Ginkgolic Acid Inhibits Coronavirus Strain 229E Infection of Human Epithelial Lung Cells
Abstract
:1. Introduction
2. Results
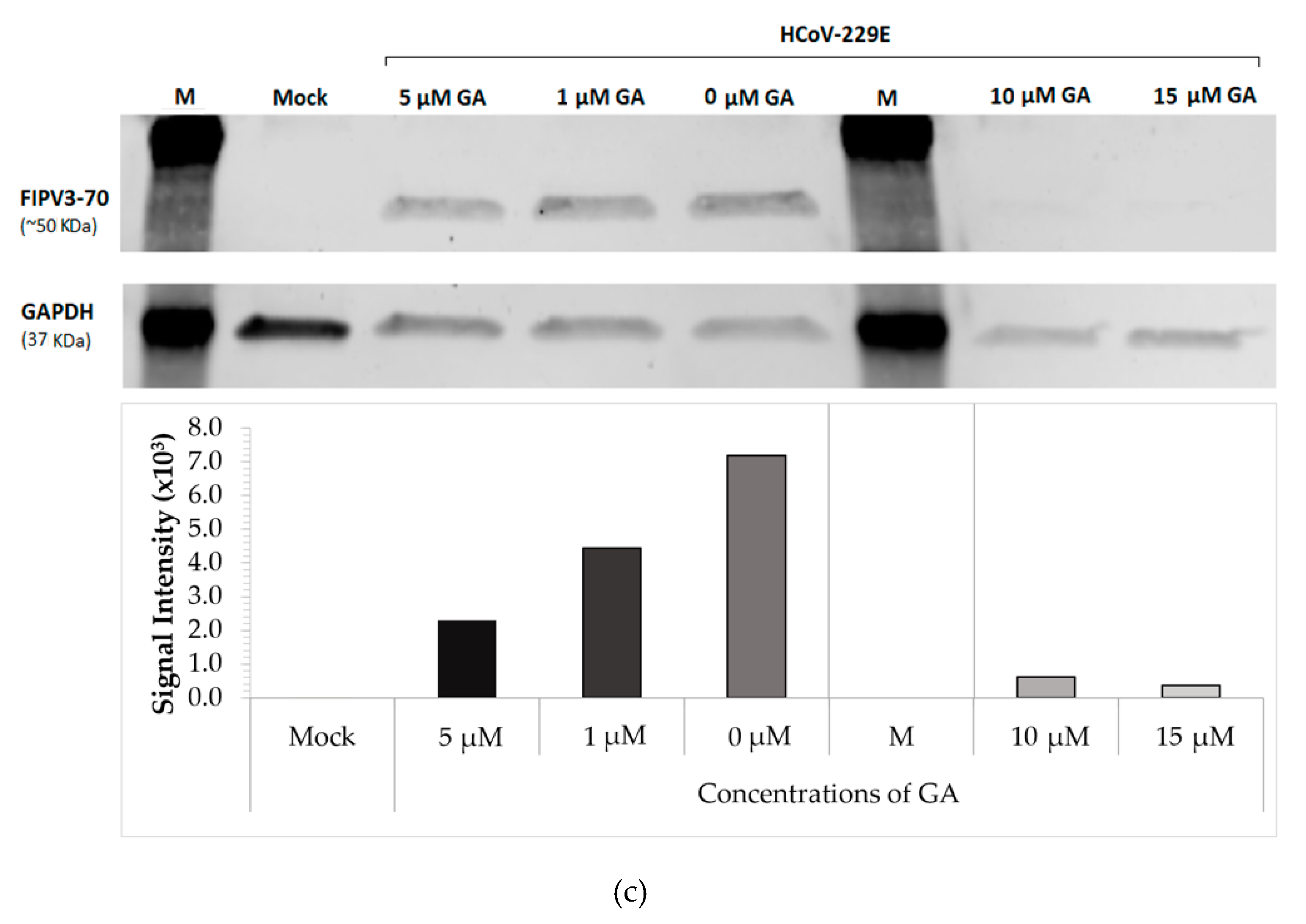
2.1. GA Demonstrates Antiviral Activity against HCoV-229E in a Dose-Dependent Manner
2.2. Administration of GA Protects MRC-5 Cells Post-Infection
3. Discussion
4. Materials and Methods
4.1. Cell Line, Virus, and Compound
4.2. Treatments of HCoV-229E Infected MRC-5 Cells with Ginkgolic Acid
4.3. qRT-PCR
4.4. Western Blot
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bakowski, M.A.; Beutler, N.; Wolff, K.C.; Kirkpatrick, M.G.; Chen, E.; Nguyen, T.-T.H.; Riva, L.; Shaabani, N.; Parren, M.; Ricketts, J.; et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021, 12, 3309. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encycl. Virol. 2021, 428–440. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johns Hopkins Coronavirus Resource Center (JHCRC). COVID-19 Map—Johns Hopkins Coronavirus Resource Center. 2021. Available online: https://coronavirus.jhu.edu/map.html (accessed on 18 August 2021).
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021. [Google Scholar] [CrossRef]
- Montazeri Aliabadi, H.; Totonchy, J.; Mahdipoor, P.; Parang, K.; Uludağ, H. Suppression of Human Coronavirus 229E Infection in Lung Fibroblast Cells via RNA Interference. Front. Nanotechnol. 2021, 3, 34. [Google Scholar] [CrossRef]
- Chassagne, F.; Huang, X.; Lyles, J.T.; Quave, C.L. Validation of a 16th Century Traditional Chinese Medicine Use of Ginkgo biloba as a Topical Antimicrobial. Front. Microbiol. 2019, 10, 775. [Google Scholar] [CrossRef]
- Woelkart, K.; Feizlmayr, E.; Dittrich, P.; Beubler, E.; Pinl, F.; Suter, A.; Bauer, R. Pharmacokinetics of bilobalide, ginkgolide A and B after administration of three different Ginkgo biloba L. preparations in humans. Phytother. Res. 2010, 24, 445–450. [Google Scholar] [CrossRef]
- Bampali, E.; Germer, S.; Bauer, R.; Kulić, Ž. HPLC-UV/HRMS methods for the unambiguous detection of adulterations of Ginkgo biloba leaves with Sophora japonica fruits on an extract level. Pharm. Biol. 2021, 59, 438–443. [Google Scholar] [CrossRef]
- López-Gutiérrez, N.; Romero-González, R.; Vidal, J.L.M.; Frenich, A.G. Quality control evaluation of nutraceutical products from Ginkgo biloba using liquid chromatography coupled to high resolution mass spectrometry. J. Pharm. Biomed. Anal. 2016, 121, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Tredici, P.D. Ginkgos and people-a thousand years of interaction. Arnoldia 1991, 51, 2–15. [Google Scholar]
- Ma, J.; Duan, W.; Han, S.; Lei, J.; Xu, Q.; Chen, X.; Jiang, Z.; Nan, L.; Li, J.; Chen, K.; et al. Ginkgolic acid suppresses the development of pancreatic cancer by inhibiting pathways driving lipogenesis. Oncotarget 2015, 6, 20993–21003. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.M.; Ye, Y.R.; Wang, P.; Chen, J.; Guo, T. Study on anti-bacterium activities of extract of Ginkgo biloba leaves (EGbs) and Ginkgolic Acids (GAs). Food Sci. 2004, 25, 68–71. [Google Scholar]
- Lü, J.M.; Yan, S.; Jamaluddin, S.; Weakley, S.M.; Liang, Z.; Siwak, E.B.; Yao, Q.; Chen, C. Ginkgolic acid inhibits HIV protease activity and HIV infection in vitro. Med. Sci. Monit. 2012, 18, BR293–BR298. [Google Scholar] [CrossRef] [Green Version]
- Qiao, L.; Zheng, J.; Jin, X.; Wei, G.; Wang, G.; Sun, X.; Li, X. Ginkgolic acid inhibits the invasiveness of colon cancer cells through AMPK activation. Oncol Lett. 2017, 14, 5831–5838. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, Y.G.; Ryu, S.Y.; Cho, M.H.; Lee, J. Ginkgolic acids and Ginkgo biloba extract inhibit Escherichia coli O157:H7 and Staphylococcus aureus biofilm formation. Int. J. Food Microbiol. 2014, 174, 47–55. [Google Scholar] [CrossRef]
- Borenstein, R.; Hanson, B.A.; Markosyan, R.M.; Gallo, E.S.; Narasipura, S.D.; Bhutta, M.; Shechter, O.; Lurain, N.S.; Cohen, F.S.; Al-Harthi, L.; et al. Ginkgolic acid inhibits fusion of enveloped viruses. Sci. Rep. 2020, 10, 4746. [Google Scholar] [CrossRef]
- Bhutta, M.S.; Shechter, O.; Gallo, E.S.; Martin, S.D.; Jones, E.; Doncel, G.F.; Borenstein, R. Ginkgolic Acid Inhibits Herpes Simplex Virus Type 1 Skin Infection and Prevents Zosteriform Spread in Mice. Viruses 2021, 13, 86. [Google Scholar] [CrossRef]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Wu, C.; Fan, G.; Tang, Z.; Cao, F. The antibacterial activity and mechanism of ginkgolic acid C15:1. BMC Biotechnol. 2017, 17, 5. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Li, J.; Lu, D.; Li, J.; Liu, M.; He, Y.; Williams, B.; Li, J.; Yang, T. Ginkgolic acid, a sumoylation inhibitor, promotes adipocyte commitment but suppresses adipocyte terminal differentiation of mouse bone marrow stromal cells. Sci. Rep. 2018, 8, 2545. [Google Scholar] [CrossRef] [PubMed]
- Ayres, J.S. A metabolic handbook for the COVID-19 pandemic. Nat. Metab. 2020, 2, 572–585. [Google Scholar] [CrossRef]
- Gassen, N.C.; Papies, J.; Bajaj, T.; Emanuel, J.; Dethloff, F.; Chua, R.L.; Trimpert, J.; Heinemann, N.; Niemeyer, C.; Weege, F.; et al. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat. Commun. 2021, 12, 3818. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, M.S.; Gallo, E.S.; Borenstein, R. Multifaceted Role of AMPK in Viral Infections. Cells 2021, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Q.; Xiao, H.; Tam, J.P.; Liu, D.X. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 2005, 579, 2387–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, A.; Li, M.; Liu, Y.; Zhao, W.; Gao, D. Ginkgolic acid exerts an anti-inflammatory effect in human umbilical vein endothelial cells induced by ox-LDL. Pharmazie 2018, 73, 408–412. [Google Scholar] [CrossRef]
- Rajendran, P.; Rengarajan, T.; Nandakumar, N.; Palaniswami, R.; Nishigaki, Y.; Nishigaki, I. Kaempferol, a potential cytostatic and cure for inflammatory disorders. Eur. J. Med. Chem. 2014, 86, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Goldie, M.; Dolan, S. Bilobalide, a unique constituent of Ginkgo biloba, inhibits inflammatory pain in rats. Behav. Pharmacol. 2013, 24, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Son, K.H.; Chang, H.W.; Kang, S.S.; Kim, H.P. Effects of anti-inflammatory biflavonoid, ginkgetin, on chronic skin inflammation. Biol. Pharm. Bull. 2006, 29, 1046–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.D.; Baric, R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Li, R.; Pan, W.; Huang, W.; Liu, B.; Xie, Y.; Wang, Z.; Li, C.; Jiang, H.; Huang, J.; et al. Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-κB) signaling pathway. Phytomedicine 2020, 78, 153296. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cui, Q.; Cooper, L.; Zhang, P.; Lee, H.; Chen, Z.; Wang, Y.; Liu, X.; Rong, L.; Du, R. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhu, G.H.; Wang, H.N.; Hu, Q.; Chen, L.L.; Guan, X.Q.; Li, H.L.; Chen, H.Z.; Tang, H.; Ge, G.B. Discovery of naturally occurring inhibitors against SARS-CoV-2 3CLpro from Ginkgo biloba leaves via large-scale screening. Fitoterapia 2021, 152, 104909. [Google Scholar] [CrossRef]
- Lambert, F.; Jacomy, H.; Marceau, G.; Talbot, P.J. Titration of human coronaviruses, HcoV-229E and HCoV-OC43, by an indirect immunoperoxidase assay. Methods Mol. Biol. 2008, 454, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Szretter, K.J.; Balish, A.L.; Katz, J.M. Influenza: Propagation, quantification, and storage. Curr. Protoc. Microbiol. 2006, 3, 15G.1.1–15G.1.22. [Google Scholar] [CrossRef]
- Milewska, A.; Kaminski, K.; Ciejka, J.; Kosowicz, K.; Zeglen, S.; Wojarski, J.; Nowakowska, M.; Szczubiałka, K.; Pyrc, K. HTCC: Broad Range Inhibitor of Coronavirus Entry. PLoS ONE 2016, 11, e0156552. [Google Scholar] [CrossRef] [Green Version]


| Name | Melting Temperature | Sequences |
|---|---|---|
| Sense | 54.9 °C | 5′-GTTGTGGCCAATGGTGTTAAAG-3′ |
| Antisense | 55.2 °C | 5′-AGTGTTGCCTGACTCTTTGG-3′ |
| Probe | 60.6 °C | /56-FAM/ACAATTTGCTGAGCTTGTGCCGTC/36-TAMSp/ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhutta, M.S.; Sausen, D.G.; Gallo, E.S.; Dahari, H.; Doncel, G.F.; Borenstein, R. Ginkgolic Acid Inhibits Coronavirus Strain 229E Infection of Human Epithelial Lung Cells. Pharmaceuticals 2021, 14, 980. https://doi.org/10.3390/ph14100980
Bhutta MS, Sausen DG, Gallo ES, Dahari H, Doncel GF, Borenstein R. Ginkgolic Acid Inhibits Coronavirus Strain 229E Infection of Human Epithelial Lung Cells. Pharmaceuticals. 2021; 14(10):980. https://doi.org/10.3390/ph14100980
Chicago/Turabian StyleBhutta, Maimoona S., Daniel G. Sausen, Elisa S. Gallo, Harel Dahari, Gustavo F. Doncel, and Ronen Borenstein. 2021. "Ginkgolic Acid Inhibits Coronavirus Strain 229E Infection of Human Epithelial Lung Cells" Pharmaceuticals 14, no. 10: 980. https://doi.org/10.3390/ph14100980
APA StyleBhutta, M. S., Sausen, D. G., Gallo, E. S., Dahari, H., Doncel, G. F., & Borenstein, R. (2021). Ginkgolic Acid Inhibits Coronavirus Strain 229E Infection of Human Epithelial Lung Cells. Pharmaceuticals, 14(10), 980. https://doi.org/10.3390/ph14100980







