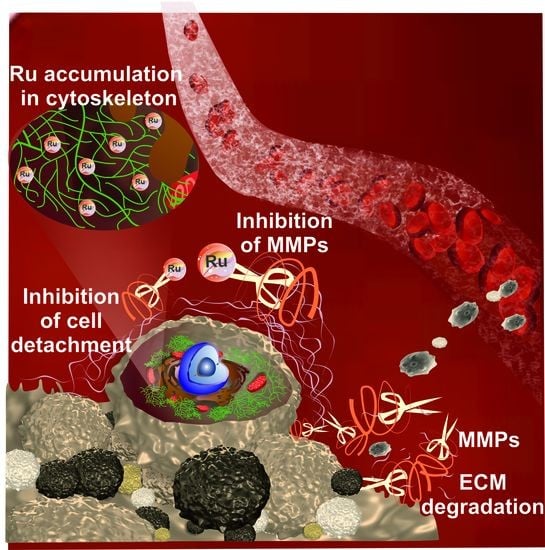
Inhibition of Matrix Metalloproteinases and Cancer Cell Detachment by Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-phenanthroline Ligands—New Candidates for Antimetastatic Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cytotoxicity and Uptake
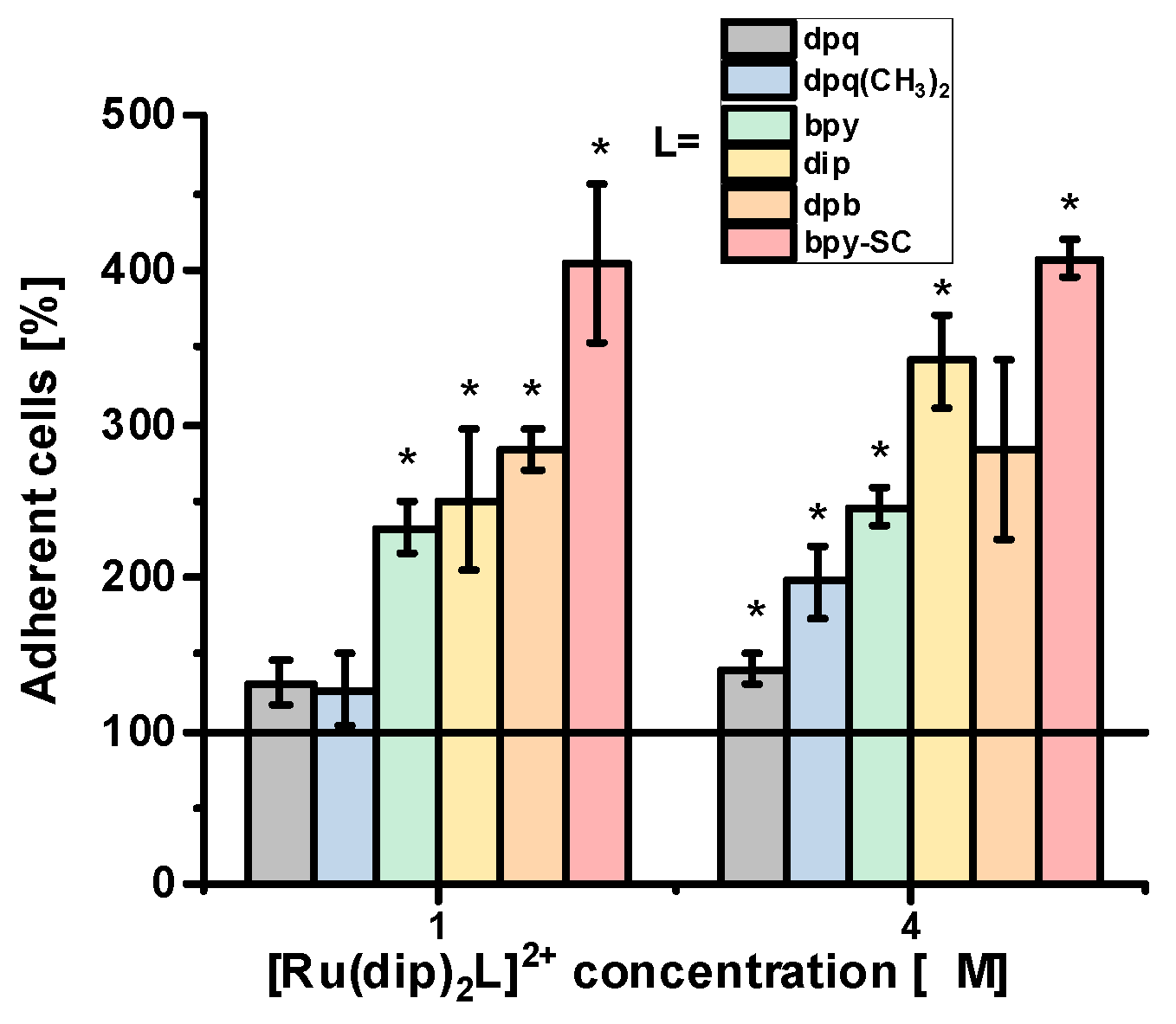
2.2. Effect on Adherence Properties of Cells
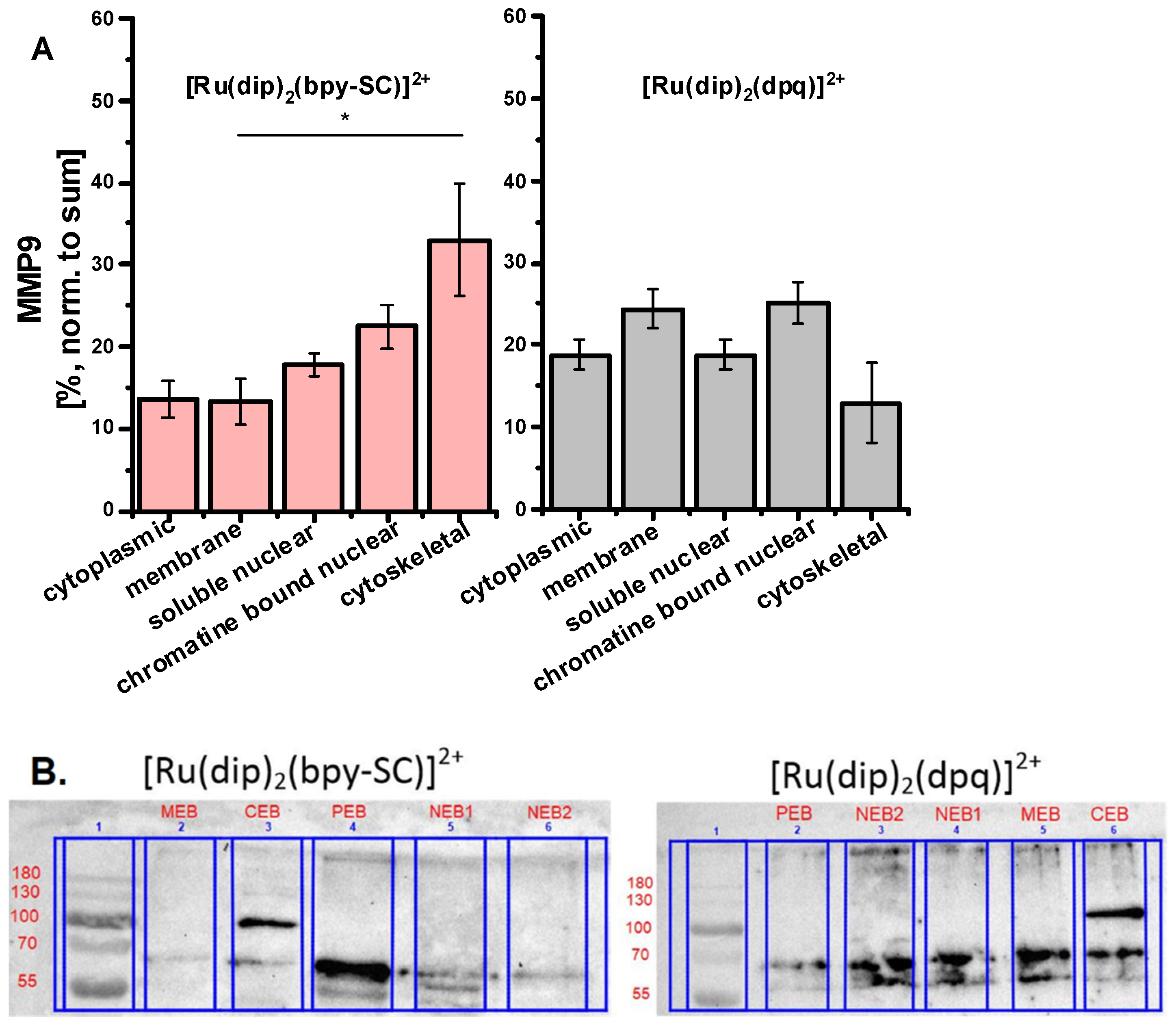
2.3. MMP-2 and MMP-9 Inhibition
3. Materials and Methods
3.1. Materials
3.2. Cell Culture and Cytotoxicity Assay
3.3. Cellular Uptake
3.4. Trypsin Resistance Assay
3.5. Subcellular Fractionation
3.6. Western Blot Analysis
3.7. MMP-2 and MMP-9 Inhibition
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitra, S.; Ganguli, S.; Chakrabarti, J. Introduction. Cancer Noncoding RNAs 2018, 1, 1–23. [Google Scholar] [CrossRef]
- Stoletov, K.; Bond, D.; Hebron, K.; Raha, S.; Zijlstra, A.; Lewis, J.D. Metastasis as a Therapeutic Target in Prostate Cancer: A Conceptual Framework. Am. J. Clin. Exp. Urol. 2014, 2, 45. [Google Scholar]
- Qian, C.-N.; Mei, Y.; Zhang, J. Cancer Metastasis: Issues and Challenges. Chin. J. Cancer 2017, 36, 38. [Google Scholar] [CrossRef] [PubMed]
- al Husaini, H.; Wheatley-Price, P.; Clemons, M.; Shepherd, F. Prevention and Management of Bone Metastases in Lung Cancer: A Review. J. Thorac. Oncol. 2009, 4, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Targeting Metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef]
- Verma, R.P.; Hansch, C. Matrix Metalloproteinases (MMPs): Chemical-Biological Functions and (Q)SARs. Bioorg. Med. Chem. 2007, 15, 2223–2268. [Google Scholar] [CrossRef]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lai, H.; Xiong, Z.; Chen, B.; Chen, T. Functionalization and Cancer-Targeting Design of Ruthenium Complexes for Precise Cancer Therapy. Chem. Commun. 2019, 55, 9904–9914. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Jin, S.; Muhammad, N.; Guo, Z. Stimuli-Responsive Therapeutic Metallodrugs. Chem. Rev. 2019, 119, 1138–1192. [Google Scholar] [CrossRef] [PubMed]
- Poynton, F.E.; Bright, S.A.; Blasco, S.; Williams, D.C.; Kelly, J.M.; Gunnlaugsson, T. The Development of Ruthenium(Ii) Polypyridyl Complexes and Conjugates for in Vitro Cellular and in Vivo Applications. Chem. Soc. Rev. 2017, 46, 7706–7756. [Google Scholar] [CrossRef] [PubMed]
- Mazuryk, O.; Maciuszek, M.; Stochel, G.; Suzenet, F.; Brindell, M. 2-Nitroimidazole-Ruthenium Polypyridyl Complex as a New Conjugate for Cancer Treatment and Visualization. J. Inorg. Biochem. 2014, 134, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zheng, W.; Chen, T. Ruthenium Polypyridyl Complex Inhibits Growth and Metastasis of Breast Cancer Cells by Suppressing FAK Signaling with Enhancement of TRAIL-Induced Apoptosis. Sci. Rep. 2015, 5, 9157. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Yu, G.; Zhang, S.; Li, Y.; Wu, Q.; Huang, X.; Mei, W. Nucleus-Enriched Ruthenium Polypyridine Complex Acts as a Potent Inhibitor to Suppress Triple-Negative Breast Cancer Metastasis In Vivo. Comput. Struct. Biotechnol. J. 2019, 17, 21–30. [Google Scholar] [CrossRef]
- Wang, J.Q.; Kou, J.F.; Zhao, Z.Z.; Qiu, K.Q.; Chao, H. Anthraquinone-Bridged Diruthenium(II) Complexes Inhibit Migration and Invasion of Human Hepatocarcinoma MHCC97-H Cells. Inorg. Chem. Front. 2017, 4, 1003–1012. [Google Scholar] [CrossRef]
- Wan, D.; Lai, S.H.; Zeng, C.C.; Zhang, C.; Tang, B.; Liu, Y.J. Ruthenium(II) Polypyridyl Complexes: Synthesis, Characterization and Anticancer Activity Studies on BEL-7402 Cells. J. Inorg. Biochem. 2017, 173, 1–11. [Google Scholar] [CrossRef]
- Jiang, G.B.; Zhang, W.Y.; He, M.; Gu, Y.Y.; Bai, L.; Wang, Y.J.; Yi, Q.Y.; Du, F. Design and Synthesis of New Ruthenium Polypyridyl Complexes with Potent Antitumor Activity in Vitro. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 220, 117132. [Google Scholar] [CrossRef]
- Gurgul, I.; Mazuryk, O.; Łomzik, M.; Gros, P.C.; Rutkowska-Zbik, D.; Brindell, M. Unexplored Features of Ru(Ii) Polypyridyl Complexes-towards Combined Cytotoxic and Antimetastatic Activity. Metallomics 2020, 12, 784–793. [Google Scholar] [CrossRef]
- Bergamo, A.; Sava, G. Linking the Future of Anticancer Metal-Complexes to the Therapy of Tumour Metastases. Chem. Soc. Rev. 2015, 44, 8818–8835. [Google Scholar] [CrossRef]
- Alessio, E. Thirty Years of the Drug Candidate NAMI-A and the Myths in the Field of Ruthenium Anticancer Compounds: A Personal Perspective. Eur. J. Inorg. Chem. 2017, 2017, 1549–1560. [Google Scholar] [CrossRef]
- Mazuryk, O.; Suzenet, F.; Kieda, C.; Brindell, M. The Biological Effect of the Nitroimidazole Derivative of a Polypyridyl Ruthenium Complex on Cancer and Endothelial Cells. Metallomics 2015, 7, 553–566. [Google Scholar] [CrossRef]
- Gava, B.; Zorzet, S.; Spessotto, P.; Cocchietto, M.; Sava, G. Inhibition of B16 Melanoma Metastases with the Ruthenium Complex Imidazolium Trans-Imidazoledimethylsulfoxide-Tetrachlororuthenate and Down-Regulation of Tumor Cell Invasion. J. Pharmacol. Exp. Ther. 2006, 317, 284–291. [Google Scholar] [CrossRef]
- Łomzik, M.; Mazuryk, O.; Rutkowska-Zbik, D.; Stochel, G.; Gros, P.C.; Brindell, M. New Ruthenium Compounds Bearing Semicarbazone 2-Formylopyridine Moiety: Playing with Auxiliary Ligands for Tuning the Mechanism of Biological Activity. J. Inorg. Biochem. 2017, 175, 80–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickerson, M.; Sun, Y.; Howerton, B.; Glazer, E. Modifying Charge and Hydrophilicity of Simple Ru(II) Polypyridyl Complexes Radically Alters Biological Activities: Old Complexes, Surprising New Tricks. Inorg. Chem. 2014, 53, 10370–10377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, G.; Meredith, D.; ap Gwynn, I.; Richards, R. Focal Adhesion Quantification—A New Assay of Material Biocompatibility? Review. Eur. Cells Mater. 2005, 9, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Allen, J.; Mason, R.; Kamimura, T.; Zhang, S. TGF-Beta1 Induces Human Alveolar Epithelial to Mesenchymal Cell Transition (EMT). Respir. Res. 2005, 6, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araya, J.; Maruyama, M.; Sassa, K.; Fujita, T.; Hayashi, R.; Matsui, S.; Kashii, T.; Yamashita, N.; Sugiyama, E.; Kobayashi, M. Ionizing Radiation Enhances Matrix Metalloproteinase-2 Production in Human Lung Epithelial Cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 280, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Malara, A.; Ligi, D.; di Buduo, C.A.; Mannello, F.; Balduini, A. Sub-Cellular Localization of Metalloproteinases in Megakaryocytes. Cells 2018, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Mazuryk, O.; Janczy-Cempa, E.; Kuśnierz, J.; Rutkowska-Zbik, D.; Machnicka, A.; Stochel, G.; Brindell, M. Relevance of Electron Transfer Pathway in Photodynamic Activity of Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-Phenantroline Ligands under Normoxic and Hypoxic Conditions. Dalton Transactions (Under review). The purity and identity were confirmed by HPLC and HRMS. For [Ru(dip)2(dpq)]Cl2: Yield 50%, HRMS for [Ru(dip)2(dpq)]2+ calculated m/z = 524.930, found m/z = 525.139. For [Ru(dip)2(dpq(CH3)2)]Cl2: Yield 77%, HRMS for [Ru(dip)2(dpq(CH3)2)]2+ calculated m/z = 539.005, found m/z = 539.153. For [Ru(dip)2(dpb)]Cl2: Yield 52%, HRMS for [Ru(dip)2(dpb)]2+ calculated m/z = 549.950, found m/z = 550.147.
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: New York, NY, USA, 2006. [Google Scholar]
- Lin, Y.; Ukaji, T.; Koide, N.; Umezawa, K. Inhibition of Late and Early Phases of Cancer Metastasis by the NF-ΚB Inhibitor DHMEQ Derived from Microbial Bioactive Metabolite Epoxyquinomicin: A Review. Int. J. Mol. Sci. 2018, 19, 729. [Google Scholar] [CrossRef] [Green Version]
- Gandalovičová, A.; Rosel, D.; Fernandes, M.; Veselý, P.; Heneberg, P.; Čermák, V.; Petruželka, L.; Kumar, S.; Sanz-Moreno, V.; Brábek, J. Migrastatics—Anti-Metastatic and Anti-Invasion Drugs: Promises and Challenges. Trends Cancer 2017, 3, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]

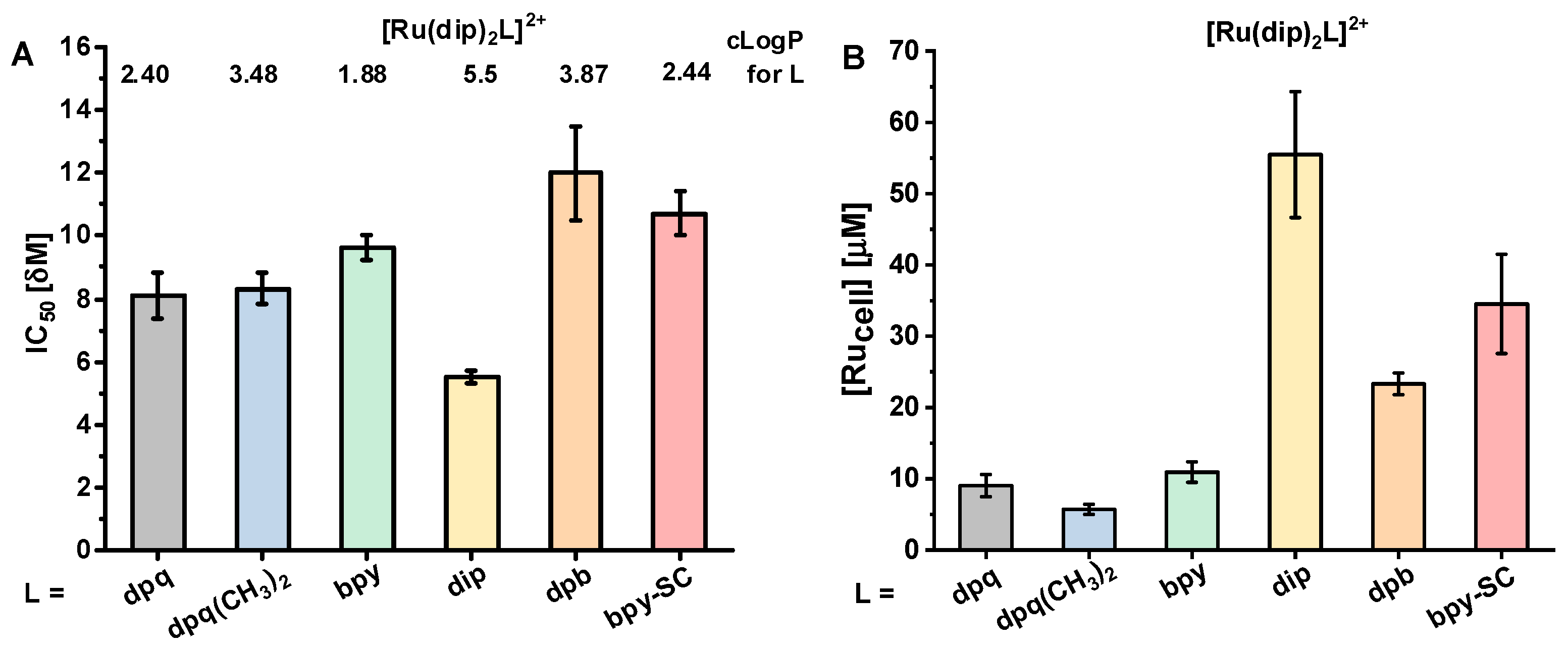

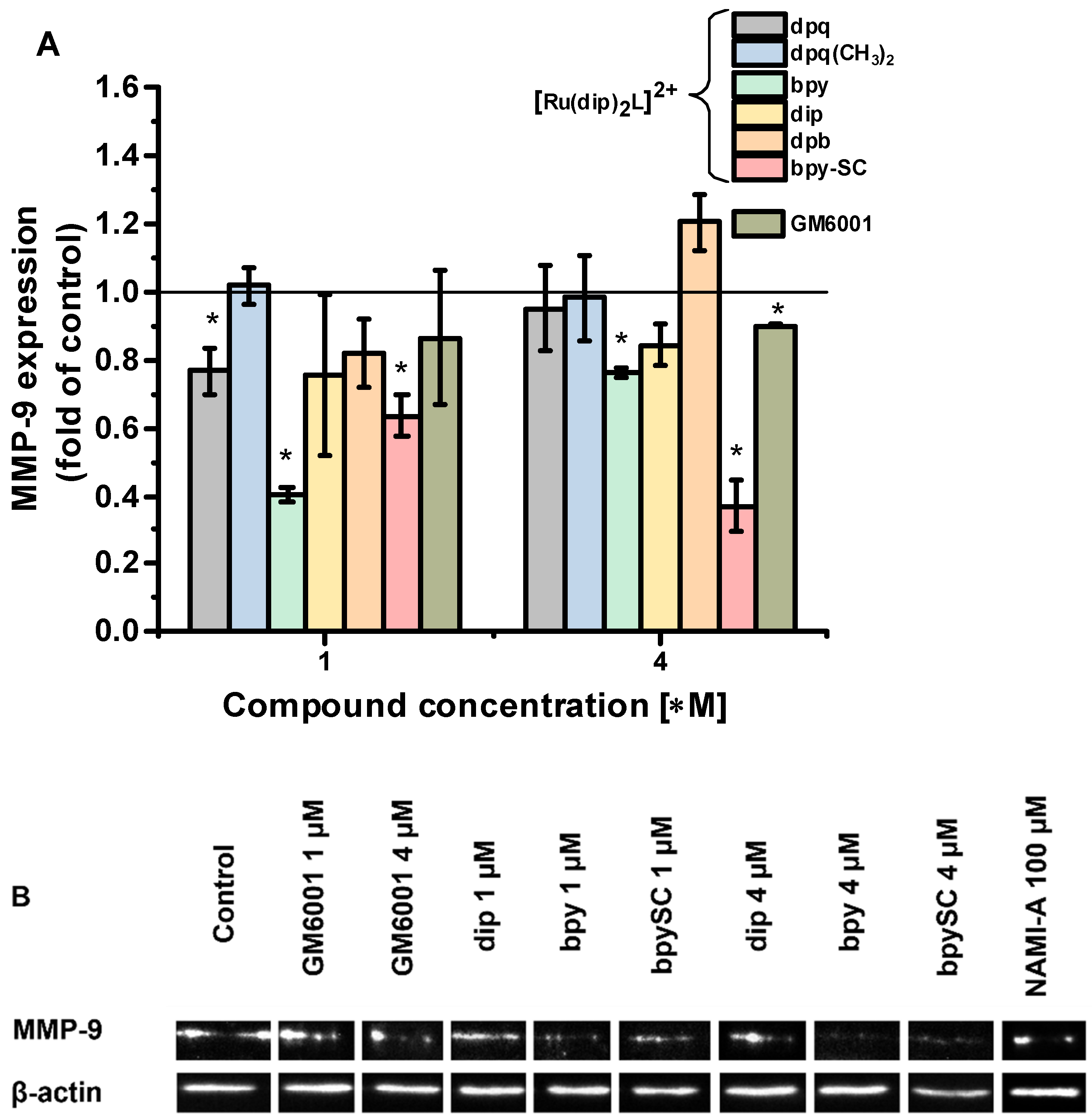
| Compound | IC50 for MMP-2 [µM] | IC50 for MMP-9 [µM] |
|---|---|---|
| [Ru(dip)2(dpq)]2+ | n.i. | n.i. |
| [Ru(dip)2(dpq(CH3)2]2+ | n.i. | 16.8 ± 9.4 |
| [Ru(dip)2(bpy)]2+ | 22.5 ± 2.0 | 6.2 ± 2.8 |
| [Ru(dip)3]2+ | 8.3 ± 1.1 | 4.1 ± 1.6 |
| [Ru(dip)2(dpb)]2+ | 13.1 ± 4.6 | 9.0 ± 3.7 |
| [Ru(dip)2(bpy-SC)]2+ | 8.0 ± 1.1 | 5.9 ± 1.0 |
| GM6001 | 34 1 | 0.6 1 |
| NAMI-A | n.i. | n.i. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajda-Morszewski, P.; Gurgul, I.; Janczy-Cempa, E.; Mazuryk, O.; Łomzik, M.; Brindell, M. Inhibition of Matrix Metalloproteinases and Cancer Cell Detachment by Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-phenanthroline Ligands—New Candidates for Antimetastatic Agents. Pharmaceuticals 2021, 14, 1014. https://doi.org/10.3390/ph14101014
Gajda-Morszewski P, Gurgul I, Janczy-Cempa E, Mazuryk O, Łomzik M, Brindell M. Inhibition of Matrix Metalloproteinases and Cancer Cell Detachment by Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-phenanthroline Ligands—New Candidates for Antimetastatic Agents. Pharmaceuticals. 2021; 14(10):1014. https://doi.org/10.3390/ph14101014
Chicago/Turabian StyleGajda-Morszewski, Przemysław, Ilona Gurgul, Ewelina Janczy-Cempa, Olga Mazuryk, Michał Łomzik, and Małgorzata Brindell. 2021. "Inhibition of Matrix Metalloproteinases and Cancer Cell Detachment by Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-phenanthroline Ligands—New Candidates for Antimetastatic Agents" Pharmaceuticals 14, no. 10: 1014. https://doi.org/10.3390/ph14101014
APA StyleGajda-Morszewski, P., Gurgul, I., Janczy-Cempa, E., Mazuryk, O., Łomzik, M., & Brindell, M. (2021). Inhibition of Matrix Metalloproteinases and Cancer Cell Detachment by Ru(II) Polypyridyl Complexes Containing 4,7-Diphenyl-1,10-phenanthroline Ligands—New Candidates for Antimetastatic Agents. Pharmaceuticals, 14(10), 1014. https://doi.org/10.3390/ph14101014