Three-Component Synthesis of 2-Amino-3-cyano-4H-chromenes, In Silico Analysis of Their Pharmacological Profile, and In Vitro Anticancer and Antifungal Testing
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Instruments
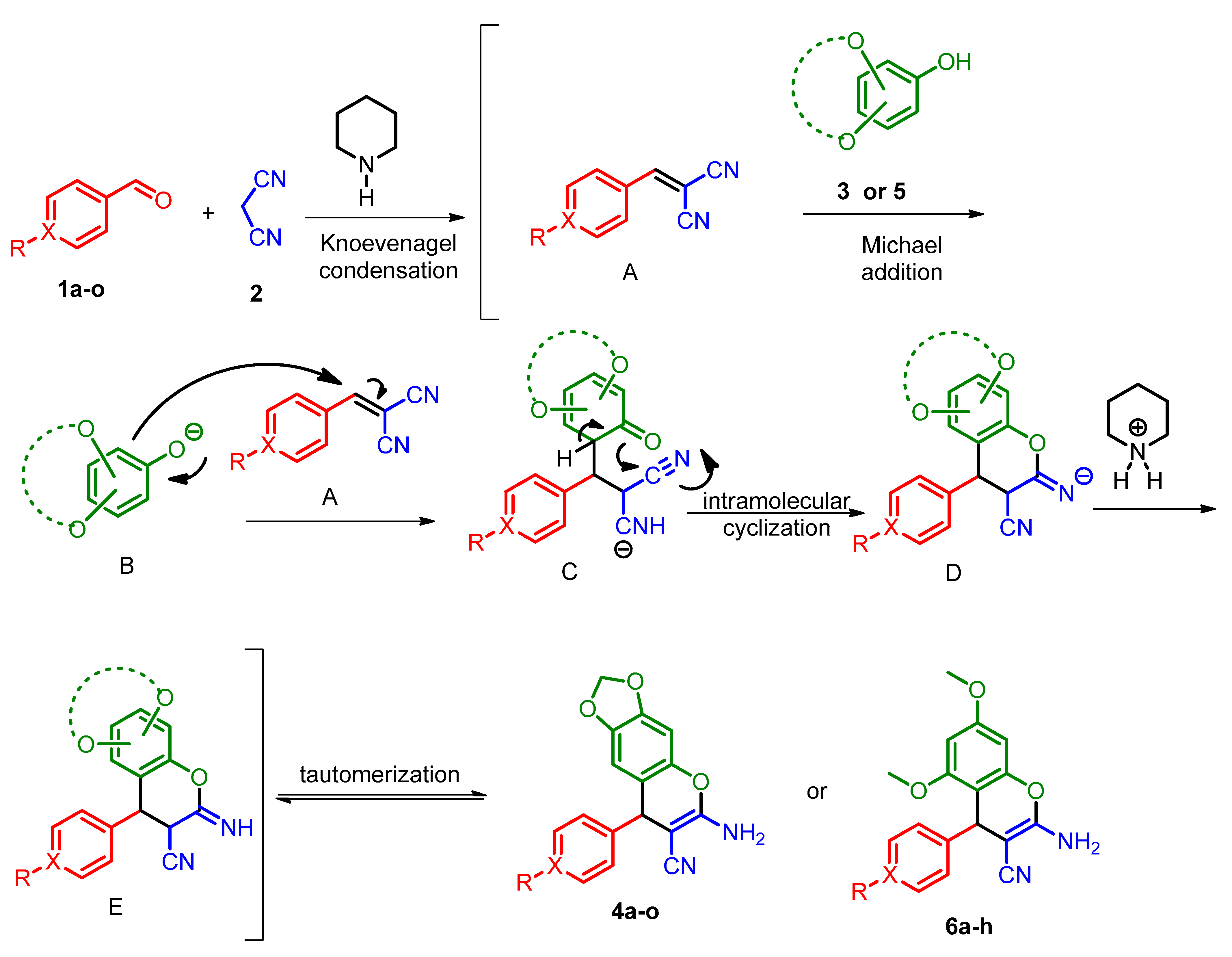
2.2. General Procedure for the Preparation of 2-Amino-3-cyano-4H-chromenes 4a–o and 6a–h
2.2.1. 2-Amino-3-cyano-4-phenyl-6,7-methylendioxy-4H-chromene (4a)
2.2.2. 2-Amino-3-cyano-4-(4-fluorophenyl)-6,7-methylendioxy-4H-chromene (4b)
2.2.3. 2-Amino-3-cyano-4-(4-chlorophenyl)-6,7-methylendioxy-4H-chromene (4c)
2.2.4. 2-Amino-4-(4-bromophenyl)-3-cyano-6,7-methylendioxy-4H-chromene (4d)
2.2.5. 2-Amino-3-cyano-4-(4-cyanophenyl)-6,7-methylendioxy-4H-chromene (4e)
2.2.6. 2-Amino-3-cyano-6,7-methylendioxy-4-(4-nitrophenyl)-4H-chromene (4f)
2.2.7. 2-Amino-3-cyano-6,7-methylendioxy-4-(p-tolyl)-4H-chromene (4g)
2.2.8. 2-Amino-3-cyano-6,7-methylendioxy-4-(4-methoxyphenyl)-4H-chromene (4h)
2.2.9. 2-Amino-3-cyano-6,7-methylendioxy-4-(pyridin-4-yl)-4H-chromene (4i)
2.2.10. 2-Amino-3-cyano-4-(3-fluorophenyl)-6,7-methylendioxy-4H-chromene (4j)
2.2.11. 2-Amino-3-cyano-4-(3-cyanophenyl)-6,7-methylendioxy-4H-chromene (4k)
2.2.12. 2-Amino-3-cyano-4-(2-fluorophenyl)-6,7-methylendioxy-4H-chromene (4l)
2.2.13. 2-Amino-3-cyano-4-(2-chlorophenyl)-6,7-methylendioxy-4H-chromene (4m)
2.2.14. 2-Amino-3-cyano-4-(2-bromophenyl)-6,7-methylendioxy-4H-chromene (4n)
2.2.15. 2-Amino-3-cyano-4-(2-nitrophenyl)-6,7-methylendioxy-4H-chromene (4o)
2.2.16. 2-Amino-3-cyano-5,7-dimethoxy-4-phenyl-4H-chromene (6a)
2.2.17. 2-Amino-3-cyano-5,7-dimethoxy-4-(4-fluorophenyl)-4H-chromene (6b)
2.2.18. 2-Amino-3-cyano-5,7-dimethoxy-4-(4-chlorophenyl)-4H-chromene (6c)
2.2.19. 2-Amino-4-(4-bromophenyl)-3-cyano-5,7-dimethoxy-4H-chromene (6d)
2.2.20. 2-Amino-3-cyano-4-(4-cyanophenyl)-5,7-dimethoxy-4H-chromene (6e)
2.2.21. 2-Amino-3-cyano-5,7-dimethoxy-4-(4-nitrophenyl)-4H-chromene (6f)
2.2.22. 2-Amino-3-cyano-5,7-dimethoxy-4-(p-tolyl)-4H-chromene (6g)
2.2.23. 2-Amino-3-cyano-5,7-dimethoxy-4-(4-methoxyphenyl)-4H-chromene (6h)
2.3. In-Silico Analysis of 2-Amino-3-cyano-4-aryl-6,7-methylendioxy-4H-chromenes 4a–o and 2-Amino-3-cyano-5,7-dimethoxy-4-aryl-4H-chromenes 6a–h
2.3.1. Multiple Sequence Alignment and Generation of 3D CYP51 from Candida spp. through Homology Modeling
2.3.2. Evaluation and Validation of the 3D Model of CYP51 from Candida spp.
2.3.3. Molecular Docking on CYP51
2.3.4. Molecular Docking on Topoisomerase I
2.4. Antifungal Activity Assays
2.5. Cytotoxicity Assay
3. Results and Discussion
3.1. Chemistry
3.2. In Silico Analysis of Physicochemical, Pharmacokinetic, Drug-likeness and Toxicological Properties of 2-Amino-3-cyano-4H-chromenes 4a–o and 6a–h
3.3. Multiple Sequence Alignment of CYP51 Enzymes of Candida spp.
3.4. Generating 3D Models of CYP51 Enzymes of Candida spp.
3.5. Molecular Docking Studies
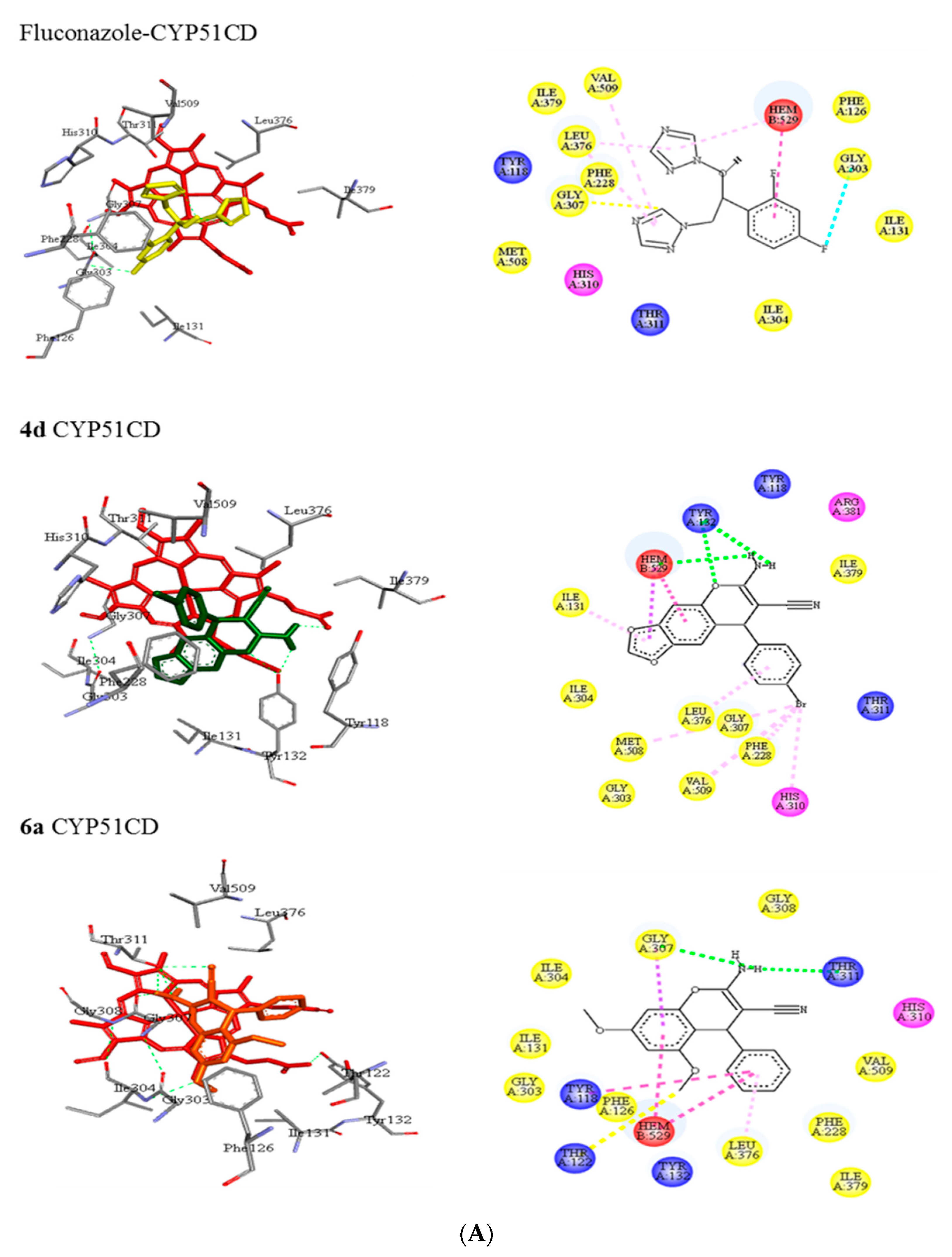
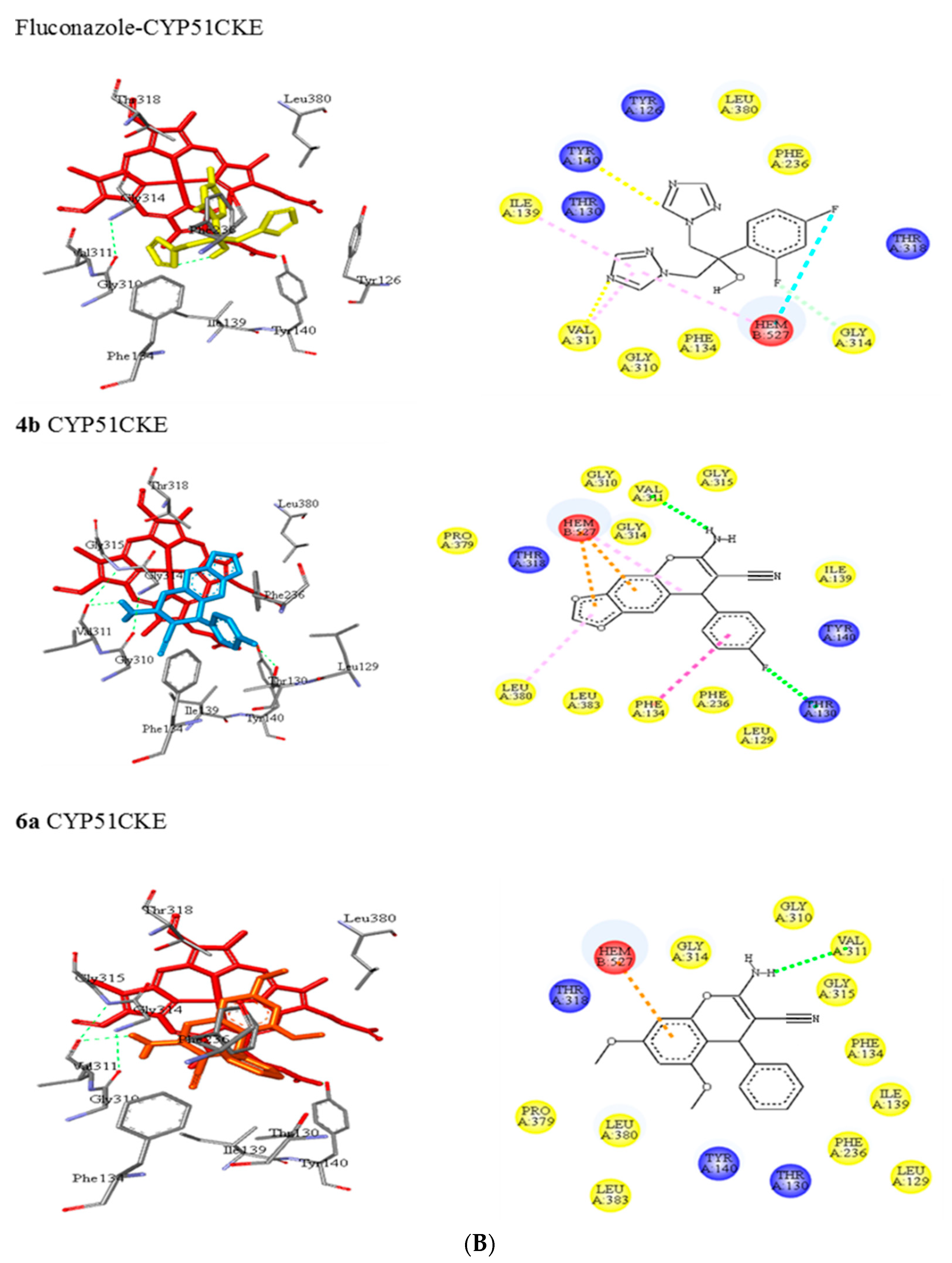
3.5.1. Molecular Docking of 2-Amino-3-cyano-4H-chromenes 4a–o and 6a–h and Fluconazole (8) with CYP51 from Candida spp.
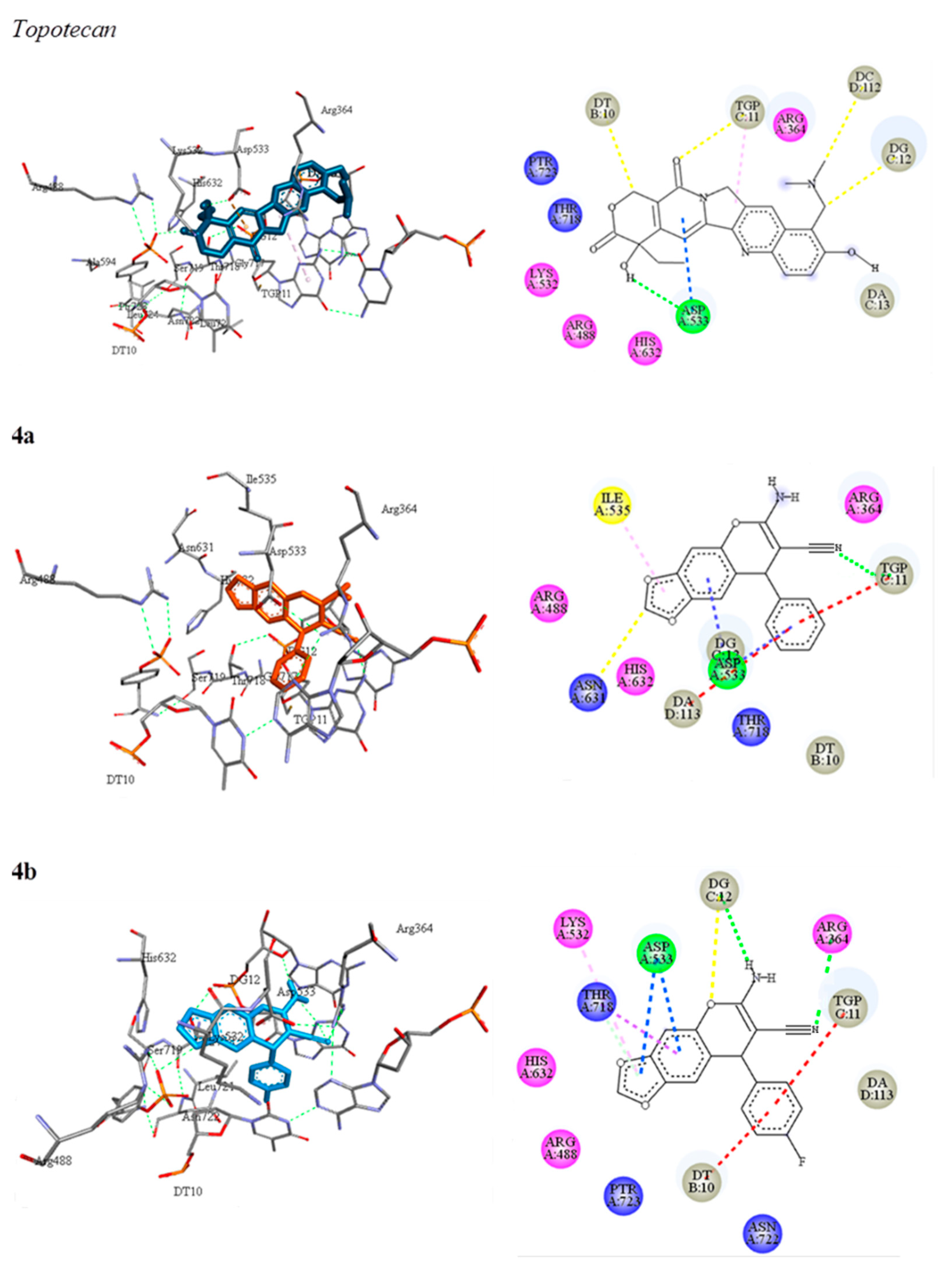
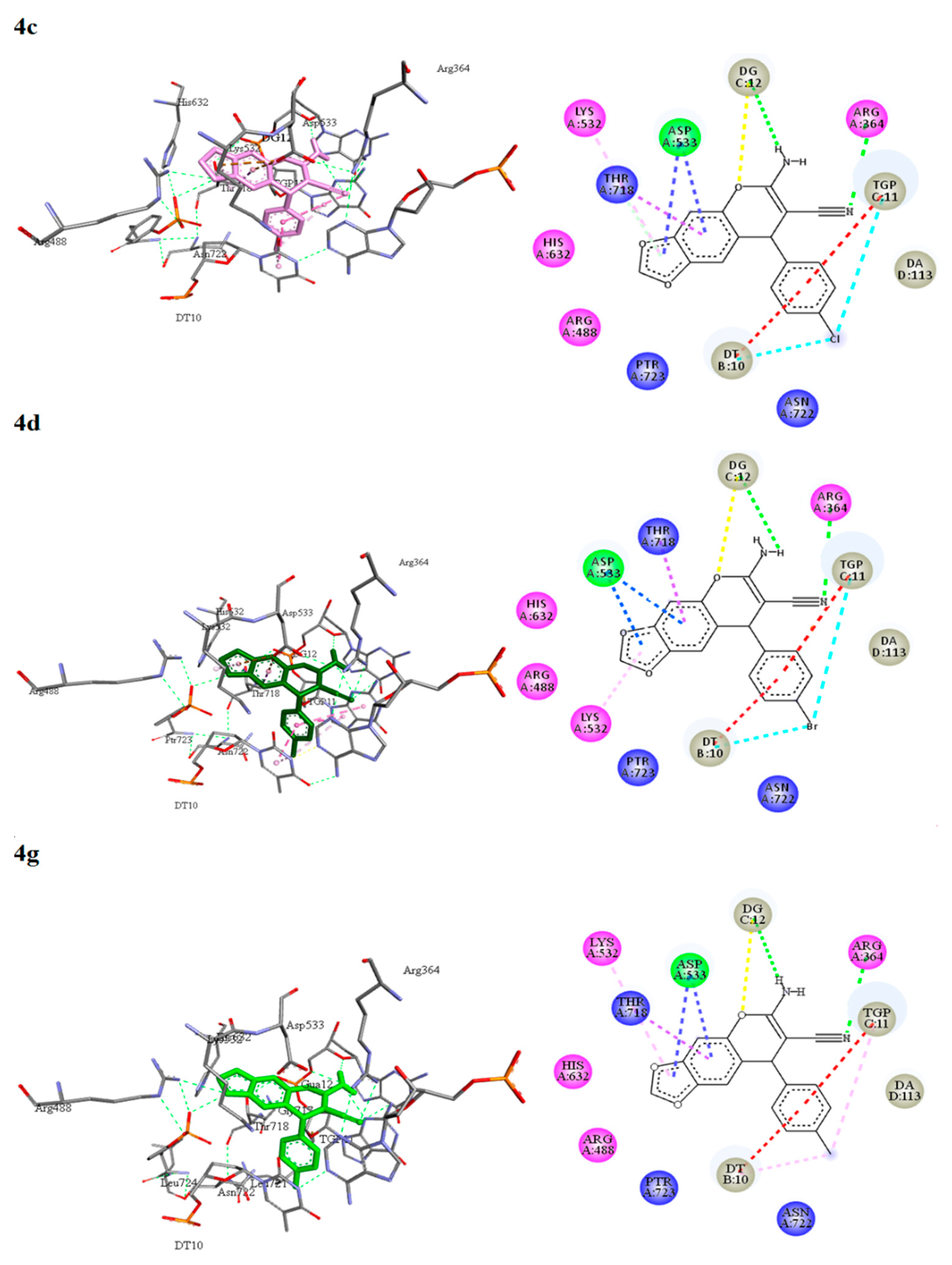
3.5.2. Molecular Docking of 2-Amino-3-cyano-4H-chromenes 4a–o, 6a–h and Topotecan (7) on Topoisomerase I
3.6. Antifungal Activity
3.7. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patil, S.A.; Wang, J.; Li, X.S.; Chen, J.; Jones, T.S.; Hosni-Ahmed, A.; Patil, R.; Seibel, W.L.; Li, W.; Miller, D.D. New substituted 4H-chromenes as anticancer agents. Bioorg. Med. Chem. Lett. 2012, 22, 4458–4461. [Google Scholar] [CrossRef]
- Thanh, N.D.; Hai, D.S.; Bich, V.T.N.; Hien, P.T.T.; Duyen, N.T.K.; Mai, N.T.; Dung, T.T.; Toan, V.N.; Van, H.T.K.; Dang, L.H.; et al. Efficient click chemistry towards novel 1H-1,2,3-triazole-tethered 4H-chromene-d-glucose conjugates: Design, synthesis, and evaluation of in vitro antibacterial, MRSA and antifungal activities. Eur. J. Med. Chem. 2019, 167, 454–471. [Google Scholar] [CrossRef]
- Subbareddy, C.V.; Sundarrajan, S.; Mohanapriya, A.; Subashini, R.; Shanmugam, R. Synthesis, antioxidant, antibacterial, solvatochromism and molecular docking studies of indolyl-4H-chromene-phenylprop-2-en-1-one derivatives. J. Mol. Liq. 2018, 251, 296–307. [Google Scholar] [CrossRef]
- Martin, E.F.; Mbaveng, A.T.; de Moraes, M.H.; Kuete, V.; Biavatti, M.W.; Steindel, M.; Efferth, T.; Sandjo, L.P. Prospecting for cytotoxic and antiprotozoal 4-aryl-4H-chromenes and 10-aryldihydropyrano[2,3-f]chromenes. Arch. Pharm. (Weinh.) 2018, 351, e1800100. [Google Scholar] [CrossRef]
- Shestopalov, A.M.; Litvinov, Y.M.; Rodinovskaya, L.A.; Malyshev, O.R.; Semenova, M.N.; Semenov, V.V. Polyalkoxy substituted 4H-chromenes: Synthesis by domino reaction and anticancer activity. ACS Comb. Sci. 2012, 14, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Singla, R.; Jaitak, V. Coumarins as anticancer agents: A review on synthetic strategies, mechanism of action and SAR studies. Eur. J. Med. Chem. 2015, 101, 476–495. [Google Scholar] [CrossRef]
- Hassanpour, S.H.; Dehghani, M. Review of cancer from perspective of molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Pommier, Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem. Rev. 2009, 109, 2894–2902. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.; Gao, K.; Chu, L.; Zhang, R.; Yang, J.; Zheng, J. Aurora kinases: Novel therapy targets in cancers. Oncotarget 2017, 8, 23937–23954. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, S.; Moulder, S.; Keyomarsi, K.; Layman, R.M. Inhibiting CDK in cancer therapy: Current evidence and future directions. Target. Oncol. 2018, 13, 21–38. [Google Scholar] [CrossRef]
- Katsetos, C.D.; Draber, P. Tubulins as therapeutic targets in cancer: From bench to bedside. Curr. Pharm. Des. 2012, 18, 2778–2792. [Google Scholar] [CrossRef]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.; Zhivotovsky, B. Caspases and cancer. Cell Death Differ. 2011, 18, 1441–1449. [Google Scholar] [CrossRef]
- Becher, R.; Wirsel, S.G.R. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef]
- Wiederhold, N.P. The antifungal arsenal: Alternative drugs and future targets. Int. J. Antimicrob. Agents 2018, 51, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.R.; Zhao, F.; Wang, S.; Lv, S.; Mou, Y.; Yao, C.L.; Zhou, Y.; Li, F.Q. Molecular mechanism of azoles resistant Candida albicans in a patient with chronic mucocutaneous candidiasis. BMC. Infect. Dis. 2020, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nakib, T.A.; Bezjak, V.; Rashid, S.; Fullam, B.; Meegan, M.J. The synthesis and antifungal activity of 2-amino-4-aryl-4H,5H-[1]benzothiopyrano[4,3-b]pyran-3-carbonitriles and 2-alkoxy-4-aryl-5H-[1]benzothiopyrano[4,3-b]pyridine-3-carbonitriles. Eur. J. Med. Chem. 1991, 26, 221–230. [Google Scholar] [CrossRef]
- Fouda, A.M.; Assiri, M.A.; Mora, A.; Ali, T.E.; Afifi, T.H.; El-Agrody, A.M. Microwave synthesis of novel halogenated β-enaminonitriles linked 9-bromo-1H-benzo[f]chromene moieties: Induces cell cycle arrest and apoptosis in human cancer cells via dual inhibition of topoisomerase I and II. Bioorg. Chem. 2019, 93, 103289. [Google Scholar] [CrossRef]
- Hermanson, D.L.; Das, S.G.; Li, Y.; Xing, C. Overexpression of Mcl-1 confers multidrug resistance, whereas topoisomerase IIβ downregulation introduces mitoxantrone-specific drug resistance in acute myeloid leukemia. Mol. Pharmacol. 2013, 84, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Prakash Rao, H.S.; Adigopula, L.N.; Ramalingam, G.; Lone, J.A.; Ramadas, K. Design, synthesis, anticancer properties and in silico evaluation of C(4)-N-heteroaryl-4H-chromenes. Chem. Select. 2018, 3, 13161–13166. [Google Scholar] [CrossRef]
- Kemnitzer, W.; Drewe, J.; Jiang, S.; Zhang, H.; Wang, Y.; Zhao, J.; Jia, S.; Herich, J.; Labreque, D.; Storer, R.; et al. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based high-throughput screening assay. 1. Structure-activity relationships of the 4-aryl group. J. Med. Chem. 2004, 47, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Verma, C.K. A combination of pharmacophore modeling, molecular docking and virtual screening study reveals 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)-4H-chromen-4-one as a potential anti-cancer agent of COT kinase. Indian J. Pharm. Educ. Res. 2018, 52, 699–706. [Google Scholar] [CrossRef]
- Frasinyuk, M.S.; Zhang, W.; Wyrebek, P.; Yu, T.; Xu, X.; Sviripa, V.M.; Bondarenko, S.P.; Xie, Y.; Ngo, H.X.; Morris, A.J.; et al. Developing antineoplastic agents that target peroxisomal enzymes: Cytisine-linked isoflavonoids as inhibitors of hydroxysteroid 17-beta-dehydrogenase-4 (HSD17B4). Org. Biomol. Chem. 2017, 36, 7623–7629. [Google Scholar] [CrossRef] [PubMed]
- Quan-Wang, J.Q.; Yang, M.D.; Chen, X.; Wang, Y.; Chen, L.Z.; Cheng, X.; Liu, X.H. Discovery of new chromen-4-one derivatives as telomerase inhibitors through regulating expression of dyskerin. J. Enzym. Inhib. Med. Chem. 2018, 33, 1199–1211. [Google Scholar] [CrossRef]
- Brasil, E.M.; Canavieira, L.M.; Cardoso, E.T.C.; Silva, E.O.; Lameira, J.O.; Nascimento, J.L.M.; Eifler-Lima, V.L.; Macchi, B.M.; Sriram, D.; Bernhardt, P.V.; et al. Inhibition of tyrosinase by 4H-chromene analogs: Synthesis, kinetic studies, and computational analysis. Chem. Biol. Drug Des. 2017, 90, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wang, M.; Miller, K.D.; Hutchins, G.D.; Zheng, Q.H. Synthesis of carbon-11-labeled 4-aryl-4H-chromens as new PET agents for imaging of apoptosis in cancer. Appl. Radiat. Isot. 2010, 68, 110–116. [Google Scholar] [CrossRef]
- Kemnitzer, W.; Drewe, J.; Jiang, S.; Zhang, H.; Crogan-Grundy, C.; Labreque, D.; Bubenick, M.; Attardo, G.; Denis, R.; Lamothe, S.; et al. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell-and caspase-based HTS assay. Part 5: Modifications of the 2- and 3-positions. J. Med. Chem. 2008, 51, 417–423. [Google Scholar] [CrossRef]
- Kidwai, M.; Saxena, S.; Rahaman Khan, M.K.; Thukral, S.S. Aqua mediated synthesis of substituted 2-amino-4H-chromenes and in vitro study as antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 4295–4298. [Google Scholar] [CrossRef]
- Datta, B.; Pasha, M.A. Glycine catalyzed convenient synthesis of 2-amino-4H-chromenes in aqueous medium under sonic condition. Ultrason. Sonochem. 2012, 19, 725–728. [Google Scholar] [CrossRef]
- Elinson, M.N.; Dorofeev, A.S.; Feducovich, S.K.; Gorbunov, S.V.; Nasybullin, R.F.; Miloserdov, F.M.; Nikishin, G.I. The implication of electrocatalysis in MCR strategy: Electrocatalytic multicomponent transformation of cyclic 1,3-Diketones, Aldehydes and Malononitrile into substituted 5,6,7,8-tetrahydro-4H-chromenes. Eur. J. Org. Chem. 2006, 4335–4339. [Google Scholar] [CrossRef]
- Nikpassand, M.; Fekri, L.Z.; Ahmadi, P. Grinding synthesis of 2-amino-4H-chromenes using 3,3-(butane-1,4-diyl) bis (1,2-dimethyl-1H-imidazole-3-ium) Br-can as a novel reagent. J. Chil. Chem. Soc. 2017, 62, 3399–3402. [Google Scholar] [CrossRef]
- Sánchez, A.; Hernández, F.; Cruz, P.C.; Alcaraz, Y.; Tamariz, J.; Delgado, F.; Vázquez, M.A. Infrared irradiation-assisted multicomponent synthesis of 2-amino-3-cyano-4H-pyran derivatives. J. Mex. Chem. Soc. 2012, 56, 121–127. [Google Scholar] [CrossRef]
- El-Agrody, A.M.; Fouda, A.M.; Khattab, E.S.A.E.H. Synthesis, antitumor activity of 2-amino-4H-benzo[h]chromene derivatives, and structure–activity relationships of the 3- and 4-positions. Med. Chem. Res. 2013, 22, 6105–6120. [Google Scholar] [CrossRef]
- Saikia, M.; Saikia, L. Sulfonic acid-functionalized MIL-101(Cr) as a highly efficient heterogeneous catalyst for one-pot synthesis of 2-amino-4H-chromenes in aqueous medium. RSC Adv. 2016, 6, 15846–15853. [Google Scholar] [CrossRef]
- Ahmad, A.; Silva, L.F., Jr. Synthesis of chromanes and 4H-chromenes: Exploring the oxidation of 2H-chromenes and dihydro-1-benzoxepines by hypervalent iodine (III). Synthesis 2012, 44, 3671–3677. [Google Scholar] [CrossRef]
- Mondal, J.; Modak, A.; Nandi, M.; Uyama, H.; Bhaumik, A. Triazine functionalized ordered mesoporous organosilica as a novel organocatalyst for the facile one-pot synthesis of 2-amino-4H-chromenes under solvent-free conditions. RSC Adv. 2012, 2, 11306–11317. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, W.; Lu, J.; Gao, K.; Liao, X.; Chen, X. One-pot synthesis of tetrahydro-4H-chromenes by supramolecular catalysis in water. RSC Adv. 2015, 5, 79405–79412. [Google Scholar] [CrossRef]
- Azarifar, A.; Nejat-Yami, R.; Al Kobaisi, M.; Azarifar, D. Magnetic La0.7Sr0.3MnO3 nanoparticles: Recyclable and efficient catalyst for ultrasound-accelerated synthesis of 4H-chromenes, and 4H-pyrano[2,3-c]pyrazoles. J. Iran. Chem. Soc. 2013, 10, 439–446. [Google Scholar] [CrossRef]
- Funabiki, K.; Komeda, T.; Kubota, Y.; Matsui, M. Brönsted acid ionic liquid-catalyzed direct benzylation, allylation and propargylation of 1,3-dicarbonyl compounds with alcohols as well as one-pot synthesis of 4H-chromenes. Tetrahedron 2009, 65, 7457–7463. [Google Scholar] [CrossRef]
- Heravi, M.M.; Daraie, M. Heterogeneous catalytic three-component one-pot synthesis of novel 8H-[1,3]dioxolo[4,5-g]chromenes by basic alumina in water. Monatsh. Chem. 2014, 145, 1479–1482. [Google Scholar] [CrossRef]
- Wu, B.; Gao, X.; Yan, Z.; Chen, M.W.; Zhou, Y.G. C-H Oxidation/Michael addition/cyclization cascade for enantioselective synthesis of functionalized 2-amino-4H-chromenes. Org. Lett. 2015, 17, 6134–6137. [Google Scholar] [CrossRef]
- Ballini, R.; Bosica, G.; Conforti, M.L.; Maggi, R.; Mazzacani, A.; Righi, P.; Sartori, G. Three-component process for the synthesis of 2-amino-2-chromenes in aqueous media. Tetrahedron 2001, 57, 1395–1398. [Google Scholar] [CrossRef]
- Jabbarzare, S.; Ghashang, M. Preparation of 2-amino-5,7-dimethoxy-4-aryl/alkyl-4H-chromene-3-carbonitriles using Na2O-Al2O3-P2O5 glass-ceramic system. Chin. Chem. Lett. 2015, 26, 1385–1388. [Google Scholar] [CrossRef]
- Sander, T.; Freyss, J.; Von Korff, M.; Rufener, C. 2015 data warrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Sharma, S.; Ciufo, S.; Starchenko, E.; Darji, D.; Chlumsky, L.; Karsch-Mizrachi, I.; Schoch, C.L. The NCBI bioCollections database. Database 2018, 2018, bay006. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 2000, 132, 221–241. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Dassault Systèmes BIOVIA. Discovery Studio Modeling Environment; Release 2017; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK—A program to check the stereochemical quality of protein structures. J. App. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Auto-mated docking with selective receptorflexiblity. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Sagatova, A.A.; Keniya, M.V.; Wilson, R.K.; Monk, B.C.; Tyndall, J.D. Structural insights into binding of the antifungal drug fluconazole to Saccharomyces cerevisiae lanosterol 14α-demethylase. Antimicrob. Agents Chemother. 2015, 59, 4982–4989. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Cousins, K.R. Computer review of ChemDraw Ultra 12.0. J. Am. Chem. Soc. 2011, 133, 8388. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B. Gaussian 98 Inc., Version A.6, Wallingford CT. 2004. Available online: http://gaussian.com/g03citation/ (accessed on 26 October 2021).
- Staker, B.L.; Feese, M.D.; Cushman, M.; Pommier, Y.; Zembower, D.; Stewart, L.; Burgin, A.B. Structures of three classes of anticancer agents bound to the human topoisomerase I-DNA covalent complex. J. Med. Chem. 2005, 48, 2336–2345. [Google Scholar] [CrossRef]
- Irwin, J.J.; Shoichet, B.K. ZINC–A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef]
- Cuenca-Estrella, M.; Lee-Yang, W.; Ciblak, M.A.; Arthington-Skaggs, B.A.; Mellado, E.; Warnock, D.W.; Rodríguez-Tudela, J.L. Comparative evaluation of NCCLS M27-A and EUCAST broth microdilution procedures for antifungal susceptibility testing of Candida Species. Antimicrob. Agents Chemother. 2002, 46, 3644–4647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gómez, E.; Contreras-Ordoñez, G.; Ramírez-Apan, T. Synthesis, characterization and in vitro cytotoxicity of pentacoordinated Tin(IV) complexes derived from amino alcohols. Chem. Pharm. Bull. 2006, 54, 54–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gómez-García, O.; Gómez, E.; Monzón-González, C.; Ramírez-Apan, T.; Álvarez-Toledano, C. An efficient strategy for the synthesis of 1-(trifluoromethylsulfonamido)propan-2-yl esters and the evaluation of their cytotoxic activity. Chem. Pharm. Bull. 2017, 65, 248–252. [Google Scholar] [CrossRef]
- Vraka, C.; Nics, L.; Wagner, K.H.; Hacker, M.; Wadsak, W.; Mitterhauser, M. 2017 Log P, a yesterday’s value? Nucl. Med. Biol. 2017, 50, 1–10. [Google Scholar] [CrossRef]
- Trapani, A.; Lopedota, A.; Denora, N.; Laquintana, V.; Franco, M.; Latrofa, A.; Trapani, G.; Liso, G. A rapid screening tool for estimating the potential of 2-hydroxypropyl-beta-cyclodextrin complexation for solubilization purposes. Int. J. Pharm. 2005, 295, 163–175. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Schultes, S.; De Graaf, C.; Haaksma, E.E.J.; De Esch, I.J.P.; Leurs, R.; Krämer, O. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov. Today Technol. 2010, 7, e157–e162. [Google Scholar] [CrossRef] [PubMed]
- Perola, E. An analysis of the binding efficiencies of drugs and their leads in successful drug discovery programs. J. Med. Chem. 2010, 53, 2986–2997. [Google Scholar] [CrossRef]
- Rackham, M.D.; Brannigan, J.A.; Rangachari, K.; Meister, S.; Wilkinson, A.J.; Holder, A.A.; Leatherbarrow, R.J.; Tate, E.W. Design, and synthesis of high affinity inhibitors of Plasmodium falciparum and plasmodium vivax N-myristoyltransferases directed by ligand efficiency dependent lipophilicity (LELP). J. Med. Chem. 2014, 57, 2773–2788. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I. Property distribution of drug-related chemical databases. J. Comput.-Aided Mol. Des. 2000, 14, 251–264. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Potts, R.O.; Guy, R.H. Predicting skin permeability. Pharm. Res. 1992, 9, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chang, Z.; Pan, Z.; Fu, Z.Q.; Wang, X. Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. USA 2008, 105, 13883–13888. [Google Scholar] [CrossRef] [PubMed]
- Leiva, K.; Werner, N.; Sepúlveda, D.; Barahona, S.; Baeza, M.; Cifuentes, V.; Alcaíno, J. Identification and functional characterization of the CYP51 gene from the yeast xanthophyllomyces dendrorhous that is involved in ergosterol biosynthesis. BMC Microbiol. 2015, 15, 1–12. [Google Scholar] [CrossRef]
- Zhang, J.; Li, L.; Lv, Q.; Yan, L.; Wang, Y.; Jiang, Y. The fungal CYP51s: Their functions, structures, related drug resistance, and inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef] [PubMed]
- Gómez-García, O.; Andrade-Pavón, D.; Campos-Aldrete, E.; Ballinas-Indilí, R.; Méndez-Tenorio, A.; Villa-Tanaca, L.; Álvarez-Toledano, C. Synthesis, molecular docking, and antimycotic evaluation of some 3-acylimidazo[1,2-a]pyrimidines. Molecules 2018, 23, 599. [Google Scholar] [CrossRef] [PubMed]
- Davood, A.; Rahimi, A.; Iman, M.; Azerang, P.; Sardar-I., S.; Mahboubi, A. Design and synthesis of new antifungals based on N-un-substituted azoles as 14α-demethylase inhibitor. Curr. Comput.-Aided Drug. Des. 2021, 17, 235–243. [Google Scholar] [CrossRef]
- Che, X.; Sheng, C.; Wang, W.; Cao, Y.; Xu, Y.; Ji, H.; Dong, G.; Miao, Z.; Yao, J.; Zhang, W. New azoles with potent antifungal activity: Design, synthesis and molecular docking. Eur. J. Med. Chem. 2009, 44, 4218–4226. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Zhang, W.; Zhou, Y.; Zhang, M.; Zhu, J.; Song, Y.; Lü, J.; Zhu, J. A three-dimensional model of lanosterol 14-α-demethylase of Candida albicans and its interaction with azole antifungals. J. Med. Chem. 2000, 43, 2493–2505. [Google Scholar] [CrossRef]
- Chen, S.H.; Sheng, C.Q.; Xu, X.H.; Jiang, Y.Y.; Zhang, W.N.; He, C. Identification of Y118 amino acid residue in Candida albicans sterol 14-alpha-demethylase associated with the enzyme activity and selective antifungal activity of azole analogues. Biol. Pharm. Bull. 2007, 30, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Ohi, M.; Aoyama, Y.; Nitahara, Y.; Chung, S.K.; Yoshida, Y. Effects of Y132H and F145L substitutions on the activity, azole resistance and spectral properties of Candida albicans sterol 14-demethylase P450 (CYP51): A live example showing the selection of altered P450 through interaction with environmental compounds. J. Biochem. 2005, 137, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Wang, W.; Che, X.; Dong, G.; Wang, S.; Ji, H.; Miao, Z.; Yao, J.; Zhang, W. Improved model of lanosterol 14-alpha-demethylase by ligand-supported homology modeling: Validation by virtual screening and azole optimization. ChemMedChem 2010, 5, 390–397. [Google Scholar] [CrossRef]
- Rocha-Del Castillo, E.; Gómez-García, O.; Andrade-Pavón, D.; Villa-Tanaca, L.; Ramírez-Apan, T.; Nieto-Camacho, A.; Gómez, E. Dibutyltin(IV) complexes derived from L-DOPA: Synthesis, molecular docking, cytotoxic and antifungal activity. Chem. Pharm. Bull. 2018, 66, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Pavón, D.; Gómez-Garcia, O.; Álvarez-Toledano, C. Exploring the binding mode of triflamide derivatives at the active site of Topo I and Topo II enzymes: In silico analysis and precise molecular docking. J. Chem. Sci. 2020, 132, 50. [Google Scholar] [CrossRef]
- Okasha, R.M.; Albalawi, F.F.; Afifi, T.H.; Fouda, A.M.; Al-Dies, A.A.M.; El-Agrody, A.M. Structural characterization and antimicrobial activities of 7H-Benzo[h]chromeno[2,3-d]pyrimidine and 14H-benzo[h]chromeno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidine Derivatives. Molecules 2016, 21, 1450. [Google Scholar] [CrossRef]
 | |||||
|---|---|---|---|---|---|
| Entry | Solvent | Base (Mol Equiv) | Temperature (°C) | Time (h) | Yield (%) b |
| 1 | EtOH | Et3N (0.2) | rt | 20 | 36 |
| 2 | EtOH | piperidine (0.2) | rt | 20 | 64 |
| 3 | EtOH | K2CO3 (0.2) | rt | 20 | 12 |
| 4 | EtOH | piperidine (2.0) | rt | 20 | 55 |
| 5 | H2O | piperidine (0.2) | rt | 20 | 34 |
| 6 | dioxane | piperidine (0.2) | rt | 20 | 16 |
| 7 | MeCN | piperidine (0.2) | rt | 20 | 50 |
| 8 | EtOH | piperidine (0.2) | 78 | 4 | 42 |
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Entry | Aldehyde | R | R1 | R2 | X | Phenol | Product | Yield (%) b |
| 1 | 1a | H | H | H | C | 3 | 4a | 74 |
| 2 | 1b | F | H | H | C | 3 | 4b | 64 |
| 3 | 1c | Cl | H | H | C | 3 | 4c | 61 |
| 4 | 1d | Br | H | H | C | 3 | 4d c | 71 |
| 5 | 1e | CN | H | H | C | 3 | 4e c | 68 |
| 6 | 1f | NO2 | H | H | C | 3 | 4f | 64 |
| 7 | 1g | Me | H | H | C | 3 | 4g | 84 |
| 8 | 1h | OMe | H | H | C | 3 | 4h | 48 |
| 9 | 1i | H | H | H | N | 3 | 4i c | 55 |
| 10 | 1j | H | F | H | C | 3 | 4j c | 57 |
| 11 | 1k | H | CN | H | C | 3 | 4k c | 70 |
| 12 | 1l | H | H | F | C | 3 | 4l c | 84 |
| 13 | 1m | H | H | Cl | C | 3 | 4m c | 51 |
| 14 | 1n | H | H | Br | C | 3 | 4n c | 68 |
| 15 | 1o | H | H | NO2 | C | 3 | 4o c | 70 |
| 16 | 1a | H | H | H | C | 5 | 6a | 57 |
| 17 | 1b | F | H | H | C | 5 | 6b c | 45 |
| 18 | 1c | Cl | H | H | C | 5 | 6c | 43 |
| 19 | 1d | Br | H | H | C | 5 | 6d c | 47 |
| 20 | 1e | CN | H | H | C | 5 | 6e c | 46 |
| 21 | 1f | NO2 | H | H | C | 5 | 6f | 45 |
| 22 | 1g | Me | H | H | C | 5 | 6g | 44 |
| 23 | 1h | OMe | H | H | C | 5 | 6h | 30 |
 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | X | R | R1 | R2 | MW | Log P | Log S | PSA | LE | LLE | LELP | H-A | H-D |
| (g/mol) | |||||||||||||
| 4a | C | H | H | H | 292.293 | 2.5 | −3.95 | 77.5 | 0.407 | 4.041 | 6.118 | 5 | 1 |
| 4b | C | F | H | H | 310.283 | 2.82 | −4.11 | 77.5 | 0.388 | 3.914 | 6.682 | 6 | 1 |
| 4c | C | Cl | H | H | 326.738 | 3.09 | −4.54 | 77.5 | 0.386 | 3.386 | 8.011 | 5 | 1 |
| 4d | C | Br | H | H | 371.189 | 3.21 | −4.86 | 77.5 | 0.383 | 3.212 | 8.391 | 5 | 1 |
| 4e | C | CN | H | H | 317.303 | 2.83 | −3.89 | 101.29 | 0.371 | 4.169 | 6.269 | 6 | 1 |
| 4f | C | NO2 | H | H | 337.29 | 1.76 | −4.01 | 123.32 | 0.355 | 6.627 | −0.437 | 8 | 1 |
| 4g | C | Me | H | H | 306.32 | 2.837 | −4.25 | 77.5 | 0.388 | 3.676 | 7.302 | 5 | 1 |
| 4h | C | OMe | H | H | 322.319 | 2.5 | −4.02 | 86.73 | 0.371 | 4.068 | 6.53 | 6 | 1 |
| 4i | N | - | H | H | 293.276 | 1.77 | −3.28 | 90.39 | 0.407 | 5.04 | 3.663 | 6 | 1 |
| 4j | C | F | H | H | 310.283 | 2.81 | −4.11 | 77.5 | 0.512 | 5.992 | 5.065 | 5 | 1 |
| 4k | C | H | CN | H | 317.303 | 2.27 | −3.89 | 101.29 | 0.371 | 4.169 | 6.269 | 6 | 1 |
| 4l | C | H | H | F | 310.283 | 2.81 | −4.11 | 77.5 | 0.388 | 3.914 | 6.682 | 5 | 1 |
| 4m | C | H | H | Cl | 326.738 | 3.03 | −4.54 | 77.5 | 0.386 | 3.386 | 8.011 | 5 | 1 |
| 4n | C | H | H | Br | 371.189 | 3.11 | −4.86 | 77.5 | 0.383 | 3.212 | 8.391 | 5 | 1 |
| 4o | C | H | H | NO2 | 337.29 | 1.77 | −4.01 | 123.32 | 0.355 | 6.627 | −0.437 | 8 | 1 |
| 7 | - | - | - | - | 421.452 | 0.461 | −1.959 | 103.2 | 0.377 | 8.061 | 1.223 | 8 | 2 |
 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | R | MW | Log P | Log S | PSA | LE | LLE | LELP | H-A | H-D |
| (g/mol) | ||||||||||
| 6a | H | 308.336 | 2.65 | −3.98 | 77.5 | 0.388 | 4.269 | 5.772 | 5 | 1 |
| 6b | F | 326.326 | 2.96 | −4.14 | 77.5 | 0.37 | 4.143 | 6.318 | 5 | 1 |
| 6c | Cl | 342.781 | 3.19 | −4.57 | 77.5 | 0.369 | 3.617 | 7.706 | 5 | 1 |
| 6d | Br | 387.232 | 3.26 | −4.89 | 77.5 | 0.366 | 3.445 | 8.095 | 5 | 1 |
| 6e | CN | 333.346 | 2.43 | −3.92 | 101.29 | 0.355 | 4.399 | 5.844 | 6 | 1 |
| 6f | NO2 | 353.101 | 1.92 | −4.04 | 123.32 | 0.34 | 6.858 | −1.194 | 8 | 1 |
| 6g | Me | 322.363 | 2.99 | −4.28 | 103.2 | 0.371 | 3.906 | 6.968 | 5 | 1 |
| 6h | OMe | 338.362 | 2.65 | −4.05 | 86.73 | 0.355 | 4.298 | 6.116 | 6 | 1 |
| 7 | - | 421.452 | 0.461 | −1.959 | 103.2 | 0.377 | 8.061 | 1.223 | 8 | 2 |
| Compound | Interacting Residues | Interactions | |
|---|---|---|---|
| Polar | Hydrophobic | ||
| CD a | Tyr118, Phe126, Ile131, Phe228, Gly303, Ile304, Gly307, His310, Thr311, Leu376, Ile379, Met508, Val509 | N…..H-O (Gly307) | halogen (Gly303) π-alkyl (Leu376) π-alkyl (Val509) |
| 8 | |||
| 4d | Tyr118, Ile131, Tyr132, Phe228, Gly303, Ile304, Gly307, His310, Thr311, Leu376, Ile379, Arg381, Met508, Val509 | N-H…..O (Tyr132) | π-alkyl (Ile131) π-alkyl (Phe228) π-alkyl (Gly307) π-alkyl (His310) π-alkyl (Leu376) π-alkyl (Met508) π-alkyl (Val509) |
| 6a | Tyr118, Thr122, Phe126, Ile131, Tyr132, Phe228, Gly303, Ile304, Gly307, Gly308, His310, Thr311, Leu376, Ile379, Val509 | C-H…..O (Thr122) N-H…..O (Gly307) N…..H-O (Thr311) | π-π T-shaped (Tyr118) π-sigma (Gly307) π-alkyl (Leu376) |
| CKE a | Tyr126, Thr130, Phe134, Ile139, Tyr140, Phe236, Gly310, Val311, Gly314, Thr318, Leu380 | C-H…..O (Tyr140) N…..C-H (Val311) | π-alkyl (Ile139) π-alkyl (Val311) |
| 8 | |||
| 4b | Leu129, Thr130, Phe134, Ile139, Tyr140, Phe236, Gly310, Val311, Gly314, Gly315, Thr318, Pro379, Leu380, Leu383 | N-H…..O (Val311) | π-π T-shaped (Phe134) π-alkyl (Leu380) |
| 6a | Leu129, Thr130, Phe134, Ile139, Tyr140, Phe236, Gly310, Val311, Gly314, Gly315, Thr318, Pro379, Leu380, Leu383 | N-H…..O (Val311) | - |
| 6d | Tyr126, Ile139, Tyr140, Phe231, Gly305, Val306, Gly309, Gly310, His312, Thr313, Leu376, Ile379, His473, Met510, Val511 | N-H…..O (Tyr140) C-H…..O (Gly309) | π-alkyl (Ile139) π-alkyl (Val306) π-sigma (Leu376) |
| CP a | Thr122, Phe126, Ile131, Tyr132, Gln142, Lys143, Phe228, Leu300, Gly303, Val304, Gly307, Gly308, Thr311 | C-H…..O (Gly303) O…..H-C (Gly304) | π-sigma (Ile131) halogen (Gly303) halogen (Val304) halogen (Gly307) |
| 8 | |||
| 4a | Leu121, Thr122, Phe126, Ile131, Tyr132, Leu139, Gln142, Lys143, Phe228, Leu300, Gly303, Val304, Met306, Gly307, Gly308, Gln309, Thr311 | N-H…..O (Val304) O…..H-C (Val304) | π-alkyl (Ile131) π-sigma (Ile131) |
| 4e | Tyr118, Ile131, Tyr132, Phe228, Leu300, Gly303, Val304, Gly307, Gly308, His310, Thr311, Leu376, His462, Met502, Val503 | N-H…..O (Tyr132) C-H…..O (Thr311) C-H…..N (Met502) | π-alkyl (Ile131) |
| Compound | Binding Energy ΔG (Kcal/mol) | Interacting Residues | Interactions | |
|---|---|---|---|---|
| Polar | Hydrophobic | |||
| 7 | −9.56 | Arg364, Arg488, Lys532, Asp533, His632, Gly717, Thr718, Ser719, Ptr723. DT10, TGP11, DG12, DC112, DA113. | C-H......O (DT10) O...H-C (TGP11) C-H....N (DG12) C-H..O (DC112) O-H..O (Asp533) | π-alkyl (TGP11). π-anion (Asp533) |
| 4a | −8.42 | Arg364, Arg488, Asp533, Ile535, Asn631, His632, Thr718. DT10, TGP11, DG12, DA113. | N......H-N (TGP11) C-H......O (Asn631) | π-anion (Asp533) π-π stacked (TGP11, DA113) |
| 4b | −9.27 | Arg364, Arg488, Lys532, Asp533, His632, Thr718, Asn722, Ptr723. DT10, TGP11, DG12, DA113. | N-H......O (DG12) O......H-C (DG12) N...H-N (Arg364) | π-alkyl (Lys532). π-anion (Asp533) π-sigma (Thr718) π-π stacked (DT10, TGP11) |
| 4c | −9.86 | Arg364, Arg488, Lys532, Asp533, His632, Thr718, Asn722, Ptr723. DT10, TGP11, DG12, DA113. | N-H......O (DG12) O......H-C (DG12) N......H-N (Arg364) | π-alkyl (Lys532). π-anion (Asp533) π-sigma (Thr718) π-π stacked (DT10, TGP11) halogen (DT10, TGP11) |
| 4d | −10.05 | Arg364, Arg488, Lys532, Asp533, His632, Thr718, Asn722, Ptr723. DT10, TGP11, DG12, DA113. | N-H......O (DG12) O......H-C (DG12) N......H-N (Arg364) | π-alkyl (Lys532). π-anion (Asp533) π-sigma (Thr718) π-π stacked (DT10, TGP11) halogen (DT10, TGP11) |
| 4g | −9.75 | Arg364, Arg488, Lys532, Asp533, His632, Thr718, Asn722, Ptr723, DT10, TGP11, DG12, DA113. | N-H......O (DG12) O......H-C (DG12) N......H-N (Arg364) | π-alkyl (Lys532, DT10, TGP11). π-anion (Asp533) π-sigma (Thr718) π-π stacked (TGP11, DA113) |
| Compound | MIC50 (µg/mL) | |||||
|---|---|---|---|---|---|---|
| CA a | CD a | CG a | CKE a | CK a | CP a | |
| 8 | 0.500 | 0.500 | 4.000 | 0.500 | 4.000 | 2.000 |
| 4a | 0.125 | 0.250 | 0.063 | 0.125 | 0.125 | 0.500 |
| 4b | 0.125 | 0.125 | 0.063 | 0.063 | 0.125 | 0.500 |
| 4c | 0.250 | 0.063 | 0.125 | 0.063 | 0.125 | 0.125 |
| 4d | 0.250 | 0.063 | 0.063 | 0.063 | 0.25 | 1.000 |
| 4e | 0.125 | 0.063 | 0.125 | 0.125 | 0.125 | 0.250 |
| 4f | 0.063 | 0.250 | 0.125 | 0.125 | 0.125 | 0.063 |
| 4g | 0.25 | 0.125 | 0.063 | 0.125 | 0.250 | 0.250 |
| 4h | 0.063 | 0.063 | 0.063 | 0.063 | 0.250 | 0.063 |
| 4i | 0.125 | 0.250 | 0.125 | 0.063 | 0.250 | 0.063 |
| 6a | 0.125 | 0.125 | 0.250 | 0.063 | 0.125 | 0.125 |
| 6b | 0.125 | 0.063 | 0.250 | 0.063 | 0.500 | 0.063 |
| 6c | 0.125 | 0.063 | 0.063 | 0.063 | 0.500 | 0.125 |
| 6d | 0.063 | 0.063 | 0.063 | 0.063 | 0.125 | 0.063 |
| 6e | 0.063 | 0.063 | 0.250 | 0.063 | 0.250 | 0.063 |
| 6f | 0.125 | 0.063 | 0.250 | 0.125 | 0.250 | 0.250 |
| 6g | 0.125 | 0.063 | 0.125 | 0.125 | 0.25 | 0.25 |
| 6h | 0.125 | 0.063 | 0.125 | 0.125 | 0.125 | 0.125 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feliciano, A.; Gómez-García, O.; Escalante, C.H.; Rodríguez-Hernández, M.A.; Vargas-Fuentes, M.; Andrade-Pavón, D.; Villa-Tanaca, L.; Álvarez-Toledano, C.; Ramírez-Apan, M.T.; Vázquez, M.A.; et al. Three-Component Synthesis of 2-Amino-3-cyano-4H-chromenes, In Silico Analysis of Their Pharmacological Profile, and In Vitro Anticancer and Antifungal Testing. Pharmaceuticals 2021, 14, 1110. https://doi.org/10.3390/ph14111110
Feliciano A, Gómez-García O, Escalante CH, Rodríguez-Hernández MA, Vargas-Fuentes M, Andrade-Pavón D, Villa-Tanaca L, Álvarez-Toledano C, Ramírez-Apan MT, Vázquez MA, et al. Three-Component Synthesis of 2-Amino-3-cyano-4H-chromenes, In Silico Analysis of Their Pharmacological Profile, and In Vitro Anticancer and Antifungal Testing. Pharmaceuticals. 2021; 14(11):1110. https://doi.org/10.3390/ph14111110
Chicago/Turabian StyleFeliciano, Alberto, Omar Gómez-García, Carlos H. Escalante, Mario A. Rodríguez-Hernández, Mariana Vargas-Fuentes, Dulce Andrade-Pavón, Lourdes Villa-Tanaca, Cecilio Álvarez-Toledano, María Teresa Ramírez-Apan, Miguel A. Vázquez, and et al. 2021. "Three-Component Synthesis of 2-Amino-3-cyano-4H-chromenes, In Silico Analysis of Their Pharmacological Profile, and In Vitro Anticancer and Antifungal Testing" Pharmaceuticals 14, no. 11: 1110. https://doi.org/10.3390/ph14111110
APA StyleFeliciano, A., Gómez-García, O., Escalante, C. H., Rodríguez-Hernández, M. A., Vargas-Fuentes, M., Andrade-Pavón, D., Villa-Tanaca, L., Álvarez-Toledano, C., Ramírez-Apan, M. T., Vázquez, M. A., Tamariz, J., & Delgado, F. (2021). Three-Component Synthesis of 2-Amino-3-cyano-4H-chromenes, In Silico Analysis of Their Pharmacological Profile, and In Vitro Anticancer and Antifungal Testing. Pharmaceuticals, 14(11), 1110. https://doi.org/10.3390/ph14111110






