during treatment, Finland, December 2018. Eurosurveillance 2019, 24, 1900256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Shi, Q.; Hu, H.; Hong, B.; Wu, X.; Du, X.; Akova, M.; Yu, Y. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 2020, 26, 124.e1–124.e4. [Google Scholar] [CrossRef] [PubMed]
- Venditti, C.; Nisii, C.; D’Arezzo, S.; Vulcano, A.; Capone, A.; Antonini, M.; Ippolito, G.; Di Caro, A. Molecular and phenotypical characterization of two cases of antibiotic-driven ceftazidime-avibactam resistance in blaKPC-3-harboring Klebsiella pneumoniae. Infect. Drug Resist. 2019, 12, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Hemarajata, P.; Humphries, R.M. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J. Antimicrob. Chemother. 2019, 74, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef]
- Hsu, L.-Y.; Apisarnthanarak, A.; Khan, E.; Suwantarat, N.; Ghafur, A.; Tambyah, P.A. Carbapenem-Resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin. Microbiol. Rev. 2016, 30, 1–22. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Nordmann, P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int. J. Antimicrob. Agents 2015, 45, 568–585. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug-Resistant Pseudomonas Aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- Abboud, M.I.; Damblon, C.; Brem, J.; Smargiasso, N.; Mercuri, P.; Gilbert, B.; Rydzik, A.M.; Claridge, T.D.W.; Schofield, C.J.; Frère, J.-M. Interaction of Avibactam with Class B Metallo-β-Lactamases. Antimicrob. Agents Chemother. 2016, 60, 5655–5662. [Google Scholar] [CrossRef]
- Wayne, P. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically M7-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; ISBN 1562389874. [Google Scholar]
- Oh, S.; Kwak, J.; Lee, J.; Han, H.; Biek, D.; Oh, K.; Cho, Y. Serum and Iron Effects on the In Vitro Activity of Siderophore Cephalosporin GT-1. In Posters of American Society for Microbiology (ASM) Microbe 2018; ASM Microbe: Atlanta, GA, USA, 2018. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
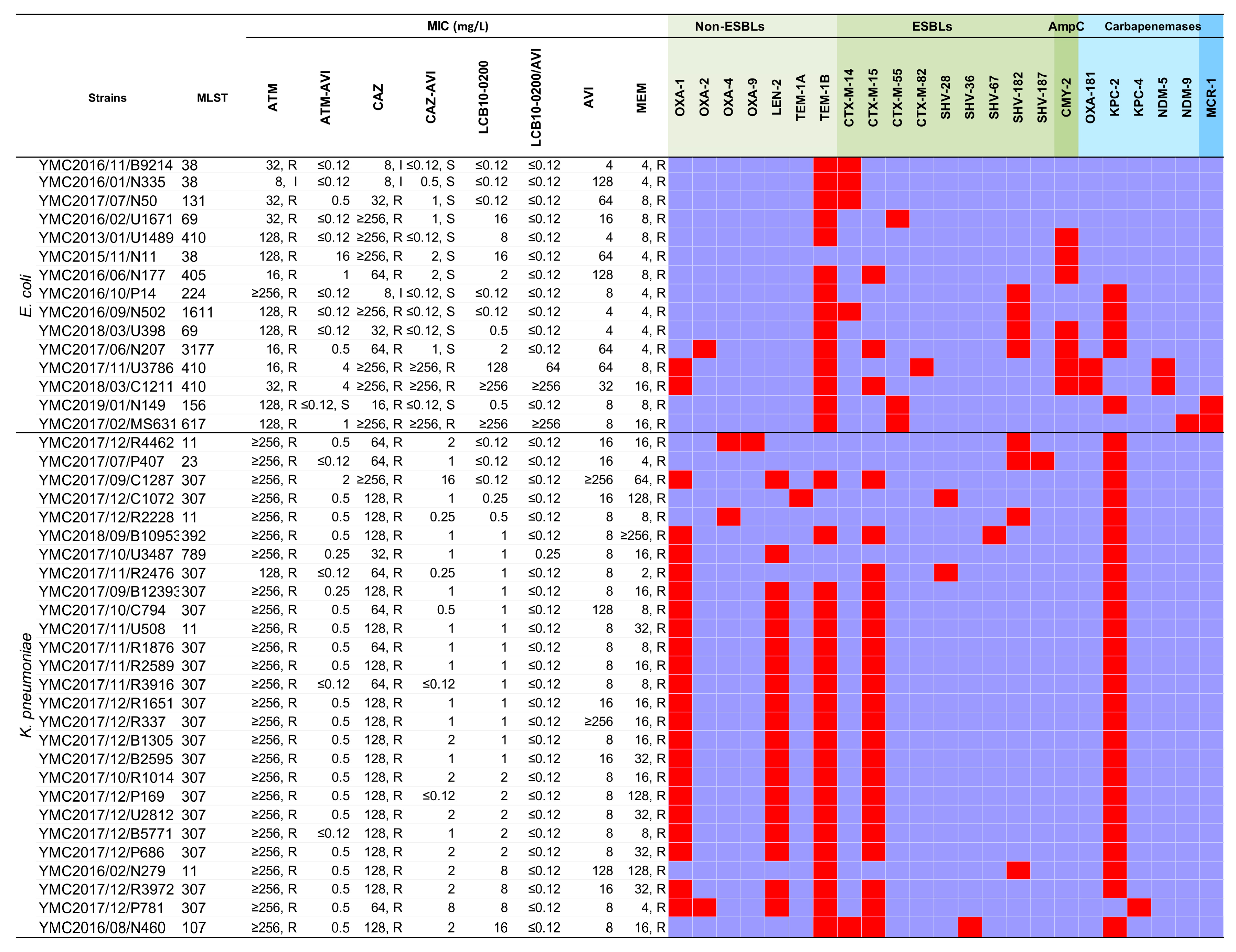

| Species (No. of Isolates, Percentages of Isolates) | MIC Data (mg/L) | MIC Interpretation (%) | ||||
|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | Susceptible | Intermediate | Resistant | |
| All isolates (93, 100%) | ||||||
| ATM | 64 | ≥256 | 2–≥256 | NA | NA | NA |
| ATM-AVI | 4 | 64 | ≤0.12–128 | NA | NA | NA |
| CAZ | 128 | ≥256 | 2–≥256 | NA | NA | NA |
| CAZ-AVI | 8 | 64 | ≤0.12–≥256 | NA | NA | NA |
| LCB10-0200 | 1 | 16 | ≤0.12–≥256 | NA | NA | NA |
| LCB10-0200/AVI | 0.5 | 16 | ≤0.12–≥256 | NA | NA | NA |
| AVI | ≥256 | ≥256 | 4–≥256 | NA | NA | NA |
| MEM | 32 | 128 | 2–≥256 | NA | NA | NA |
| Fermenting gram-negative bacilli: E. coli & K. pneumoniae isolates (42) | ||||||
| ATM | ≥256 | ≥256 | 8–≥256 | 0 | 2.4 | 97.6 |
| ATM-AVI | 0.5 | 1 | ≤0.12–16 | NA | NA | NA |
| CAZ | 128 | ≥256 | 2–≥256 | 2.4 | 4.8 | 92.9 |
| CAZ-AVI | 1 | 8 | ≤0.12–≥256 | NA | NA | NA |
| LCB10-0200 | 1 | 16 | ≤0.12–≥256 | NA | NA | NA |
| LCB10-0200/AVI | ≤0.12 | ≤0.12 | ≤0.12–≥256 | NA | NA | NA |
| AVI | 8 | 128 | 4–≥256 | NA | NA | NA |
| MEM | 8 | 64 | 2–≥256 | 0 | 2.4 | 97.6 |
| Non-fermenting gram-negative bacilli: A. baumannii & P. aeruginosa (51) | ||||||
| ATM | 32 | 128 | 2–128 | NA | NA | NA |
| ATM-AVI | 32 | 128 | 2–128 | NA | NA | NA |
| CAZ | 128 | ≥256 | 4–≥256 | 11.6 | 1.9 | 88.5 |
| CAZ-AVI | 32 | 64 | 2–≥256 | NA | NA | NA |
| LCB10-0200 | 0.5 | 16 | 0.12–≥256 | NA | NA | NA |
| LCB10-0200/AVI | 1 | 16 | 0.12–≥256 | NA | NA | NA |
| AVI | ≥256 | ≥256 | ≥256 | NA | NA | NA |
| MEM | 64 | 128 | 8–≥256 | 0 | 0 | 100 |
| E. coli isolates (15, 16%) | ||||||
| ATM | 32 | 128 | 8–≥256 | 0 | 6.7 | 93.3 |
| ATM-AVI | 0.12 | 4 | 0.12–16 | NA | NA | NA |
| CAZ | 64 | ≥256 | 2–≥256 | 0 | 20 | 80 |
| CAZ-AVI | 1 | ≥256 | ≤0.12–≥256 | 80 | 0 | 20 |
| LCB10-0200 | 2 | ≥256 | ≤0.12–≥256 | NA | NA | NA |
| LCB10-0200/AVI | ≤0.12 | ≥256 | ≤0.12–≥256 | NA | NA | NA |
| AVI | 16 | 128 | 4–128 | NA | NA | NA |
| MEM | 8 | 16 | 4–16 | 0 | 0 | 100 |
| K. pneumoniae isolates (27, 29%) | ||||||
| ATM | ≥256 | ≥256 | 128–≥256 | 0 | 0 | 100 |
| ATM-AVI | 0.5 | 0.5 | 0.12–2 | NA | NA | NA |
| CAZ | 128 | 128 | 32–≥256 | 0 | 0 | 100 |
| CAZ-AVI | 1 | 2 | ≤0.12–16 | 96.3 | NA | 3.7 |
| LCB10-0200 | 1 | 8 | ≤0.12–16 | NA | NA | NA |
| LCB10-0200/AVI | 0.12 | 0.12 | ≤0.12–0.25 | NA | NA | NA |
| AVI | 8 | 128 | 8–≥256 | NA | NA | NA |
| MEM | 8 | 128 | 2–≥256 | 0 | 3.7 | 96.3 |
| KPC-producing E. coli & K. pneumoniae isolates (32) | ||||||
| ATM | ≥256 | ≥256 | 16–≥256 | 0 | 0 | 100 |
| ATM-AVI | 0.5 | 0.5 | ≤0.12–2 | NA | NA | NA |
| CAZ | 128 | 128 | 8–≥256 | 0 | 3.1 | 96.9 |
| CAZ-AVI | 1 | 2 | ≤0.12–16 | 96.9 | 3.1 | 0 |
| LCB10-0200 | 1 | 8 | ≤0.12–16 | NA | NA | NA |
| LCB10-0200/AVI | ≤0.12 | ≤0.12 | ≤0.12–0.25 | NA | NA | NA |
| AVI | 8 | 128 | 4–≥256 | NA | NA | NA |
| MEM | 16 | 128 | 2–≥256 | 0 | 3.1 | 96.9 |
| A. baumannii isolates (25, 27%) | ||||||
| ATM | 64 | 128 | 32–128 | NA | NA | NA |
| ATM-AVI | 64 | 128 | 16–128 | NA | NA | NA |
| CAZ | ≥256 | ≥256 | 16–≥256 | 0 | 4 | 96 |
| CAZ-AVI | 32 | 64 | 4–≥256 | NA | NA | 94 |
| LCB10-0200 | 1 | 4 | 0.25–8 | NA | NA | NA |
| LCB10-0200/AVI | 1 | 4 | 0.5–4 | NA | NA | NA |
| AVI | ≥256 | ≥256 | ≥256 | NA | NA | NA |
| MEM | 64 | 128 | 32–128 | 0 | 0 | 100 |
| P. aeruginosa isolates (26, 28%) | ||||||
| ATM | 32 | 64 | 2–64 | 34.6 | 7.7 | 57.7 |
| ATM-AVI | 8 | 32 | 2–64 | NA | NA | NA |
| CAZ | 64 | ≥256 | 4–≥256 | 23.1 | NA | 76.9 |
| CAZ-AVI | 16 | 64 | 2–≥256 | 46.2 | NA | 53.8 |
| LCB10-0200 | 0.5 | 32 | 0.12–≥256 | NA | NA | NA |
| LCB10-0200/AVI | 0.5 | 32 | 0.12–≥256 | NA | NA | NA |
| AVI | ≥256 | ≥256 | ≥256 | NA | NA | NA |
| MEM | 32 | 64 | 8–≥256 | 0 | 0 | 100 |
| OXA-type producing A. baumannii & P. aeruginosa (46) | ||||||
| ATM | 32 | 128 | 2–128 | NA | NA | NA |
| ATM-AVI | 32 | 128 | 2–128 | NA | NA | NA |
| CAZ | 128 | 256 | 4–≥256 | 13 | 2.2 | 84.8 |
| CAZ-AVI | 32 | 64 | 2–≥256 | 30.4 | 10.9 | 58.7 |
| LCB10-0200 | 0.5 | 4 | ≤0.12–32 | NA | NA | NA |
| LCB10-0200/AVI | 1 | 4 | ≤0.12–32 | NA | NA | NA |
| AVI | ≥256 | ≥256 | ≥256 | NA | NA | NA |
| MEM | 32 | 128 | 8–128 | 0 | 0 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, L.P.; Park, C.S.; Pinto, N.A.; Lee, H.; Seo, H.S.; Vu, T.N.; Mai, H.; Pham, A.H.T.; Jang, E.; Cho, Y.L.; et al. In Vitro Activity of a Novel Siderophore-Cephalosporin LCB10-0200 (GT-1), and LCB10-0200/Avibactam, against Carbapenem-Resistant Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa Strains at a Tertiary Hospital in Korea. Pharmaceuticals 2021, 14, 370. https://doi.org/10.3390/ph14040370
Nguyen LP, Park CS, Pinto NA, Lee H, Seo HS, Vu TN, Mai H, Pham AHT, Jang E, Cho YL, et al. In Vitro Activity of a Novel Siderophore-Cephalosporin LCB10-0200 (GT-1), and LCB10-0200/Avibactam, against Carbapenem-Resistant Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa Strains at a Tertiary Hospital in Korea. Pharmaceuticals. 2021; 14(4):370. https://doi.org/10.3390/ph14040370
Chicago/Turabian StyleNguyen, Le Phuong, Chul Soon Park, Naina Adren Pinto, Hyunsook Lee, Hyun Soo Seo, Thao Nguyen Vu, Hung Mai, An H. T. Pham, Eris Jang, Young Lag Cho, and et al. 2021. "In Vitro Activity of a Novel Siderophore-Cephalosporin LCB10-0200 (GT-1), and LCB10-0200/Avibactam, against Carbapenem-Resistant Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa Strains at a Tertiary Hospital in Korea" Pharmaceuticals 14, no. 4: 370. https://doi.org/10.3390/ph14040370
APA StyleNguyen, L. P., Park, C. S., Pinto, N. A., Lee, H., Seo, H. S., Vu, T. N., Mai, H., Pham, A. H. T., Jang, E., Cho, Y. L., Goglin, K., Nguyen, K., White, R., D’Souza, R., Fouts, D. E., & Yong, D. (2021). In Vitro Activity of a Novel Siderophore-Cephalosporin LCB10-0200 (GT-1), and LCB10-0200/Avibactam, against Carbapenem-Resistant Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa Strains at a Tertiary Hospital in Korea. Pharmaceuticals, 14(4), 370. https://doi.org/10.3390/ph14040370






