Improved Detection Criteria for Detecting Drug-Drug Interaction Signals Using the Proportional Reporting Ratio
Abstract
1. Introduction
2. Results
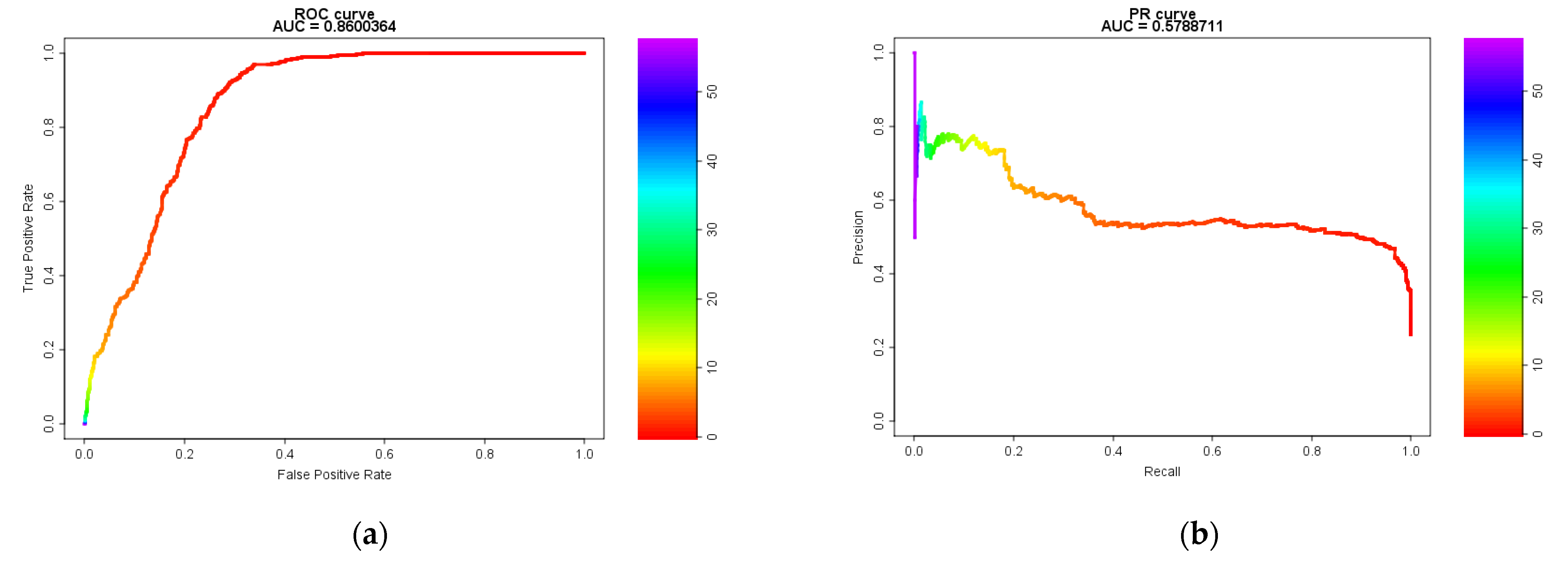
2.1. Model Evaluation Using Receiver Operating Characteristic (ROC) and Precision-Recall (PR) Curves and the Area Under the Curve (AUC)
2.2. Model Evaluation Using Machine Learning Indicators
2.3. Cohen’s Kappa Coefficient
3. Discussion
- (1)
- The true data used in this study consist of a statistics-based drug D1–drug D2 adverse event (SJS) combination rather than a pharmacology-based combination. Unfortunately, data on unknown adverse events do not exist; thus, it was necessary to use “hypothetical” true data instead of “real” true data for validation. Therefore, we used a combination of signals detected using the three algorithms as “hypothetical” true data for detecting drug-drug interaction signals.
- (2)
- It is important to compare detection trends using all adverse events recorded in the validation datasets. However, numerous combinations of drug D1–drug D2 adverse events are expected, and a study design including all such combinations is not practical. Therefore, SJS was the target adverse event in this study. Although this adverse event has been used previously in other comparative studies by our group [12,13] and other researchers [10,14], it is possible that different performance characteristics are obtained when different reference sets are used.
- (3)
- Differences in the approach adopted by regulatory authorities may result in differences in the tendency to register adverse events in the spontaneous reporting system. For example, JADER has long not accepted reports from patients, whereas the Food and Drug Administration Adverse Events Reporting System (FAERS) includes reports from non-medical professionals. It is unclear how the differences in registration tendencies would affect the results of this study [12]. However, as verified by Caster et al. [15], the statistical impact of differences in the number of cases enrolled in the spontaneous reporting system on this study might be small.
4. Materials and Methods
4.1. Data Sources
4.2. Definitions of Adverse Drug Events
4.3. “Hypothetical” True Data of Adverse Events for Comparative Verification
4.4. Statistical Models and Criteria
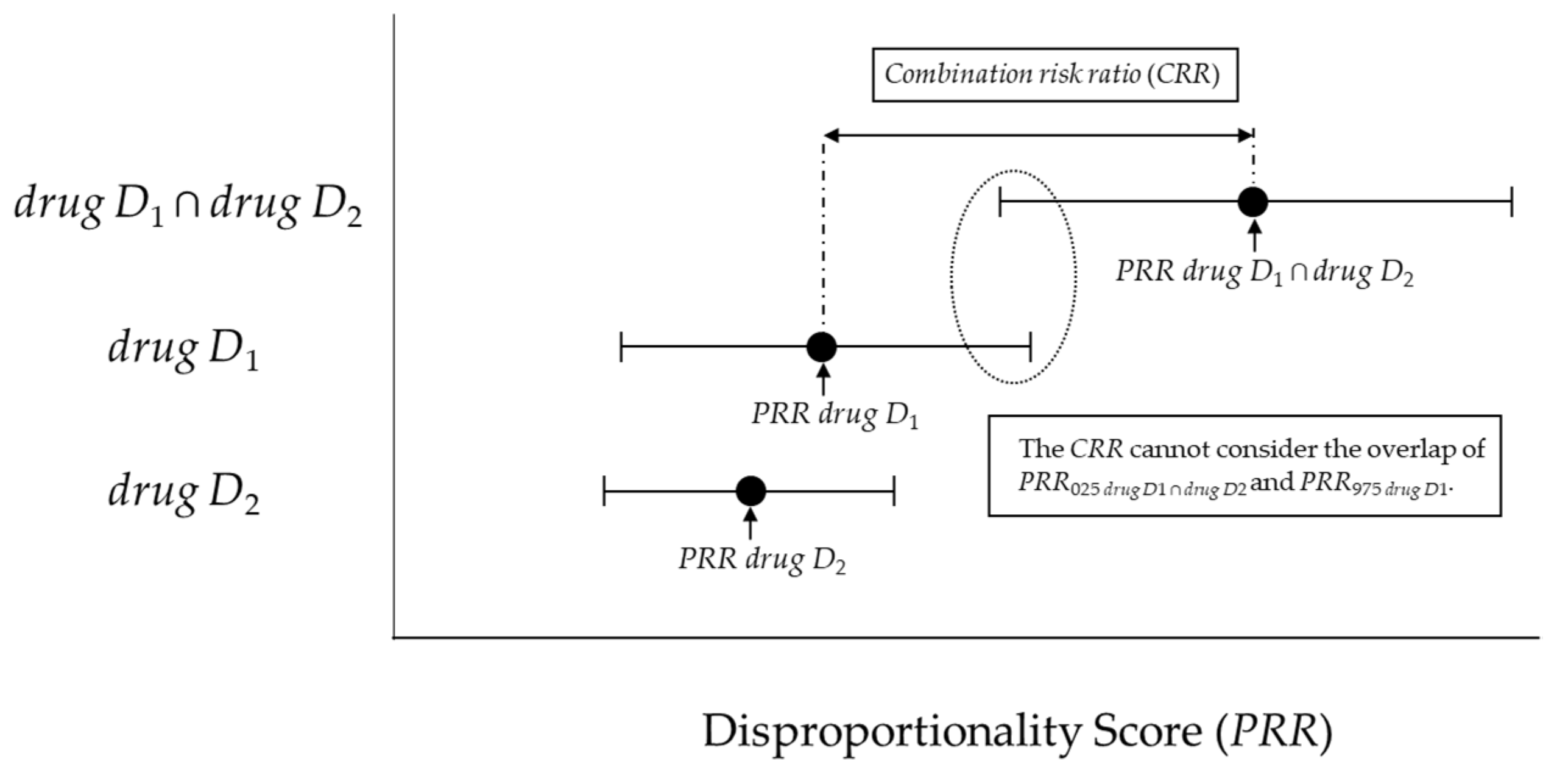
4.4.1. Previous Model (combination risk ratio) and criteria proposed by Susuta et al.
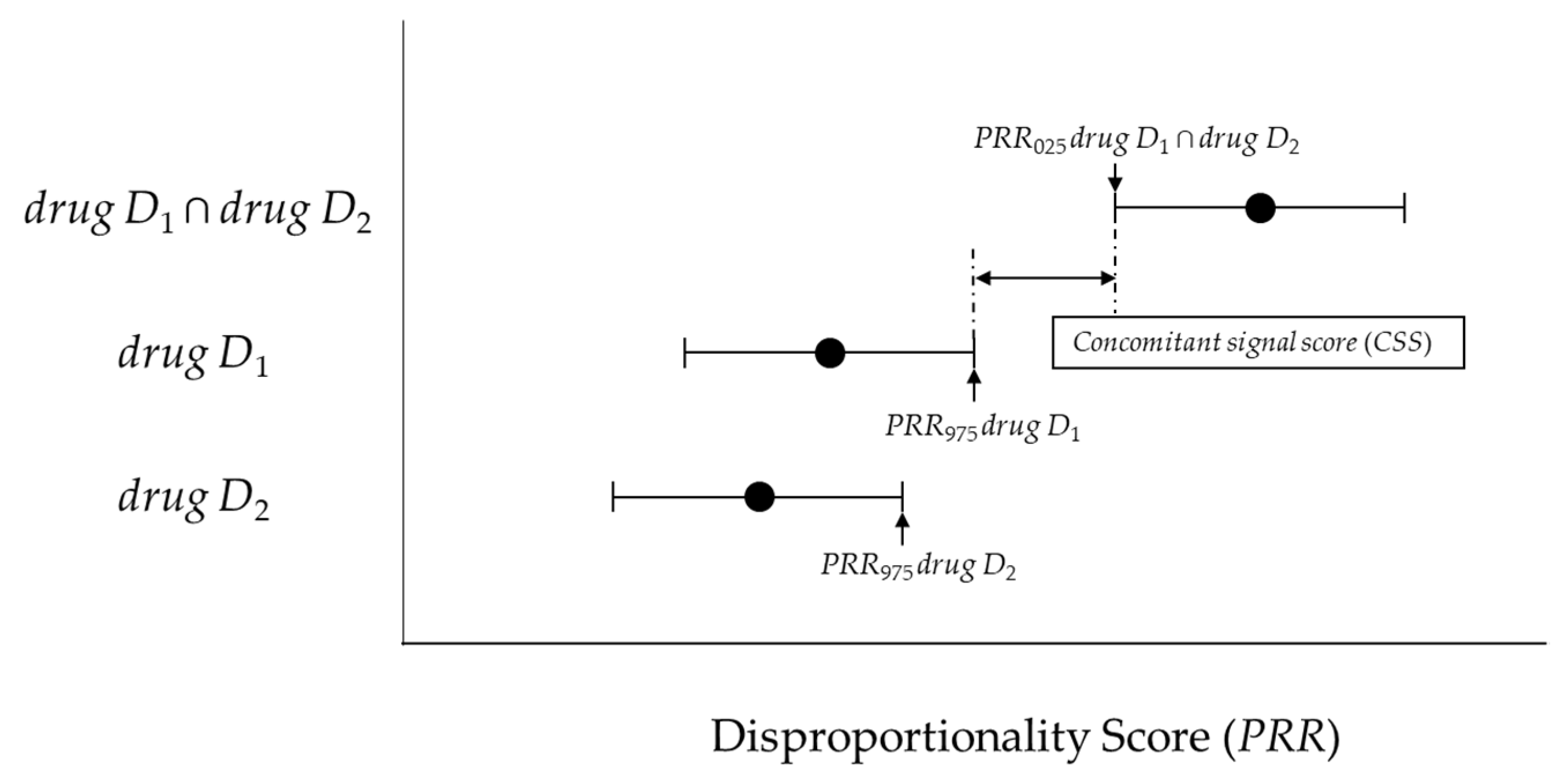
4.4.2. New Model (Concomitant Signal Score) and Criteria
4.4.3. Model (Ω Shrinkage Measure) and Criteria to be Compared.
4.5. Evaluation of Models for Detection
4.5.1. Model Evaluation Using the ROC and PR Curves and AUC
4.5.2. Using Evaluations of Classification in Machine Learning
4.5.3. Cohen’s Kappa Coefficient
4.6. Analysis Software
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iyer, S.V.; Harpaz, R.; Le Pendu, P.; Bauer-Mehren, A.; Shah, N.H. Mining clinical text for signals of adverse drug-drug interactions. J. Am. Med. Inf. Assoc. 2013, 21, 353–362. [Google Scholar] [CrossRef]
- Bate, A.; Evans, S.J. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol. Drug Saf. 2009, 18, 427–436. [Google Scholar] [CrossRef]
- Evans, S.J.; Waller, P.C.; Davis, S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol. Drug Saf. 2001, 10, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Rothman, K.J.; Lanes, S.; Sacks, S.T. The reporting odds ratio and its advantages over the proportional reporting ratio. Pharmacoepidemiol. Drug Saf. 2004, 13, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Bate, A.; Lindquist, M.; Edwards, I.R.; Olsson, S.; Orre, R.; Lansner, A.; De Freitas, R.M. A Bayesian neural network method for adverse drug reaction signal generation. Eur. J. Clin. Pharmacol. 1998, 54, 315–321. [Google Scholar] [CrossRef] [PubMed]
- DuMouchel, W. Bayesian Data Mining in Large Frequency Tables, with an Application to the FDA Spontaneous Reporting System. Am. Stat. 1999, 53, 177–190. [Google Scholar] [CrossRef]
- Noguchi, Y.; Ueno, A.; Otsubo, M.; Katsuno, H.; Sugita, I.; Kanematsu, Y.; Yoshida, A.; Esaki, H.; Tachi, T.; Teramachi, H. A new search method using association rule mining for drug-drug interaction based on spontaneous report system. Front. Pharmacol. 2019, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Vilar, S.; Friedman, C.; Hripcsak, G. Detection of drug-drug interactions through data mining studies using clinical sources, scientific literature and social media. Brief. Bioinform. 2018, 19, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Tachi, T.; Teramachi, H. Review of Statistical Methodologies for Detecting Drug-Drug Interactions Using Spontaneous Reporting Systems. Front. Pharmacol. 2019, 10, 1319. [Google Scholar] [CrossRef] [PubMed]
- Susuta, Y.; Takahashi, Y. Safety risk evaluation methodology in detecting the medicine concomitant use risk which might cause critical drug rash. Jpn. J. Pharmacoepidemiol. 2014, 19, 39–49. [Google Scholar] [CrossRef][Green Version]
- Suzuki, A.; Yuen, N.A.; Ilic, K.; Miller, R.T.; Reese, M.J.; Brown, H.R.; Ambroso, J.I.; Falls, J.G.; Hunt, C.M. Comedications alter drug-induced liver injury reporting frequency: Data mining in the WHO VigiBase™. Regul. Toxicol. Pharmacol. 2015, 72, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Tachi, T.; Teramachi, H. Subset Analysis for Screening Drug–Drug Interaction Signal Using Pharmacovigilance Database. Pharmaceutics 2020, 12, 762. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Tachi, T.; Teramachi, H. Comparison of signal detection algorithms based on frequency statistical model for drug-drug interaction using spontaneous reporting systems. Pharm. Res. 2020, 37, 86. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Koide, D.; Hirai, T. Comparison of data mining methodologies using Japanese spontaneous reports. Pharmacoepidemiol. Drug Saf. 2004, 13, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Caster, O.; Aoki, Y.; Gattepaille, L.M.; Grundmark, B. Disproportionality Analysis for Pharmacovigilance Signal Detection in Small Databases or Subsets: Recommendations for Limiting False-Positive Associations. Drug Saf. 2020, 43, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Thakrar, B.T.; Grundschober, S.B.; Doessegger, L. Detecting signals of drug-drug interactions in a spontaneous reports database. Br. J. Clin. Pharmacol. 2007, 64, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Gosho, M.; Maruo, K.; Tada, K.; Hirakawa, A. Utilization of chi-square statistics for screening adverse drug-drug interactions in spontaneous reporting systems. Eur. J. Clin. Pharmacol. 2017, 73, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Noren, G.N.; Sundberg, R.; Bate, A.; Edwards, I.R. A statistical methodology for drug-drug interaction surveillance. Stat. Med. 2008, 27, 3057–3070. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed]
- Carrington, A.M.; Fieguth, P.W.; Qazi, H.; Holzinger, A.; Chen, H.H.; Mayr, F.; Manuel, D.G. A new concordant partial AUC and partial c statistic for imbalanced data in the evaluation of machine learning algorithms. BMC Med. Inform. Decis. Mak. 2020, 20, 4. [Google Scholar] [CrossRef] [PubMed]
| Target AE | Other AEs | Total | |
|---|---|---|---|
| Concomitant use of drug D1 and drug D2 | n111 | n110 | n11+ |
| only drug D1 | n101 | n100 | n10+ |
| only drug D2 | n011 | n010 | n01+ |
| Neither drug D1 or drug D2 | n001 | n000 | n00+ |
| Total | n++1 | n++0 | n+++ |
| Analytical Model | TP | FP | TN | FN |
|---|---|---|---|---|
| Combination risk ratio (CRR) | 473 | 266 | 335 | 1 |
| Concomitant signal score (CSS) | 449 | 172 | 429 | 25 |
| Ω shrinkage measure | 451 | 106 | 495 | 23 |
| Analytical Model | Accuracy | Youden’s Index | F-Measure | Specificity | Recall (Sensitivity) | Precision (PPV) | NPV |
|---|---|---|---|---|---|---|---|
| Combination risk ratio (CRR) | 0.752 | 0.555 | 0.780 | 0.557 | 0.998 | 0.640 | 0.997 |
| Concomitant signal score (CSS) | 0.817 | 0.661 | 0.820 | 0.714 | 0.947 | 0.723 | 0.944 |
| Ω shrinkage measure | 0.880 | 0.775 | 0.875 | 0.824 | 0.951 | 0.810 | 0.956 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noguchi, Y.; Aoyama, K.; Kubo, S.; Tachi, T.; Teramachi, H. Improved Detection Criteria for Detecting Drug-Drug Interaction Signals Using the Proportional Reporting Ratio. Pharmaceuticals 2021, 14, 4. https://doi.org/10.3390/ph14010004
Noguchi Y, Aoyama K, Kubo S, Tachi T, Teramachi H. Improved Detection Criteria for Detecting Drug-Drug Interaction Signals Using the Proportional Reporting Ratio. Pharmaceuticals. 2021; 14(1):4. https://doi.org/10.3390/ph14010004
Chicago/Turabian StyleNoguchi, Yoshihiro, Keisuke Aoyama, Satoaki Kubo, Tomoya Tachi, and Hitomi Teramachi. 2021. "Improved Detection Criteria for Detecting Drug-Drug Interaction Signals Using the Proportional Reporting Ratio" Pharmaceuticals 14, no. 1: 4. https://doi.org/10.3390/ph14010004
APA StyleNoguchi, Y., Aoyama, K., Kubo, S., Tachi, T., & Teramachi, H. (2021). Improved Detection Criteria for Detecting Drug-Drug Interaction Signals Using the Proportional Reporting Ratio. Pharmaceuticals, 14(1), 4. https://doi.org/10.3390/ph14010004






