Activity of New Synthetic (2-Chloroethylthio)-1,4-naphthoquinones in Prostate Cancer Cells
Abstract
:1. Introduction
2. Results and Discussion
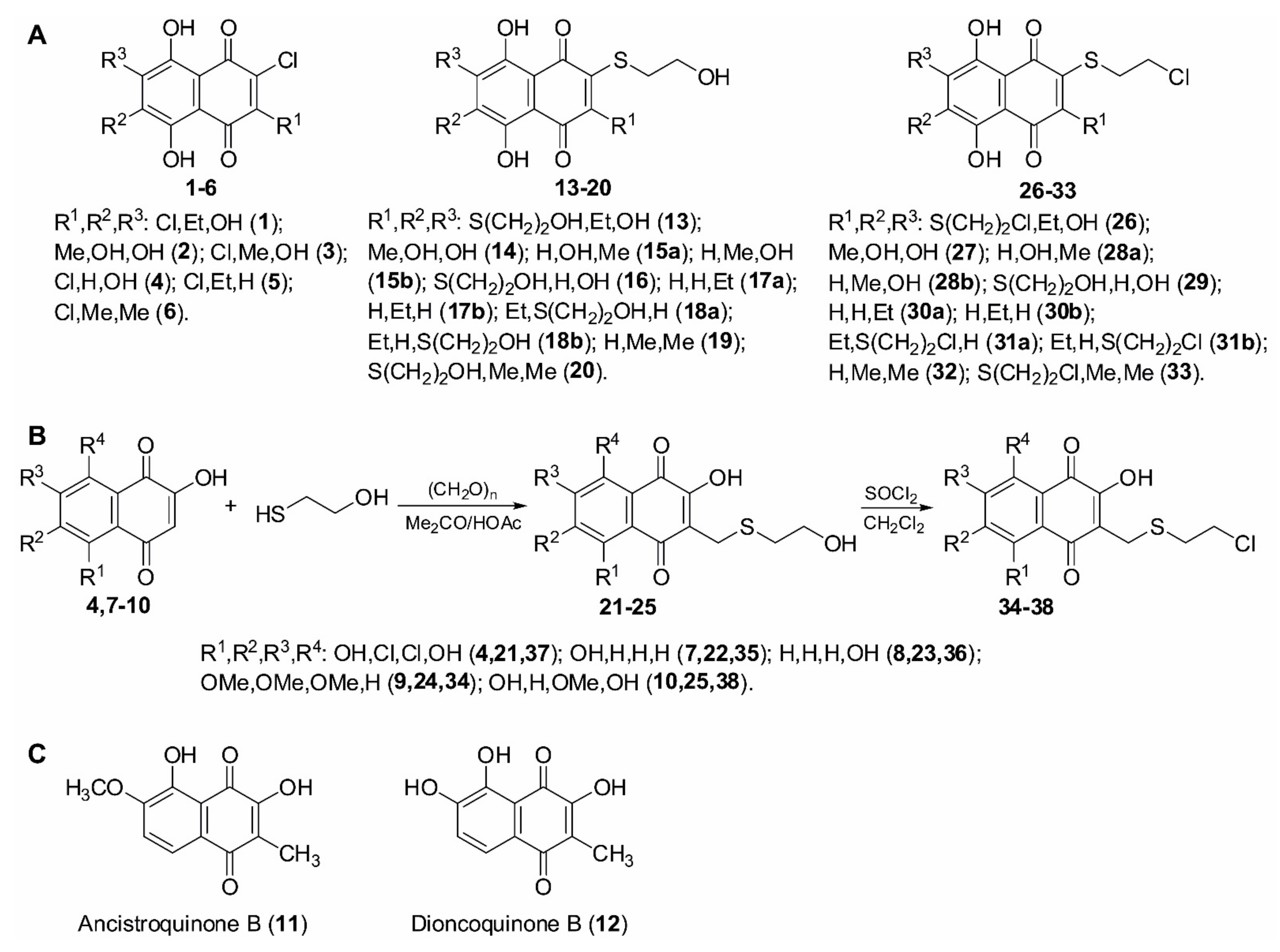
2.1. Synthesis of (2-Chloroethylthio)-1,4-naphthoquinon-2-yl Conjugates
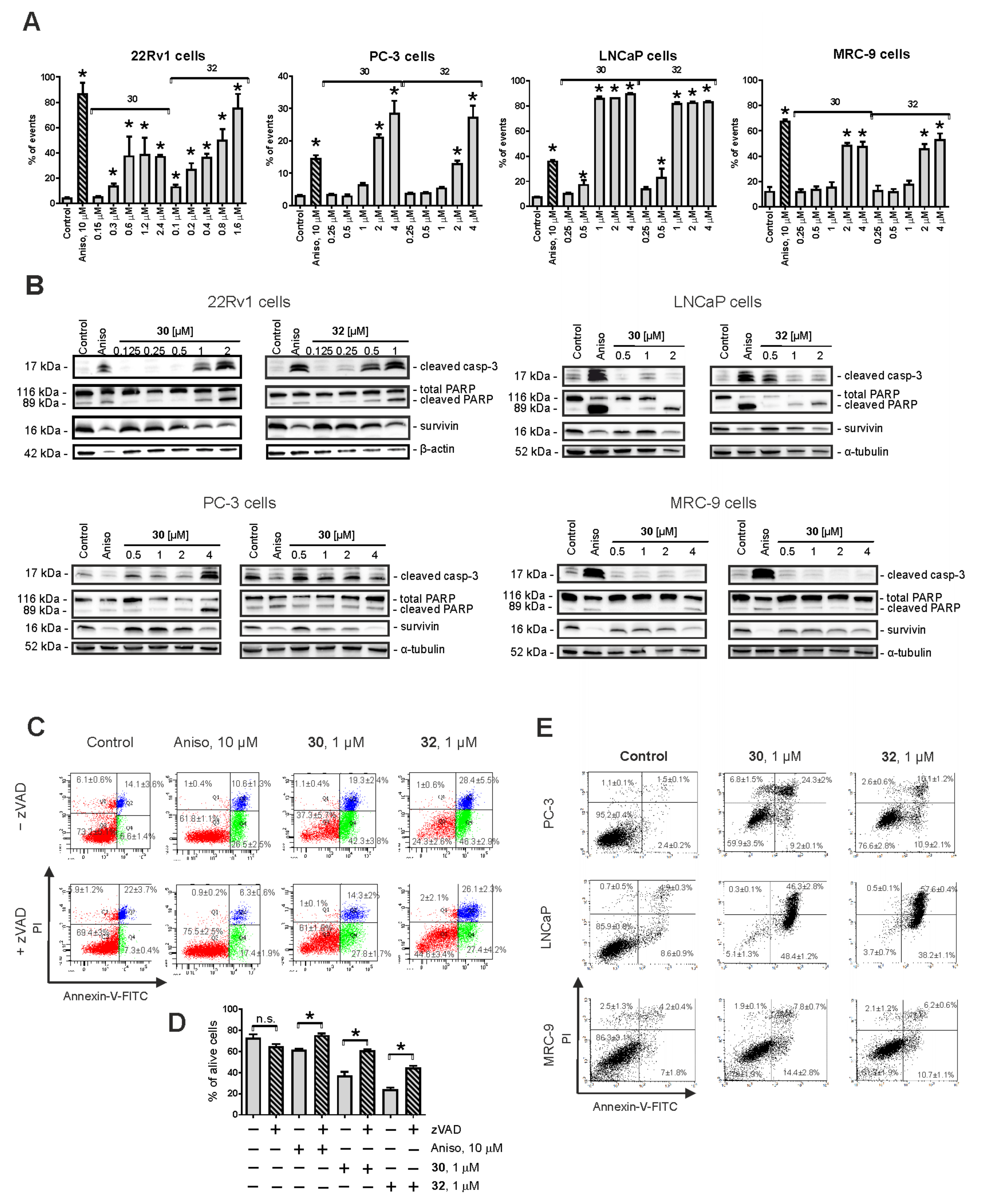
2.2. Evaluation of Cytotoxic Activity and Selectivity of the Synthesized Compounds in Human Prostate Cancer Cells
2.3. Induction of Caspase-Dependent Apoptosis by 30 and 32
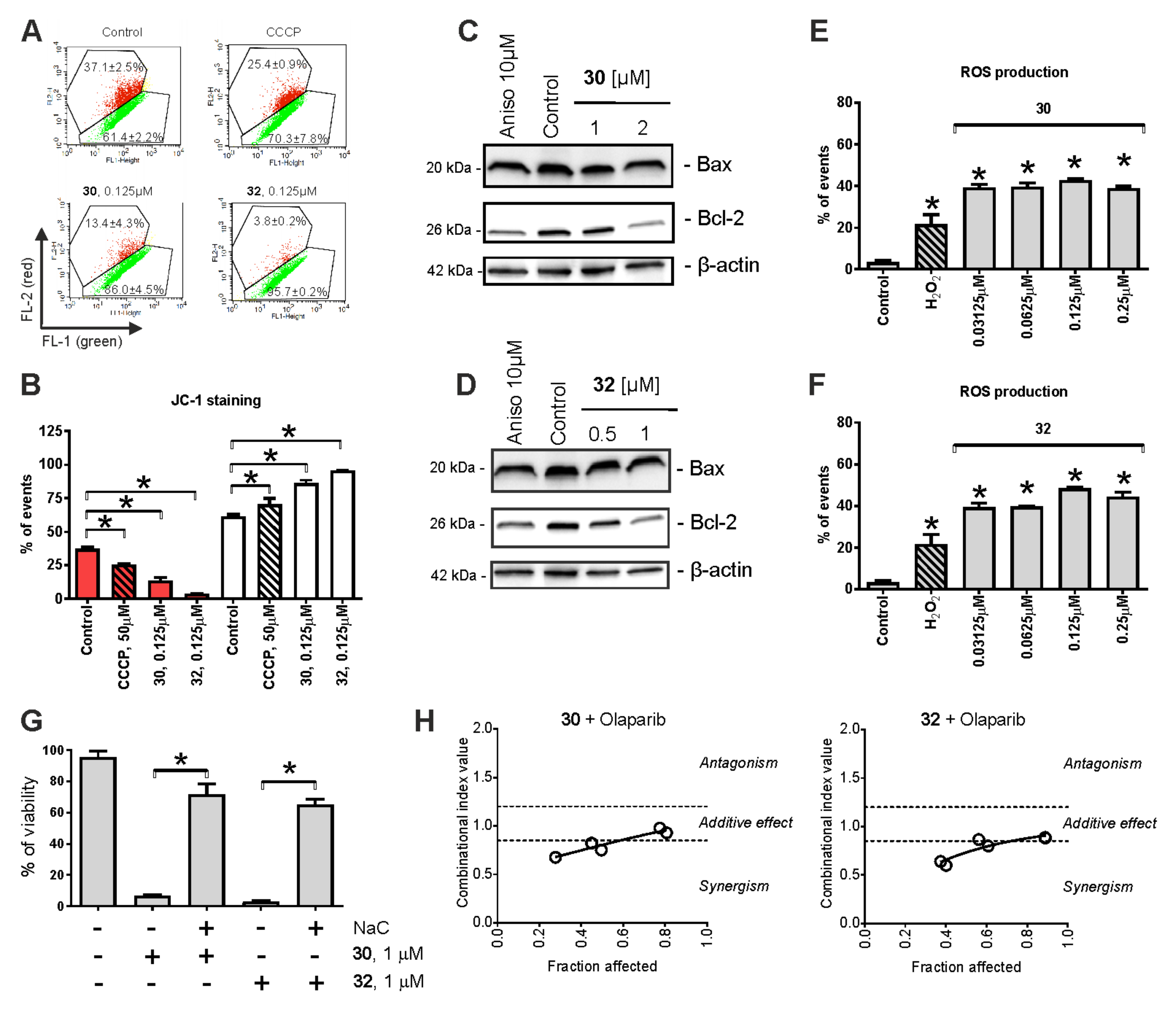
2.4. Mitochondria Targeting
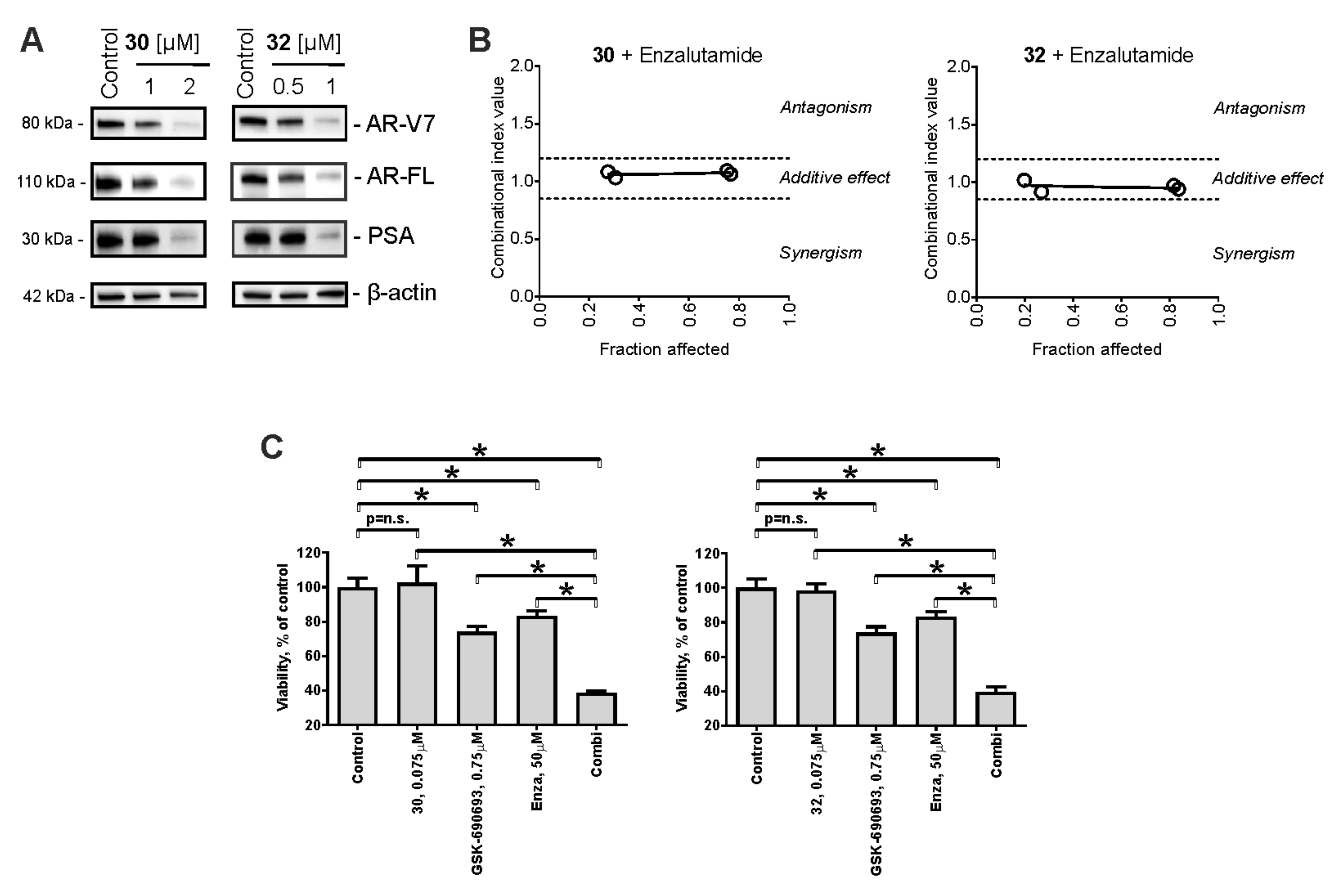
2.5. Cytotoxic Effects in Combination with Taxane and Platinum Agents
2.6. Effect on Stress-Activated Protein Kinases (SAPKs) and Other Kinases
2.7. Effect on AR Signaling
3. Materials and Methods
3.1. Chemistry
3.1.1. General Synthetic Procedures and Methods
3.1.2. General Procedure for the Synthesis of 2-Hydroxyethylthio Derivatives by Substitution of Chlorine Atoms in Chlorinated Naphthoquinones with 2-Mercaptoethanol in DMSO
3.1.3. General Procedure for the Synthesis of (2-Hydroxyethylthio)methyl Derivatives by Acid-Catalytic Condensation of 2-Hydroxynaphthoquinones with 2-Mercaptoethanol and Paraformaldehyde in Acetone
3.1.4. General Procedure for the Synthesis of 2-Chloroethylthio-1,4-naphthoquinones and (2-Chloroethylthio)methyl-1,4-naphthoquinones
3.2. Biology
3.2.1. Reagents and Antibodies
3.2.2. Cell Lines and Culture Conditions
3.2.3. In Vitro Cytotoxicity Assay (MTT Test)
3.2.4. Drug Combinational Studies
3.2.5. DNA Fragmentation and Cell Cycle Analysis
3.2.6. Detection of Apoptotic Cells by Annexin-V-FITC/PI Double Staining
3.2.7. Western Blotting
3.2.8. Evaluation of Intracellular ROS
3.2.9. Mitochondrial Membrane Potential Analysis
3.2.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ψm | mitochondrial membrane potential |
| ADC | antibody–drug conjugates |
| AR-V7 | androgen receptor splice variant V7 |
| AR-FL | androgen receptor-full length |
| AR | androgen receptor |
| ARTA | androgen receptor targeting therapy |
| CCCP | carbonyl cyanide m-chlorophenyl hydrazine |
| CI | combinational index |
| CRPC | castration-resistant prostate cancer |
| DMSO | dimethylsulfoxide |
| dsDNA | double-strand DNA |
| ssDNA | single-strand DNA |
| FACS | fluorescence-activated cell sorting |
| HDAC | histone deacetylase |
| HRR | homologous recombination repair mechanisms |
| MAPK | mitogen-activated protein kinase |
| MAPKi | MAP kinases inhibitor |
| mCRPC | metastatic castration-resistant prostate cancer |
| NaC | N-acetyl-L-cysteine |
| PCa | prostate cancer |
| PI | propidium iodide |
| PSA | prostate-specific antigen |
| ROS | reactive oxygen species |
| SAR | structure–activity relationships |
| SAPK | stress-activated protein kinase |
| zVAD | z-VAD(OMe)-fmk (pan-caspase inhibitor). |
References
- Khalaf, D.J.; Annala, M.; Taavitsainen, S.; Finch, D.L.; Oja, C.; Vergidis, J.; Zulfiqar, M.; Sunderland, K.; Azad, A.A.; Kollmannsberger, C.K.; et al. Optimal sequencing of enzalutamide and abiraterone acetate plus prednisone in metastatic castration-resistant prostate cancer: A multicentre, randomised, open-label, phase 2, crossover trial. Lancet Oncol. 2019, 20, 1730–1739. [Google Scholar] [CrossRef]
- Beyer, J.; Albers, P.; Altena, R.; Aparicio, J.; Bokemeyer, C.; Busch, J.; Cathomas, R.; Cavallin-Stahl, E.; Clarke, N.W.; Classen, J.; et al. Maintaining success, reducing treatment burden, focusing on survivorship: Highlights from the third European consensus conference on diagnosis and treatment of germ-cell cancer. Ann. Oncol. 2013, 24, 878–888. [Google Scholar] [CrossRef]
- Caffo, O.; De Giorgi, U.; Fratino, L.; Alesini, D.; Zagonel, V.; Facchini, G.; Gasparro, D.; Ortega, C.; Tucci, M.; Verderame, F.; et al. Clinical outcomes of castration-resistant prostate cancer treatments administered as third or fourth line following failure of docetaxel and other second-line treatment: Results of an Italian multicentre study. Eur. Urol. 2015, 68, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Gao, A.C. Drug resistance in castration resistant prostate cancer: Resistance mechanisms and emerging treatment strategies. Am. J. Clin. Exp. Urol. 2015, 3, 64–76. [Google Scholar] [PubMed]
- Luengo, A.; Gui, D.Y.; Vander Heiden, M.G. Targeting Metabolism for Cancer Therapy. Cell Chem. Biol. 2017, 24, 1161–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannaiyan, R.; Mahadevan, D. A comprehensive review of protein kinase inhibitors for cancer therapy. Expert Rev. Anticancer. Ther. 2018, 18, 1249–1270. [Google Scholar] [CrossRef]
- Klinakis, A.; Karagiannis, D.; Rampias, T. Targeting DNA repair in cancer: Current state and novel approaches. Cell. Mol. Life Sci. 2020, 77, 677–703. [Google Scholar] [CrossRef]
- Tebbutt, N.; Pedersen, M.W.; Johns, T.G. Targeting the ERBB family in cancer: Couples therapy. Nat. Rev. Cancer 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Hasan, M.; Leak, R.K.; Stratford, R.E.; Zlotos, D.P.; Witt-Enderby, P.A. Drug conjugates—an emerging approach to treat breast cancer. Pharmacol. Res. Perspect. 2018, 6, e00417. [Google Scholar] [CrossRef]
- Beretta, G.L.; Zaffaroni, N. Androgen Receptor-Directed Molecular Conjugates for Targeting Prostate Cancer. Front. Chem. 2019, 7, 369. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Pelageev, D.N.; Hauschild, J.; Sabutskii, Y.E.; Khmelevskaya, E.A.; Krisp, C.; Kaune, M.; Venz, S.; Borisova, K.L.; Busenbender, T.; et al. Inspired by Sea Urchins: Warburg Effect Mediated Selectivity of Novel Synthetic Non-Glycoside 1,4-Naphthoquinone-6S-Glucose Conjugates in Prostate Cancer. Mar. Drugs 2020, 18, 251. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, A.S.; Pelageev, N.D.; Hauschild, J.; Borisova, L.K.; Kaune, M.; Krisp, C.; Venz, S.; Sabutskii, E.Y.; Khmelevskaya, A.E.; Busenbender, T.; et al. Successful Targeting of the Warburg Effect in Prostate Cancer by Glucose-Conjugated 1,4-Naphthoquinones. Cancers 2019, 11, 1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wellington, K.W. Understanding cancer and the anticancer activities of naphthoquinones—A review. RSC Adv. 2015, 5, 20309–20338. [Google Scholar] [CrossRef]
- Lu, J.J.; Bao, J.L.; Wu, G.S.; Xu, W.S.; Huang, M.Q.; Chen, X.P.; Wang, Y.T. Quinones derived from plant secondary metabolites as anti-cancer agents. Anticancer Agents Med. Chem. 2013, 13, 456–463. [Google Scholar]
- Lown, J.W.; Sim, S.K.; Majumdar, K.C.; Chang, R.Y. Strand scission of DNA by bound adriamycin and daunorubicin in the presence of reducing agents. Biochem. Biophys. Res. Commun. 1977, 76, 705–710. [Google Scholar] [CrossRef]
- Pelageev, D.N.; Dyshlovoy, S.A.; Pokhilo, N.D.; Denisenko, V.A.; Borisova, K.L.; Keller-von Amsberg, G.; Bokemeyer, C.; Fedorov, S.N.; Honecker, F.; Anufriev, V.P. Quinone-carbohydrate nonglucoside conjugates as a new type of cytotoxic agents: Synthesis and determination of in vitro activity. Eur. J. Med. Chem. 2014, 77, 139–144. [Google Scholar] [CrossRef]
- Leopold, W.R.; Shillis, J.L.; Mertus, A.E.; Nelson, J.M.; Roberts, B.J.; Jackson, R.C. Anticancer activity of the structurally novel antibiotic CI-920 and its analogues. Cancer Res. 1984, 44, 1928–1932. [Google Scholar]
- Tewey, K.M.; Chen, G.L.; Nelson, E.M.; Liu, L.F. Intercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem. 1984, 259, 9182–9187. [Google Scholar] [CrossRef]
- Creech, H.J.; Preston, R.K.; Peck, R.M.; O’Connell, A.P.; Ames, B.N. Antitumor and mutagenic properties of a variety of heterocyclic nitrogen and sulfur mustards. J. Med. Chem. 1972, 15, 739–746. [Google Scholar] [CrossRef]
- Arroyo, C.M.; Schafer, R.J.; Carmichael, A.J. Reactivity of chloroethyl sulfides in the presence of a chlorinated prophylactic: A kinetic study by EPR/spin trapping and NMR techniques†**. J. Appl. Toxicol. 2000, 20, S7–S12. [Google Scholar] [CrossRef]
- Smith, S.L. War! What is it good for? Mustard gas medicine. CMAJ 2017, 189, E321–E322. [Google Scholar] [CrossRef] [Green Version]
- Bunn, P.A.; Hoffman, S.J.; Norris, D.; Golitz, L.E.; Aeling, J.L. Systemic Therapy of Cutaneous T-Cell Lymphomas (Mycosis Fungoides and the Sezary Syndrome). Ann. Intern. Med. 1994, 121, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Hudes, G. Estramustine-based chemotherapy. Semin. Urol. Oncol. 1997, 15, 13–19. [Google Scholar] [PubMed]
- Kitamura, T. Necessity of re-evaluation of estramustine phosphate sodium (EMP) as a treatment option for first-line monotherapy in advanced prostate cancer. Int. J. Urol. 2001, 8, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Ravery, V.; Fizazi, K.; Oudard, S.; Drouet, L.; Eymard, J.-C.; Culine, S.; Gravis, G.; Hennequin, C.; Zerbib, M. The use of estramustine phosphate in the modern management of advanced prostate cancer. BJU Int. 2011, 108, 1782–1786. [Google Scholar] [CrossRef]
- Pechura, C.M.; Rall, D.P. Veterans at Risk: The Health Effects of Mustard Gas and Lewisite; Pechura, C.M., Rall, D.P., Eds.; National Academy of Sciences: Washington, DC, USA, 1993. [Google Scholar]
- Bringmann, G.; Rüdenauer, S.; Irmer, A.; Bruhn, T.; Brun, R.; Heimberger, T.; Stühmer, T.; Bargou, R.; Chatterjee, M. Antitumoral and antileishmanial dioncoquinones and ancistroquinones from cell cultures of Triphyophyllum peltatum (Dioncophyllaceae) and Ancistrocladus abbreviatus (Ancistrocladaceae). Phytochemistry 2008, 69, 2501–2509. [Google Scholar] [CrossRef]
- Bringmann, G.; Zhang, G.; Hager, A.; Moos, M.; Irmer, A.; Bargou, R.; Chatterjee, M. Anti-tumoral activities of dioncoquinones B and C and related naphthoquinones gained from total synthesis or isolation from plants. Eur. J. Med. Chem. 2011, 46, 5778–5789. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.S. Targeting the androgen receptor in prostate cancer—A resilient foe. N. Engl. J. Med. 2014, 371, 1067–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Fedor, H.L.; Lotan, T.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Boudadi, K.; Antonarakis, E.S. Resistance to novel antiandrogen therapies in metastatic castration-resistant prostate cancer. Clin. Med. Insights. Oncol. 2016, 10 (Suppl. 1), 1–9. [Google Scholar] [CrossRef] [Green Version]
- Tamaki, H.; Harashima, N.; Hiraki, M.; Arichi, N.; Nishimura, N.; Shiina, H.; Naora, K.; Harada, M. Bcl-2 family inhibition sensitizes human prostate cancer cells to docetaxel and promotes unexpected apoptosis under caspase-9 inhibition. Oncotarget 2014, 5, 11399–11412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Zhu, Y.; Lou, W.; Nadiminty, N.; Chen, X.; Zhou, Q.; Shi, X.B.; deVere White, R.W.; Gao, A.C. Functional p53 determines docetaxel sensitivity in prostate cancer cells. Prostate 2013, 73, 418–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyshlovoy, S.A.; Otte, K.; Tabakmakher, K.M.; Hauschild, J.; Makarieva, T.N.; Shubina, L.K.; Fedorov, S.N.; Bokemeyer, C.; Stonik, V.A.; von Amsberg, G. Synthesis and anticancer activity of the derivatives of marine compound rhizochalin in castration resistant prostate cancer. Oncotarget 2018, 9, 16962–16973. [Google Scholar] [CrossRef]
- Marchiani, S.; Tamburrino, L.; Nesi, G.; Paglierani, M.; Gelmini, S.; Orlando, C.; Maggi, M.; Forti, G.; Baldi, E. Androgen-responsive and -unresponsive prostate cancer cell lines respond differently to stimuli inducing neuroendocrine differentiation. Int. J. Androl. 2010, 33, 784–793. [Google Scholar] [CrossRef]
- Li, X.; Fang, P.; Mai, J.; Choi, E.T.; Wang, H.; Yang, X.F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, Y.; Lim, S.; Jo, K. Single-molecule visualization of ROS-induced DNA damage in large DNA molecules. Analyst 2016, 141, 847–852. [Google Scholar] [CrossRef]
- Sachdev, E.; Tabatabai, R.; Roy, V.; Rimel, B.J.; Mita, M.M. PARP Inhibition in Cancer: An Update on Clinical Development. Target. Oncol. 2019, 14, 657–679. [Google Scholar] [CrossRef]
- Bochum, S.; Berger, S.; Martens, U.M. Olaparib. Recent Results Cancer Res. 2018, 211, 217–233. [Google Scholar] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Karanika, S.; Yang, G.; Wang, J.; Park, S.; Broom, B.M.; Manyam, G.C.; Wu, W.; Luo, Y.; Basourakos, S.; et al. Androgen receptor inhibitor-induced "BRCAness" and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 2017, 10, eaam7479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feiersinger, G.E.; Trattnig, K.; Leitner, P.D.; Guggenberger, F.; Oberhuber, A.; Peer, S.; Hermann, M.; Skvortsova, I.; Vrbkova, J.; Bouchal, J.; et al. Olaparib is effective in combination with, and as maintenance therapy after, first-line endocrine therapy in prostate cancer cells. Mol. Oncol. 2018, 12, 561–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafi, A.A.; Yen, A.E.; Weigel, N.L. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 2013, 140, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lin, A. Role of JNK activation in apoptosis: A double-edged sword. Cell Res. 2005, 15, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Sinha, K.; Das, J.; Pal, P.B.; Sil, P.C. Oxidative stress: The mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch. Toxicol. 2013, 87, 1157–1180. [Google Scholar] [CrossRef]
- Dou, Y.; Jiang, X.; Xie, H.; He, J.; Xiao, S. The Jun N-terminal kinases signaling pathway plays a “seesaw” role in ovarian carcinoma: A molecular aspect. J. Ovarian Res. 2019, 12, 99. [Google Scholar] [CrossRef] [Green Version]
- Samatar, A.A.; Poulikakos, P.I. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 2014, 13, 928–942. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crumbaker, M.; Khoja, L.; Joshua, A.M. AR Signaling and the PI3K Pathway in Prostate Cancer. Cancers 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Armstrong, A.J. Evolving standards in the treatment of docetaxel-refractory castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2011, 14, 192–205. [Google Scholar] [CrossRef] [Green Version]
- Anufriev, V.P.; Polonik, S.G.; Pokhilo, N.D.; Balanyova, N.N. Chemistry of naphthazarin derivatives. 11. Trisubstituted hydroquinone derivatives in the preparative synthesis of naphthazarins. Russ. Chem. Bull. 2003, 52, 2247–2250. [Google Scholar] [CrossRef]
- Anufriey, V.P.; Novikov, V.L.; Maximov, O.B.; Elyakov, G.B.; Levitsky, D.O.; Lebedev, A.V.; Sadretdinov, S.M.; Shvilkin, A.V.; Afonskaya, N.I.; Ruda, M.Y.; et al. Synthesis of some hydroxynaphthazarins and their cardioprotective effects under ischemia-reperfusion in vivo. Biorg. Med. Chem. Lett. 1998, 8, 587–592. [Google Scholar] [CrossRef]
- Yakubovskaya, A.Y.; Pokhilo, N.D.; Anufriev, V.F.; Anisimov, M.M. Synthesis and antimicrobial and antifungal activities of compounds of the naphthazarin series. Pharm. Chem. J. 2009, 43, 396. [Google Scholar] [CrossRef]
- Fieser, L.F.; Dunn, J.T. The Addition of Dienes to Halogenated and Hydroxylated Naphthoquinones. J. Am. Chem. Soc. 1937, 59, 1016–1021. [Google Scholar] [CrossRef]
- Li, K.; Yang, K.; Zheng, L.; Li, Y.; Wang, Q.; Lin, R.; He, D. Anti-acute myeloid leukemia activity of 2-chloro-3-alkyl-1,4-naphthoquinone derivatives through inducing mtDNA damage and GSH depletion. Biorg. Med. Chem. 2018, 26, 4191–4200. [Google Scholar] [CrossRef]
- Glazunov, V.P.; Tchizhova, A.Y.; Shestak, O.P.; Sopel"nyak, G.I.; Anufriev, V.P. Chemistry of naphthazarin derivatives. 8. Determination of structures of substituted 2-hydroxy-6(7)-methoxynaphthazarins and 7(8)-hydroxypyranonaphthazarins by IR spectroscopy. Russ. Chem. Bull. 2001, 50, 95–100. [Google Scholar] [CrossRef]
- Khmelevskaya, E.A.; Pelageev, D.N. A convenient synthetic approach to dioncoquinone B and related compounds. Tetrahedron Lett. 2019, 60, 1022–1024. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Tabakmakher, K.M.; Hauschild, J.; Shchekaleva, R.K.; Otte, K.; Guzii, A.G.; Makarieva, T.N.; Kudryashova, E.K.; Fedorov, S.N.; Shubina, L.K.; et al. Guanidine alkaloids from the marine sponge Monanchora pulchra show cytotoxic properties and prevent EGF-induced neoplastic transformation in vitro. Mar. Drugs 2016, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Dyshlovoy, S.A.; Madanchi, R.; Hauschild, J.; Otte, K.; Alsdorf, W.H.; Schumacher, U.; Kalinin, V.I.; Silchenko, A.S.; Avilov, S.A.; Honecker, F.; et al. The marine triterpene glycoside frondoside A induces p53-independent apoptosis and inhibits autophagy in urothelial carcinoma cells. BMC Cancer 2017, 17, 93. [Google Scholar] [CrossRef] [Green Version]
- Dyshlovoy, S.A.; Fedorov, S.N.; Kalinovsky, A.I.; Shubina, L.K.; Bokemeyer, C.; Stonik, V.A.; Honecker, F. Mycalamide A shows cytotoxic properties and prevents EGF-induced neoplastic transformation through inhibition of nuclear factors. Mar. Drugs 2012, 10, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Dyshlovoy, S.; Rast, S.; Hauschild, J.; Otte, K.; Alsdorf, W.; Madanchi, R.; Kalinin, V.; Silchenko, A.; Avilov, S.; Dierlamm, J.; et al. Frondoside A induces AIF-associated caspase-independent apoptosis in Burkitt’s lymphoma cells. Leuk. Lymphoma 2017, 58, 2905–2915. [Google Scholar] [CrossRef] [PubMed]

| Compound | IC50(22Rv1), [μM] | IC50(PNT2), [μM] | Selectivity Index [IC50(PNT2)/IC50(22Rv1)] |
|---|---|---|---|
| 11 | >100 | >100 | n/a |
| 12 | 89.5 ± 24.9 | >100 | n/a |
| 26 | 41.2 ± 1.18 | >100 | n/a |
| 27 | >100 | >100 | n/a |
| 28 | 93.35 ± 28.6 | >100 | n/a |
| 29 | 32.06 ± 1.66 | 6.44 ± 1.24 | 0.2 |
| 30 | 0.36 ± 0.09 | 0.49 ± 0.09 | 1.36 |
| 31 | 0.51 ± 0.18 | 0.38 ± 0.03 | 0.75 |
| 32 | 0.14 ± 0.09 | 0.77 ± 0.07 | 5.5 |
| 33 | 0.75 ± 0.16 | 0.85 ± 0.04 | 1.13 |
| 34 | >100 | >100 | n/a |
| 35 | >100 | >100 | n/a |
| 36 | >100 | >100 | n/a |
| 37 | 48.55 ± 12.2 | 28.15 ± 1.04 | 0.58 |
| 38 | >100 | >100 | n/a |
| Cell Line | IC50 [μM], 48 h | |||
|---|---|---|---|---|
| 30 | 31 | 32 | 33 | |
| Cancer Cell Lines | ||||
| 22Rv1 | 0.36 ± 0.09 | 0.51 ± 0.18 | 0.14 ± 0.09 | 0.75 ± 0.16 |
| DU145 | 0.51 ± 0.13 | 0.79 ± 0.21 | 0.71 ± 0.2 | 0.63 ± 0.05 |
| PC-3 | 0.56 ± 0.09 | 0.38 ± 0.1 | 0.58 ± 0.11 | 2.27 ± 0.19 |
| VCaP | 0.93 ± 0.04 | 0.79 ± 0.18 | 0.92 ± 0.16 | 1.57 ± 0.22 |
| LNCaP | 0.63 ± 0.19 | 0.85 ± 0.07 | 0.47 ± 0.15 | 0.85 ± 0.13 |
| Non-Cancer Cell Lines | ||||
| HEK293 | 0.24 ± 0.03 | 0.15 ± 0.01 | 0.21 ± 0.06 | 0.58 ± 0.14 |
| MRC9 | 1.62 ± 0.34 | 1.41 ± 0.52 | 1.11 ± 0.23 | 2.85 ± 0.39 |
| PNT2 | 0.49 ± 0.09 | 0.38 ± 0.03 | 0.77 ± 0.07 | 0.85 ± 0.04 |
| Antibodies | Clonality | Source | Cat. No. | Dilution | Manufacturer |
|---|---|---|---|---|---|
| anti-ERK1/2 | mAb | mouse | #9107 | 1:2000 | Cell Signaling |
| anti-JNK1/2 | mAb | rabbit | #9258 | 1:1000 | Cell Signaling |
| anti-p38 | mAb | rabbit | #9212 | 1:1000 | Cell Signaling |
| anti-phospho-ERK1/2 | mAb | rabbit | #4377 | 1:1000 | Cell Signaling |
| anti-phospho-JNK1/2 | mAb | rabbit | #4668 | 1:1000 | Cell Signaling |
| anti-phospho-p38 | mAb | rabbit | #4511 | 1:1000 | Cell Signaling |
| anti-phospho-Akt | mAb | rabbit | #4058 | 1:1000 | Cell Signaling |
| anti-Akt | pAb | rabbit | #9272 | 1:1000 | Cell Signaling |
| anti-phospho-MEK1/2 | mAb | rabbit | #2338 | 1:1000 | Cell Signaling |
| anti-MEK1/2 | pAb | rabbit | #9122 | 1:1000 | Cell Signaling |
| anti-β-Actin-HRP | pAb | goat | sc-1616 | 1:10,000 | Santa Cruz |
| anti-AR | pAb | rabbit | sc-816 | 1:200 | Santa Cruz |
| anti-AR-V7 | mAb | rabbit | 198394 | 1:1000 | abcam |
| anti-Bax | mAb | rabbit | #5023 | 1:1000 | Cell Signaling |
| anti-Bcl-2 | pAb | rabbit | #2876 | 1:1000 | Cell Signaling |
| anti-cleaved Caspase-3 | mAb | rabbit | #9664 | 1:1000 | Cell Signaling |
| anti-PARP | pAb | rabbit | #9542 | 1:1000 | Cell Signaling |
| anti-Survivin | pAb | rabbit | NB500-201 | 1:1000 | Novus |
| anti-mouse IgG-HRP | sheep | NXA931 | 1:10,000 | GE Healthcare | |
| anti-rabbit IgG-HRP | goat | #7074 | 1:5000 | Cell Signaling |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyshlovoy, S.A.; Pelageev, D.N.; Jakob, L.S.; Borisova, K.L.; Hauschild, J.; Busenbender, T.; Kaune, M.; Khmelevskaya, E.A.; Graefen, M.; Bokemeyer, C.; et al. Activity of New Synthetic (2-Chloroethylthio)-1,4-naphthoquinones in Prostate Cancer Cells. Pharmaceuticals 2021, 14, 949. https://doi.org/10.3390/ph14100949
Dyshlovoy SA, Pelageev DN, Jakob LS, Borisova KL, Hauschild J, Busenbender T, Kaune M, Khmelevskaya EA, Graefen M, Bokemeyer C, et al. Activity of New Synthetic (2-Chloroethylthio)-1,4-naphthoquinones in Prostate Cancer Cells. Pharmaceuticals. 2021; 14(10):949. https://doi.org/10.3390/ph14100949
Chicago/Turabian StyleDyshlovoy, Sergey A., Dmitry N. Pelageev, Lea S. Jakob, Ksenia L. Borisova, Jessica Hauschild, Tobias Busenbender, Moritz Kaune, Ekaterina A. Khmelevskaya, Markus Graefen, Carsten Bokemeyer, and et al. 2021. "Activity of New Synthetic (2-Chloroethylthio)-1,4-naphthoquinones in Prostate Cancer Cells" Pharmaceuticals 14, no. 10: 949. https://doi.org/10.3390/ph14100949
APA StyleDyshlovoy, S. A., Pelageev, D. N., Jakob, L. S., Borisova, K. L., Hauschild, J., Busenbender, T., Kaune, M., Khmelevskaya, E. A., Graefen, M., Bokemeyer, C., Anufriev, V. P., & von Amsberg, G. (2021). Activity of New Synthetic (2-Chloroethylthio)-1,4-naphthoquinones in Prostate Cancer Cells. Pharmaceuticals, 14(10), 949. https://doi.org/10.3390/ph14100949







