Analysis of Infection Time Courses Shows CII Levels Determine the Frequency of Lysogeny in Phage 186
Abstract
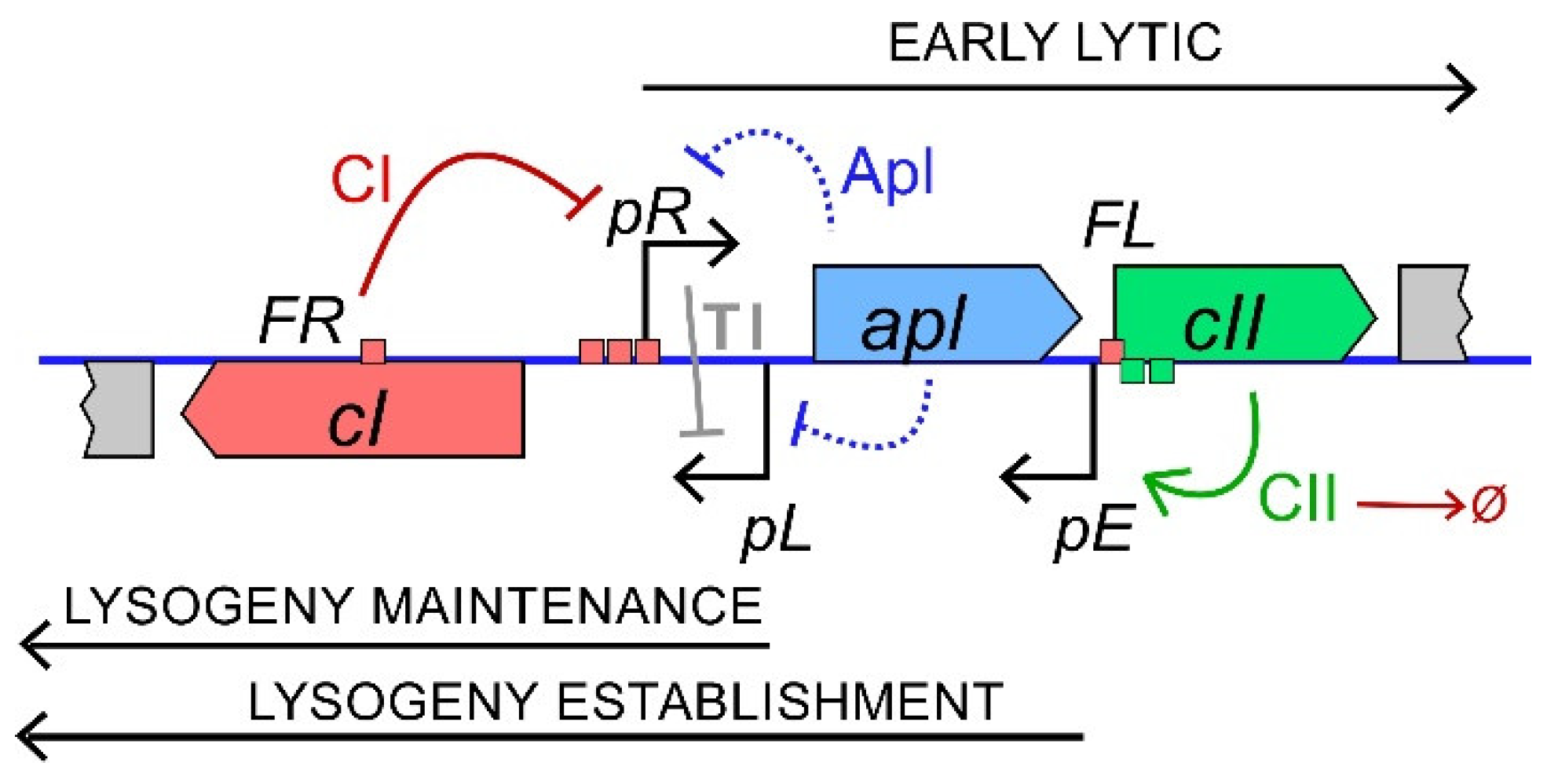
1. Introduction
2. Results
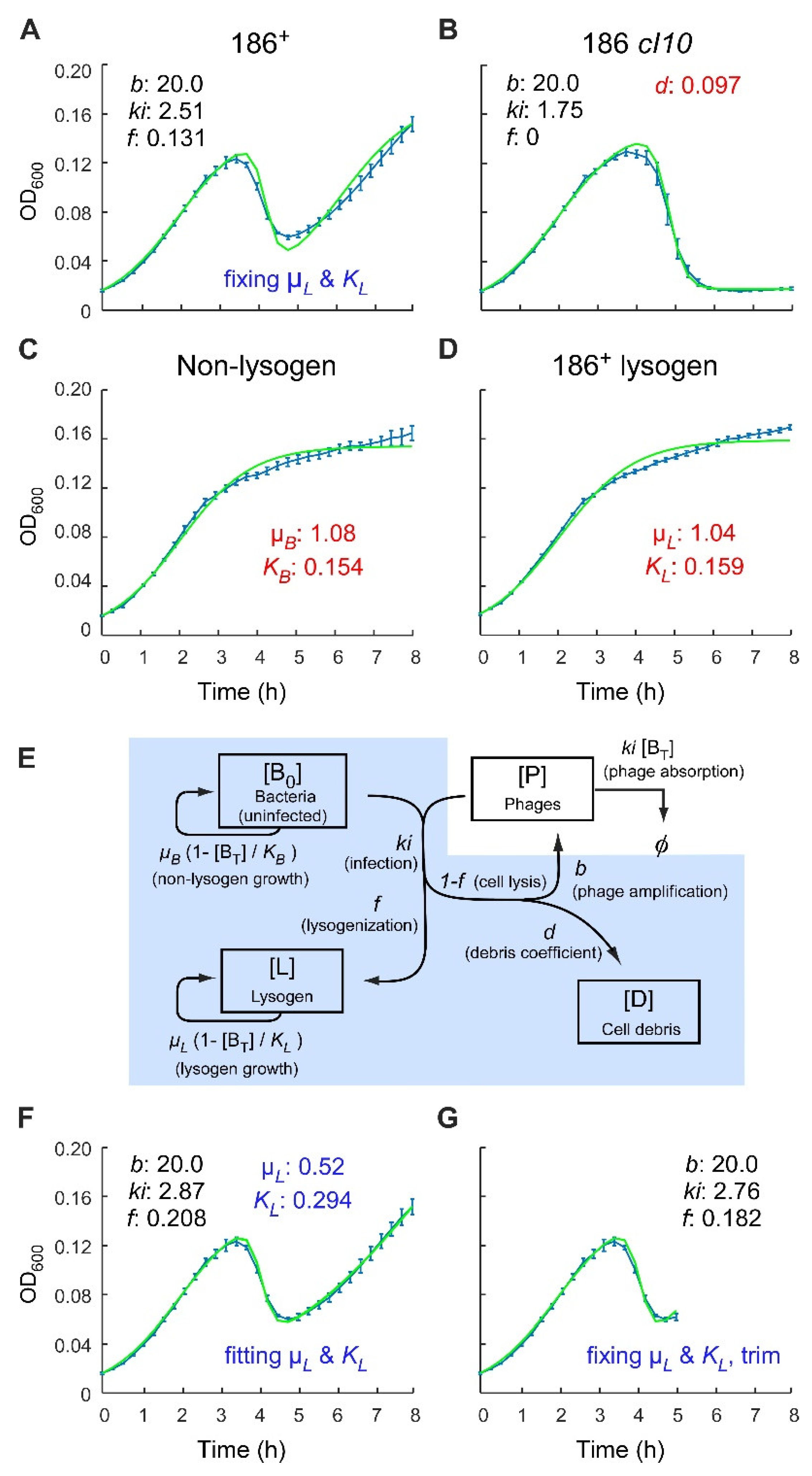
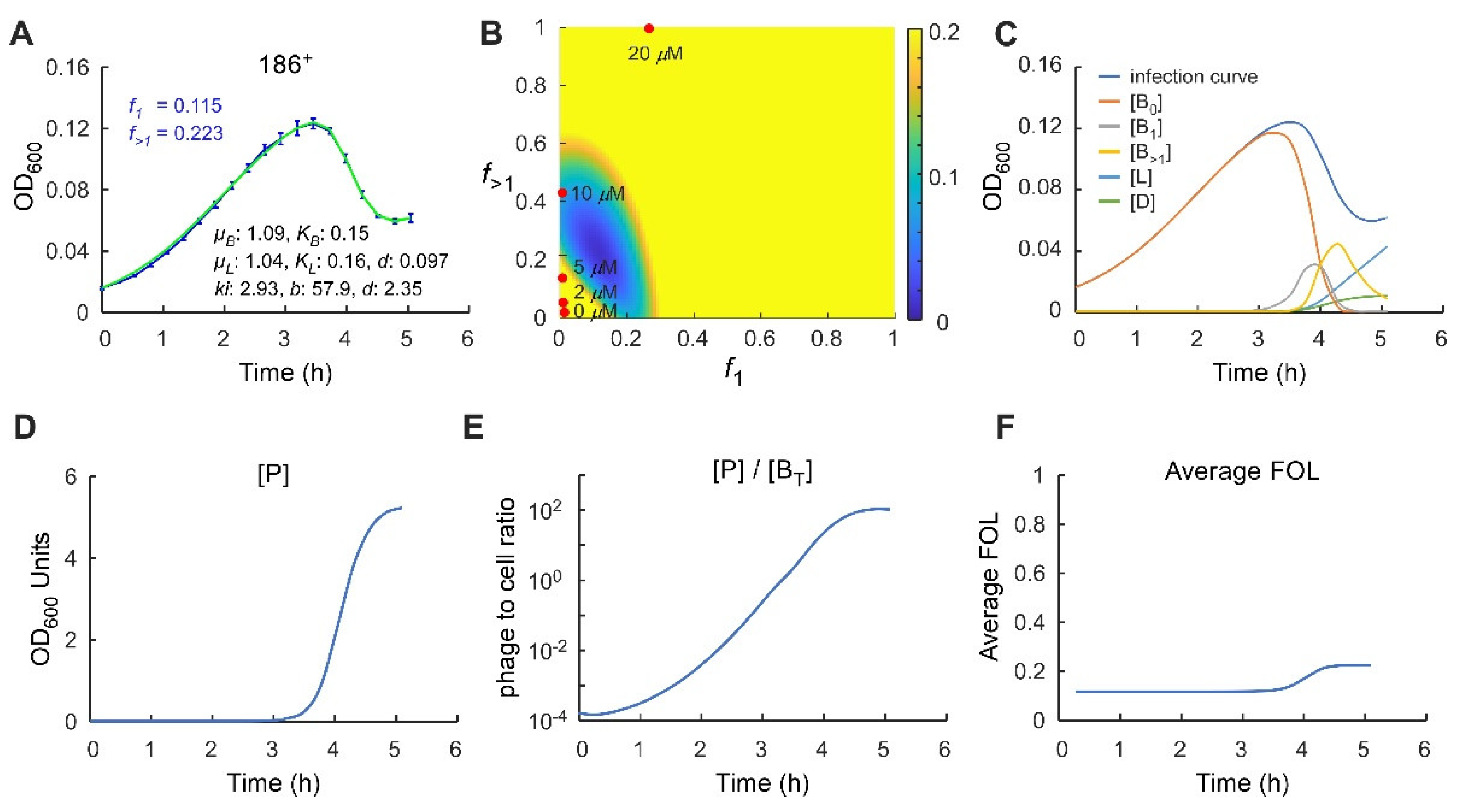
2.1. Analysis of Phage 186 Infection Time Courses with a Simple Lysogenisation Model
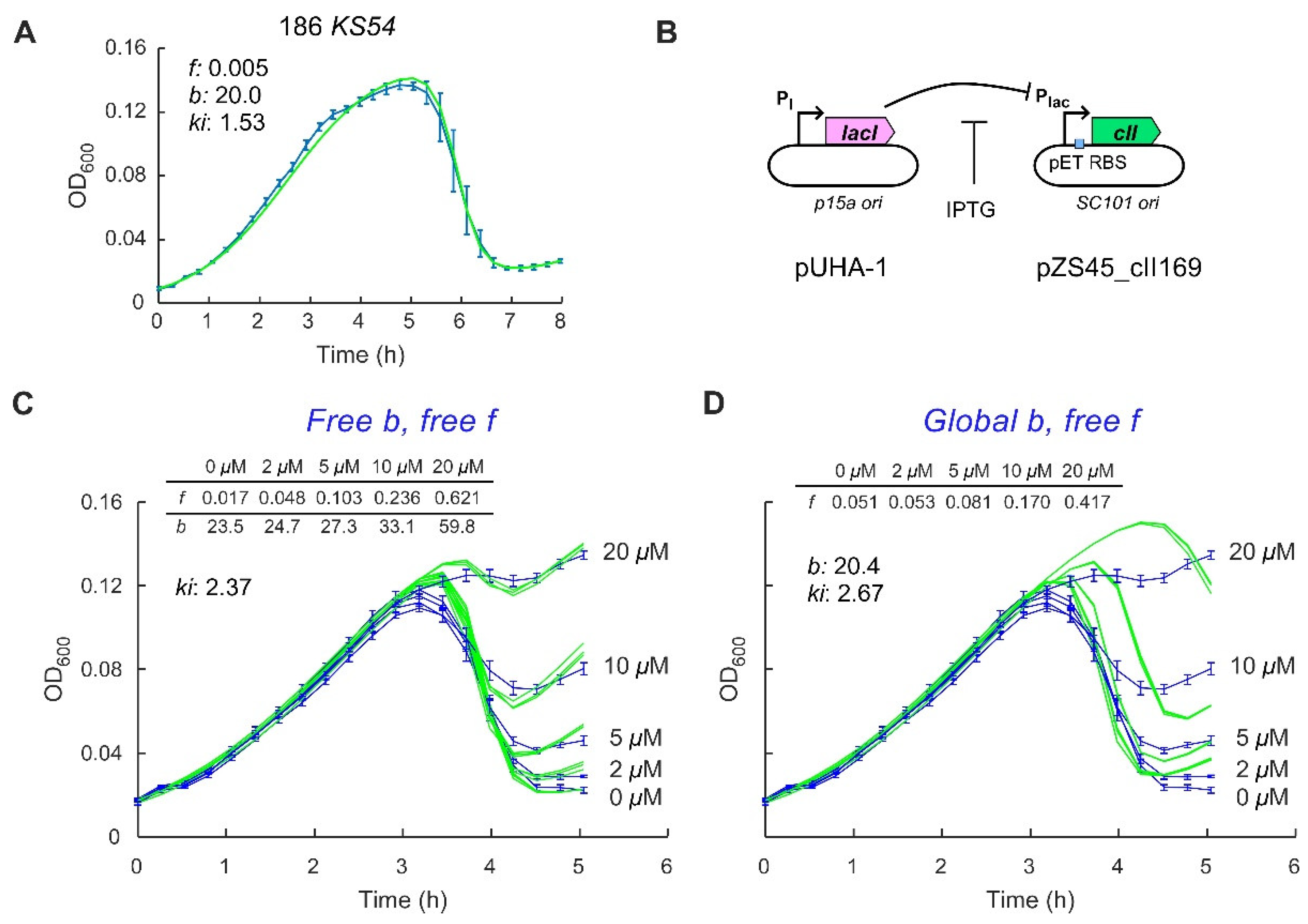
2.2. Control of the Frequency of Lysogeny by 186 CII
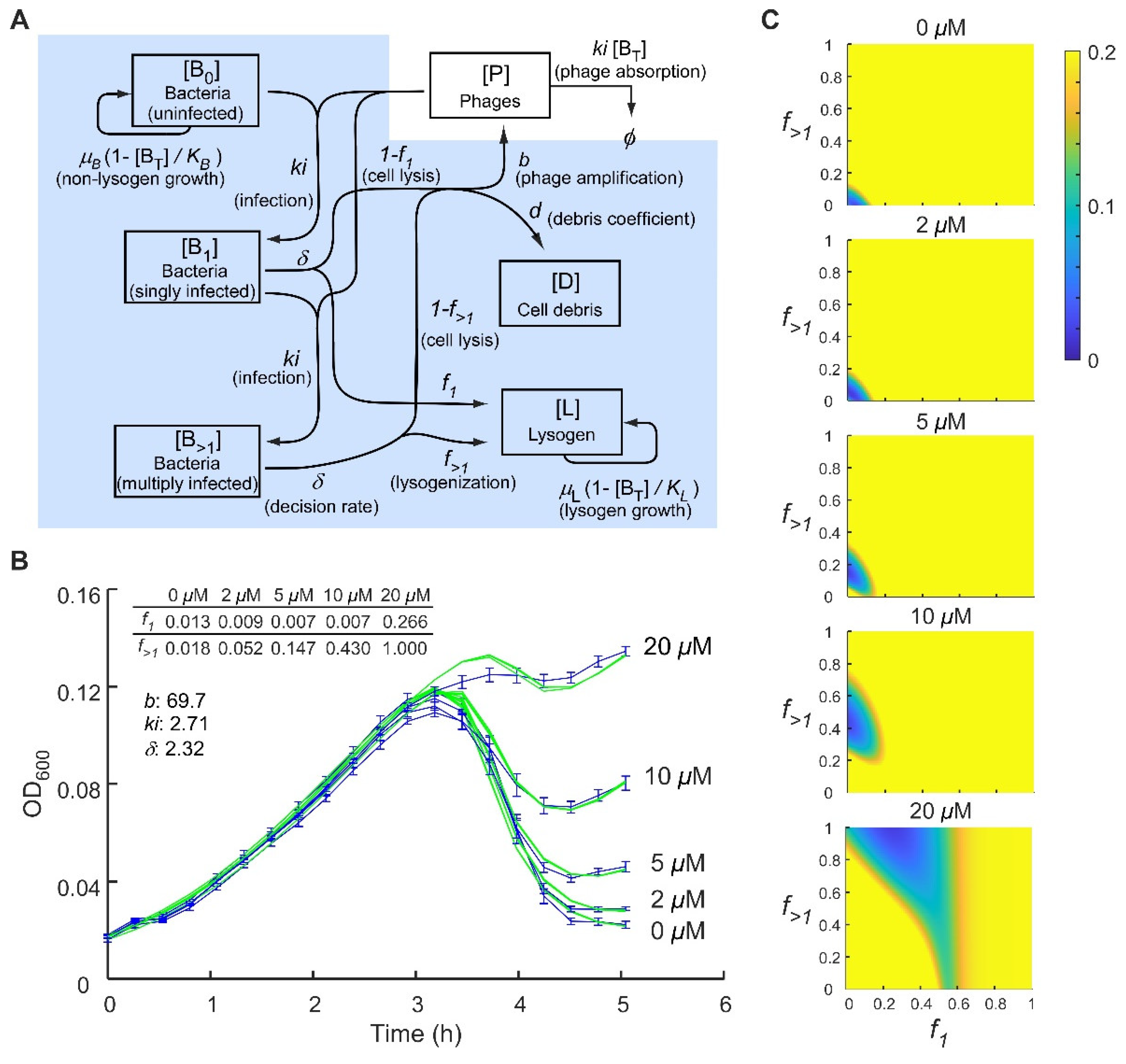
2.3. Application of a Multiple-Infection Lysogenisation Model to the CII Expression Time-Courses
2.4. Application of the Multiple-Infection Model to the Phage 186 wt Time-Course
3. Discussion
3.1. Control of the Phage 186 Lysis-Lysogeny Decision by CII
3.2. Response to the Multiplicity of Infection
4. Materials and Methods
4.1. Strains
4.2. Phage
4.3. Plasmids
4.4. Phage Titring and MOA Calculation
4.5. Microtitre Plate-Based Phage Infection Assay
4.6. Fitting Procedure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2004; Volume 3, ISBN 0879697172. [Google Scholar]
- Dodd, I.B.; Shearwin, K.E.; Perkins, A.J.; Burr, T.; Hochschild, A.; Egan, J.B. Cooperativity in long-range gene regulation by the λ CI repressor. Genes Dev. 2004, 18, 344–354. [Google Scholar] [CrossRef]
- Golding, I. Decision Making in Living Cells: Lessons from a Simple System. Annu. Rev. Biophys. 2011, 40, 63–80. [Google Scholar] [CrossRef]
- Golding, I. Single-Cell Studies of Phage λ: Hidden Treasures Under Occam’s Rug. Annu. Rev. Virol. 2016, 3, 453–472. [Google Scholar] [CrossRef]
- Golding, I. Infection by bacteriophage lambda: An evolving paradigm for cellular individuality. Curr. Opin. Microbiol. 2018, 43, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in Bacteriophage Lambda Development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, M.A.; Knight, D.M.; Das, A.; Miller, H.I.; Echols, H. Control of phage lambda development by stability and synthesis of cII protein: Role of the viral cIII and host hflA, himA and himD genes. Cell 1982, 31, 565–573. [Google Scholar] [CrossRef]
- Arkin, A.; Ross, J.; McAdams, H.H. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 1998, 149, 1633–1648. [Google Scholar] [CrossRef]
- Kobiler, O.; Rokney, A.; Friedman, N.; Court, D.L.; Stavans, J.; Oppenheim, A.B. Quantitative kinetic analysis of the bacteriophage λ genetic network. Proc. Natl. Acad. Sci. USA 2005, 102, 4470–4475. [Google Scholar] [CrossRef]
- Shotland, Y.; Shifrin, A.; Ziv, T.; Teff, D.; Koby, S.; Kobiler, O.; Oppenheim, A.B. Proteolysis of bacteriophage λ CII by Escherichia coli FtsH (HflB). J. Bacteriol. 2000, 182, 3111–3116. [Google Scholar] [CrossRef]
- Kihara, A.; Akiyama, Y.; Ito, K. Host regulation of lysogenic decision in bacteriophage λ: Transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc. Natl. Acad. Sci. USA 1997, 94, 5544–5549. [Google Scholar] [CrossRef]
- Kourilsky, P. Lysogenization by bacteriophage lambda—I. Multiple infection and the lysogenic response. MGG Mol. Gen. Genet. 1973, 122, 183–195. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, F.; Endy, D. Determination of cell fate selection during phage lambda infection. Proc. Natl. Acad. Sci. USA 2008, 105, 20705–20710. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Trinh, J.T.; Zeng, L. High-resolution studies of lysis—lysogeny decision-making in bacteriophage lambda. J. Biol. Chem. 2019, 294, 3343–3349. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.B.; Egan, J.B. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol. Microbiol. 2002, 45, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Pinkett, H.W.; Shearwin, K.E.; Stayrook, S.; Dodd, I.B.; Burr, T.; Hochschild, A.; Egan, J.B.; Lewis, M. The structural basis of cooperative regulation at an alternate genetic switch. Mol. Cell 2006, 21, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Murchland, I.M.; Ahlgren-Berg, A.; Pietsch, J.M.J.; Isabel, A.; Dodd, I.B.; Shearwin, K.E. Instability of CII is needed for efficient switching between lytic and lysogenic development in bacteriophage 186. Nucleic Acids Res. 2021, 48, 12030–12041. [Google Scholar] [CrossRef]
- Shearwin, K.E.; Egan, J.B. Establishment of lysogeny in bacteriophage 186. DNA binding and transcriptional activation by the CII protein. J. Biol. Chem. 2000, 275, 29113–29122. [Google Scholar] [CrossRef]
- Dibbens, J.A.; Gregory, S.L.; Egan, J.B. Control of gene expression in the temperate coliphage 186. X. The cl repressor directly represses transcription of the late control gene B. Mol. Microbiol. 1992, 6, 2643–2650. [Google Scholar] [CrossRef]
- Maynard, N.D.; Birch, E.W.; Sanghvi, J.C.; Chen, L.; Gutschow, M.V. A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 2010, 6, 1001017. [Google Scholar] [CrossRef]
- Lamont, I.; Richardson, H.; Carter, D.R.; Egan, J.B. Genes for the establishment and maintenance of lysogeny by the temperate coliphage 186. J. Bacteriol. 1993, 175, 5286–5288. [Google Scholar] [CrossRef][Green Version]
- Pearl, S.; Gabay, C.; Kishony, R.; Oppenheim, A.; Balaban, N.Q. Nongenetic individuality in the host-phage interaction. PLoS Biol. 2008, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Murchland, I.; Ahlgren-Berg, A.; Priest, D.G.; Dodd, I.B.; Shearwin, K.E. Promoter activation by CII, a potent transcriptional activator from bacteriophage 186. J. Biol. Chem. 2014, 289, 32094–32108. [Google Scholar] [CrossRef]
- Banuett, F.; Hoyt, M.A.; McFarlane, L.; Echols, H.; Herskowitz, I. hflB, a new Escherichia coli locus regulating lysogeny and the level of bacteriophage lambda cII protein. J. Mol. Biol. 1986, 187, 213–224. [Google Scholar] [CrossRef]
- Herman, C.; Ogura, T.; Tomoyasu, T.; Hiraga, S.; Akiyama, Y.; Ito, K.; Thomas, R.; D’Ari, R.; Bouloc, P. Cell growth and lambda phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc. Natl. Acad. Sci. USA 1993, 90, 10861–10865. [Google Scholar] [CrossRef]
- Brumby, A.M.; Lamont, I.; Dodd, I.B.; Egan, J.B. Defining the SOS operon of coliphage 186. Virology 1996, 219, 105–114. [Google Scholar] [CrossRef]
- Sinha, V.; Goyal, A.; Svenningsen, S.L.; Semsey, S.; Krishna, S. In silico evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages. Front. Microbiol. 2017, 8, 1386. [Google Scholar] [CrossRef]
- Lamont, I.; Brumby, A.M.; Egan, J.B. UV induction of coliphage 186: Prophage induction as an SOS function. Proc. Nat. Acad. Sci. USA 1989, 86, 5492–5496. [Google Scholar] [CrossRef]
- Reed, M.R.; Shearwin, K.E.; Pell, L.M.; Egan, J.B. The dual role of Apl in prophage induction of coliphage 186. Mol. Microbiol. 1997, 23, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.B.; Reed, M.R.; Egan, J.B. The Cro-like Apl repressor of coliphage 186 is required for prophage excision and binds near the phage attachment site. Mol. Microbiol. 1993, 10, 1139–1150. [Google Scholar] [CrossRef]
- Shearwin, K.E.; Brumby, A.M.; Egan, J.B. The tum protein of coliphage 186 is an antirepressor. J. Biol. Chem. 1998, 273, 5708–5715. [Google Scholar] [CrossRef] [PubMed]
- Avlund, M.; Krishna, S.; Semsey, S.; Dodd, I.B.; Sneppen, K. Minimal gene regulatory circuits for a lysis-lysogeny choice in the presence of noise. PLoS ONE 2010, 5, e15037. [Google Scholar] [CrossRef] [PubMed]
- Stokar-Avihail, A.; Tal, N.; Erez, Z.; Lopatina, A.; Sorek, R. Widespread Utilization of Peptide Communication in Phages Infecting Soil and Pathogenic Bacteria. Cell Host Microbe 2019, 25, 746–755.e5. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Skinner, S.O.; Zong, C.; Sippy, J.; Feiss, M.; Golding, I. Decision Making at a Subcellular Level Determines the Outcome of Bacteriophage Infection. Cell 2010, 141, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis–lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, N.; Agnew, D.; Krishna, S.; Dodd, I.B.; Shearwin, K.E. Analysis of Infection Time Courses Shows CII Levels Determine the Frequency of Lysogeny in Phage 186. Pharmaceuticals 2021, 14, 998. https://doi.org/10.3390/ph14100998
Hao N, Agnew D, Krishna S, Dodd IB, Shearwin KE. Analysis of Infection Time Courses Shows CII Levels Determine the Frequency of Lysogeny in Phage 186. Pharmaceuticals. 2021; 14(10):998. https://doi.org/10.3390/ph14100998
Chicago/Turabian StyleHao, Nan, Dylan Agnew, Sandeep Krishna, Ian B. Dodd, and Keith E. Shearwin. 2021. "Analysis of Infection Time Courses Shows CII Levels Determine the Frequency of Lysogeny in Phage 186" Pharmaceuticals 14, no. 10: 998. https://doi.org/10.3390/ph14100998
APA StyleHao, N., Agnew, D., Krishna, S., Dodd, I. B., & Shearwin, K. E. (2021). Analysis of Infection Time Courses Shows CII Levels Determine the Frequency of Lysogeny in Phage 186. Pharmaceuticals, 14(10), 998. https://doi.org/10.3390/ph14100998







