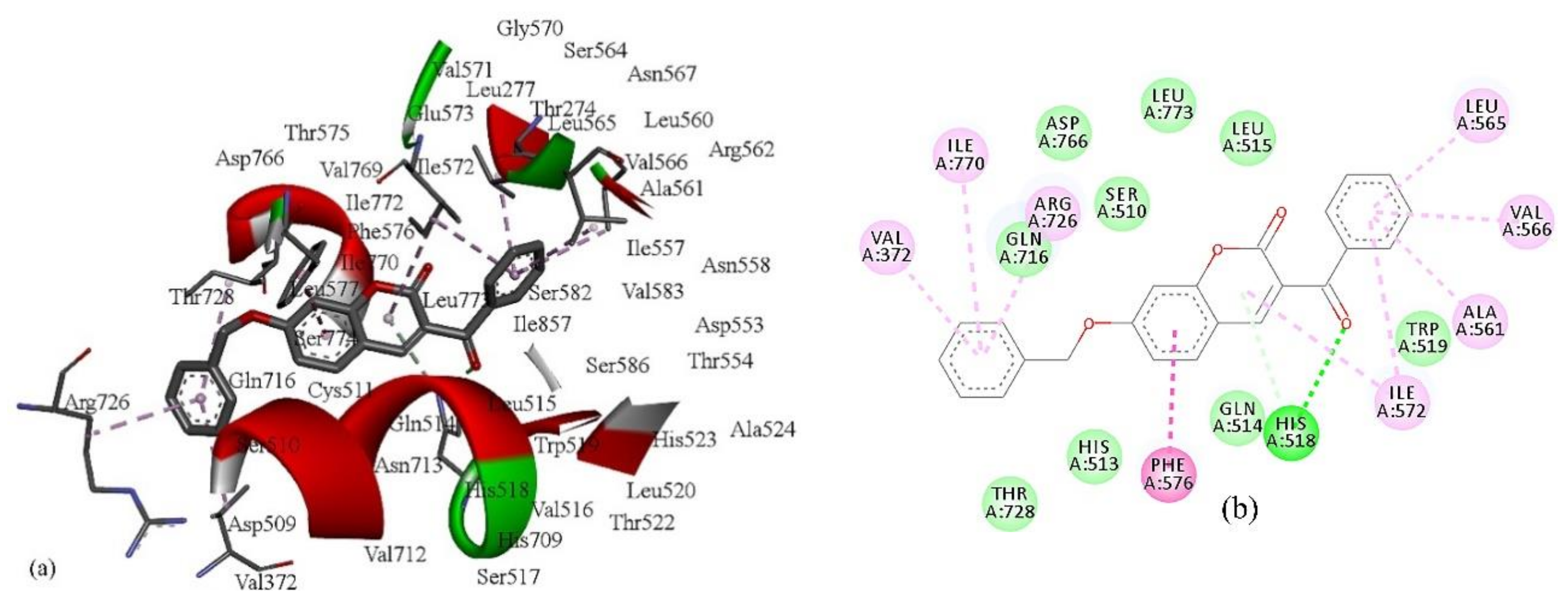
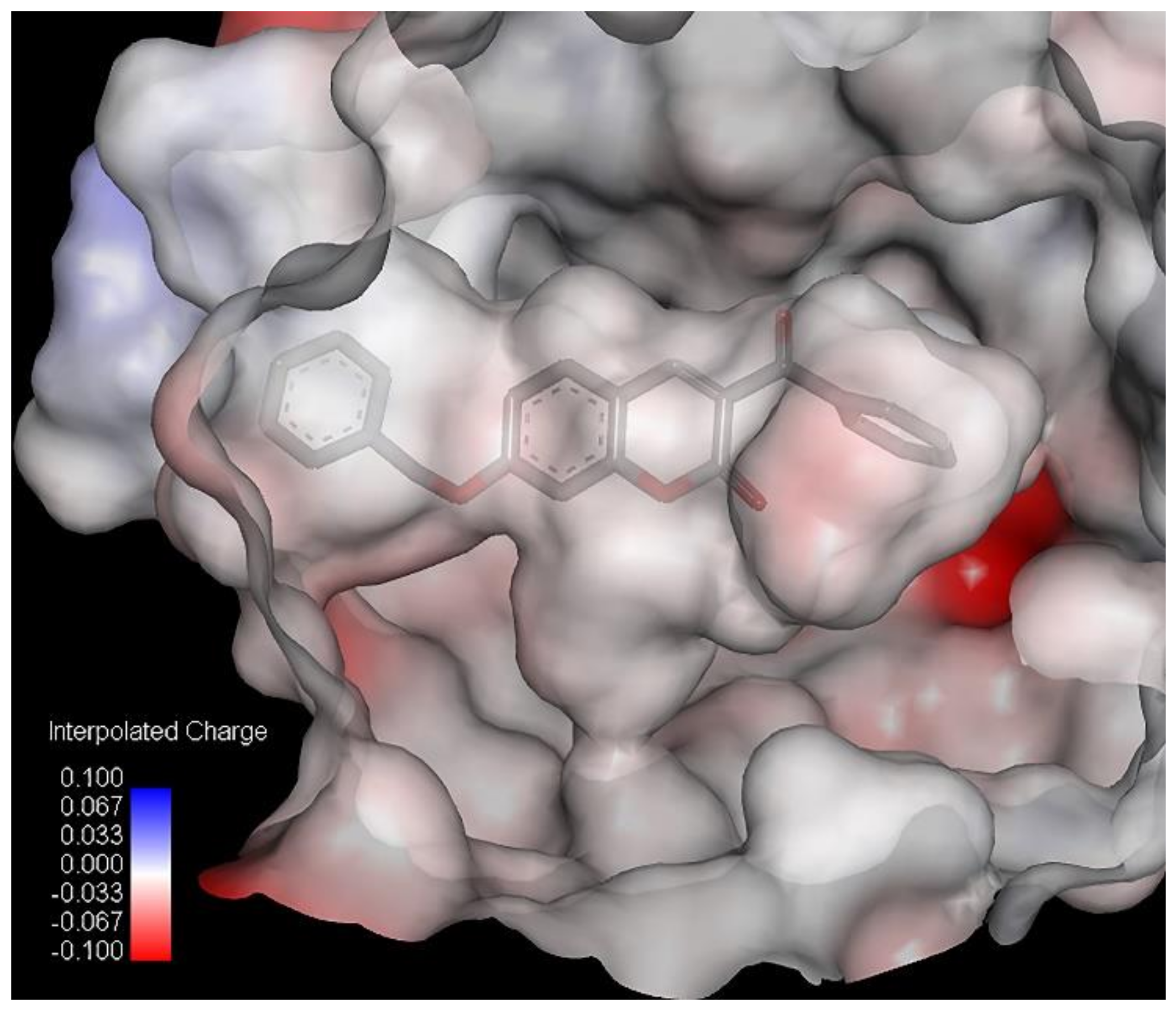
2.2. Inhibitory Activity
The synthesized coumarin derivatives were tested for inhibitory activity against soybean lipoxygenase at 100 µM concentrations in the reaction mixture, and the results are shown in
Table 1. It should be mentioned that the inhibitory activities of compounds
38 and
39 were not possible to determine since the reaction mixture was cloudy, probably due to the low solubility of the tested compounds. Five series of coumarin derivatives were synthesized in the reaction of substituted salicylaldehydes and dimethyl malonate (Series 1—
2,
12,
16,
20,
27,
30,
36), diethyl malonate (Series 2—
3,
17,
21,
24,
26,
28,
33,
37), ethyl benzoylacetate (Series 3—
5,
7,
13,
23,
25,
35,
38,
39), ethyl cyanoacetate (Series 4—
4,
6,
10,
18,
22,
29,
31,
34), and ethyl acetoacetate (Series 5—
1,
9,
11,
14,
15,
19,
32).
Substituted methyl 2-oxo-2
H-chromene-3-carboxylates (Series 1) inhibited soybean LOX-3 by 7.1–85.1%. The highest inhibition rate of 85.1% was found for methyl 6-bromo-2-oxo-2
H-chromene-3-carboxylate (
16). For the compounds of Series 2, the highest inhibition rate of 56.1% was found for ethyl 6, 8-dibromo-2-oxo-2
H-chromene-3-carboxylate (
26), followed by that for ethyl 6-bromo-2-oxo-2
H-chromene-3-carboxylate (
17) with an inhibition activity of 55.2%. 6-bromo-2-oxo-2
H-chromene-3-carbonitrile (
18) acts as an efficient LOX inhibitor (84.8%) of Series 4. When comparing all the compounds with the bromine substituent (
16,
26,
17,
18), it could be concluded that the bromine contributes to the lipoxygenase inhibition. Compound
15 also had substituted bromine in its structure but did not show as much inhibition activity as the other compounds from Series 5. The highest inhibitory activity for all the compounds was observed with 3-benzoyl-7-(benzyloxy)-2
H-chromen-2-one (
7), from Series 3 (96.6%). Series 5 had the lowest range of inhibition (11.3–48.5%) compared to other the compound Series (1–4). It could be observed that the compounds without substituents in positions 6, 7, and 8 of the coumarin core (
1,
2,
3,
4, and
5) showed very low inhibition. The 10 most active soybean LOX inhibitors among tested the coumarins are compounds
7,
16,
18,
13,
30,
26,
23,
17,
20, and
21, and all of them have a substituent in position 6 of coumarin core, except compounds from Series 3 (
7 and
13), which have substituents in position 7 of the coumarin core. Inhibitory activity against lipoxygenase exhibited by coumarin derivatives was previously reported [
29,
30]. Roussaki et al. [
31] examined twelve 3-aryl coumarins, and only half of them were found to be LOX inhibitors. Soybean LOX-3’s inhibition activities were in the range of 6–86%, and the highest inhibition rate of 86% was achieved with a compound with bromine in position 6 of the coumarin. Poor inhibition (16.1–22.6%) was achieved with the coumarins reported by Kontogiorgis and Hadjipavlou-Litina [
12]. Only three coumarins of seven showed some inhibitory activity. The inhibitory activity of coumarin (36%) was higher than that presented in
Table 1, while the value of coumarin inhibition reported by Symenidis et al. [
32] was even lower (15%). Melagraki et al. [
13] examined coumarin-3-aminoamides. Only one of the twelve tested coumarins was not active, and the inhibition was in the range of 8% to 10%.
The inhibition of lipid peroxidation was also examined for coumarin derivatives, and the results are presented in
Table 1. Lipid peroxidation inhibition was in the range of 7.0–91.0%. Compounds of Series 1, 2, 3, 4, and 5 showed lipid peroxidation inhibition ranges 16.6–86.6%, 7.0–91.0%, 42.1–86.1%, 12.9–81.0% and 23.0–69.0%, respectively. The compounds with the highest inhibition rates of 69.2–91.0% (
39,
15,
38,
30,
17,
18,
31,
7,
16, and
28) all have substituents in position 6 or 7 of coumarin. All the coumarins with bromine in position 6 (
16,
17,
38,
18, and
15) had a high rate of inhibition except compound
26 (31.1%). In addition, four compounds with the methoxy group in position 6 or 7 of the coumarin core (
39,
30,
31, and
28) are among the compounds with the highest inhibition. 3-acetyl-6-hydroxy-2
H-chromen-2-one (
19), 6-hydroxy-2-oxo-2
H-chromene-3-carbonitrile (
22), and 3-benzoyl-6-hydroxy-2
H-chromen-2-one (
23) are the compounds with hydroxyl group in position 6 of the coumarin core, and showed very similar rates of inhibition: 69.2%, 66.8%, and 66.5%, respectively. Compound
4 (2-oxo-2
H-chromene-3-carbonitrile) showed no inhibition. All the results were compared with an appropriate standard inhibitor, Trolox, that showed inhibition of 61.8%, which is in accordance with the literature. Inhibitions of 63% for Trolox were previously reported [
32,
33,
34]. Coumarin’s (
8) inhibition of 2.6% is negligible, and it is in accordance with reported data showing that coumarin is completely inactive [
35].
The interaction of the examined coumarin derivatives with the stable free radical 1-diphenyl-picrylhydrazyl (DPPH) is shown in
Table 1. The compounds showed moderate antioxidant activity in the range of 15.2 to 58.1%. Ethyl 6-hydroxy-2-oxo-2
H-chromene-3-carboxylate (
21) showed the highest antioxidant activity, followed by the three compounds (
18,
10, and
22) with the carbonitrile group in position 3 of the coumarin core. Compounds
10,
22, and
23 are coumarins with very similar rates of antioxidant activity: 39.2, 39.0, and 38.9%, respectively. All three compounds possess hydroxyl groups,
22 and
23 in position 6 and compound
10 in position 8 of the coumarin. The results are compared with the appropriate standards Trolox and NDGA.
2.3. QSAR Study
The best model obtained for lipoxygenase inhibition is:
N(training) = 29; N(test) = 8 (16, 19, 22, 24, 25, 28, 35, 37).
The statistical parameters of the obtained models are given in
Table 2. The variables in Equation (1) are listed in order of relative importance by their standardized regression coefficients (β, in brackets). However, according to the statistical results presented in
Table 1, model (1) does not satisfy the threshold for fitting and external validation parameters:
R2train > 0.60 and
R2test < 0.06. Since compound
14 from the training set has a standard residual of −2.65 (greater than 2), it has been deemed an outlier. This can be explained since it is the only compound with a 7-diethylamino group (
Table 1). After the removal of compound
14 from the training set, subsequent re-analysis produced the following improved QSAR model:
The values of the descriptors included in models (1)–(2) are given in the
Supplementary Materials Table S1. The values of log % inhibition calculated by Equation (2) for each molecule are presented in
Table 1 and
Table S1. The statistical parameters for the model (2) are presented in
Table 2. In order to exclude the possibility that the models were overfitted, the collinearity of the descriptors included in the QSAR models was evaluated with a correlation matrix (
Table 3). Low collinearity was confirmed by the values of the correlation coefficient (R), ≤ 0.7, and verified by the low values of Kxx and ΔK ≥ 0.05 (
Table 2). The model satisfied the fitting and internal validation criteria: R
2 and R
2adj ≥ 0.60 (the closer the R
2 values are to unity, the more similar the calculated values are to the experimental ones); CCC
tr ≥ 0.85; a root-mean-square error (RMSE) and mean absolute error (MAE) close to zero; and RMSE
tr < RMSE
cv (
Table 2) [
36]. The cross-validated correlation coefficient (Q
2loo = 0.06) demonstrates a satisfactory internal prediction power for model (2). Y-scrambling or response permutation/randomization testing is a technique used to check the robustness of a QSAR model. The robustness of the obtained QSAR model was affirmed by R
2yscr and Q
2yscr values < 0.2, as R
2yscr > Q
2yscr [
37]. Model (2) satisfied some of the external validation criteria: R
2ext ≥ 0.60, low RMSE and MAE, and low differences between RMSE
tr and RMSE
ex as well as between MAE
tr and MAE
ex. However, the negative values of Q
2F1, Q
2F2, and Q
2F3—lower than 0.6—and r
2m average lower than 0.6 indicate that this model is useless for external prediction [
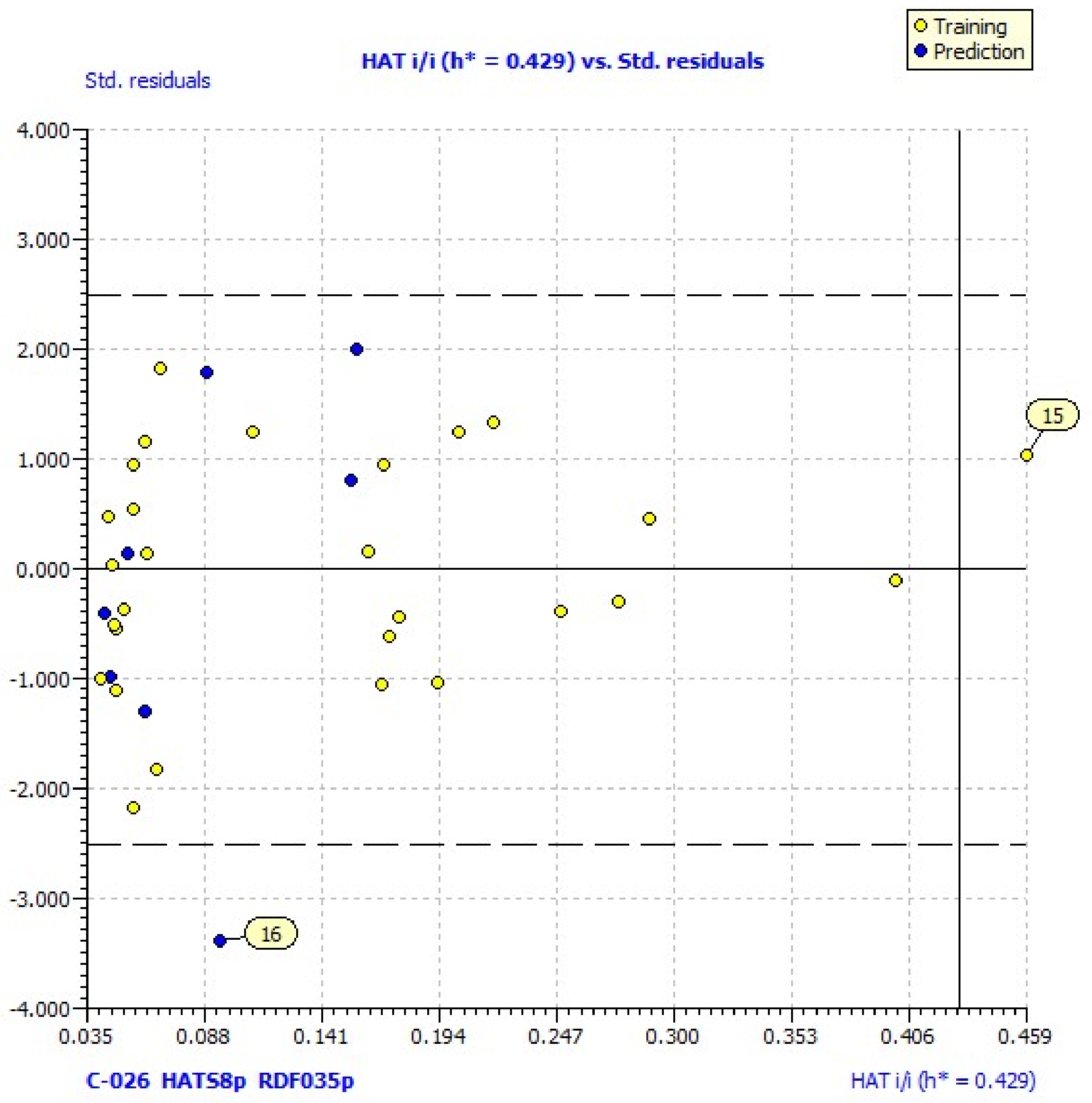
38]. Williams plots for the same models reveal one outlier (compound
16) and one compound outside of the applicability domain (
15) (
Figure 1). Its laverage (HAT = 0.583) is greater than the warning laverage (h* = 0.429); therefore, the predicted value for compound
15, which is poorly active (% LOX inh. = 11.25), must be interpreted with great care.
The largest value of the standardized regression coefficients in Equation (2) has a descriptor C-026 that belongs to the atom-centered fragments group of descriptors [
39]. Since the atom-centered fragment approach decomposes the molecule into structural pieces, these kind of descriptors take into consideration the local physicochemical properties of any part of a molecule. Thus, the descriptor
C-026 represents the occurring atomic states of R-CX-R carbon, where X could be any heteroatom (O, N, or halogens). The positive coefficient of
C-026 in Equation (2) indicates that higher values of that descriptor are favorable for lipoxygenase inhibition. For comparison, the most active compound against lipoxygenase, compound
7, has two R-CX-R carbon atoms, one from 3-benzoyl and second from a 7-benzyloxy group, and the % of LOX inhibition is 96.6%. Compound
5 has only one R-CX-R carbon atom from a 3-benzoyl group, and the % of LOX inhibition is decreased to 22.5% (
Table 1). The R-CX-R group corresponds to the presence of the alkoxy group, a structural feature that decreases the hydrophobicity of molecules [
34,
40]. According to Viswanadhan et al. [
39], the atomic hydrophobicity of the C atom in the R-CX-R group is low (log
p = −0.103). Hydrophobicity is a property that governs the interaction of the ligand (drug) molecules with the biological receptor. Thus, the oxygen atom from the benzoyl group can act as an H-bond acceptor and could form H bonds with amino acid residues.
The second variable in Equation (2) is a descriptor calculated by the Radial Distribution Function (RDF) approach [
41]. The
RDF of an ensemble of N atoms can be interpreted as the probability distribution for finding an atom in a spherical volume of radius
r. The presence of the descriptor
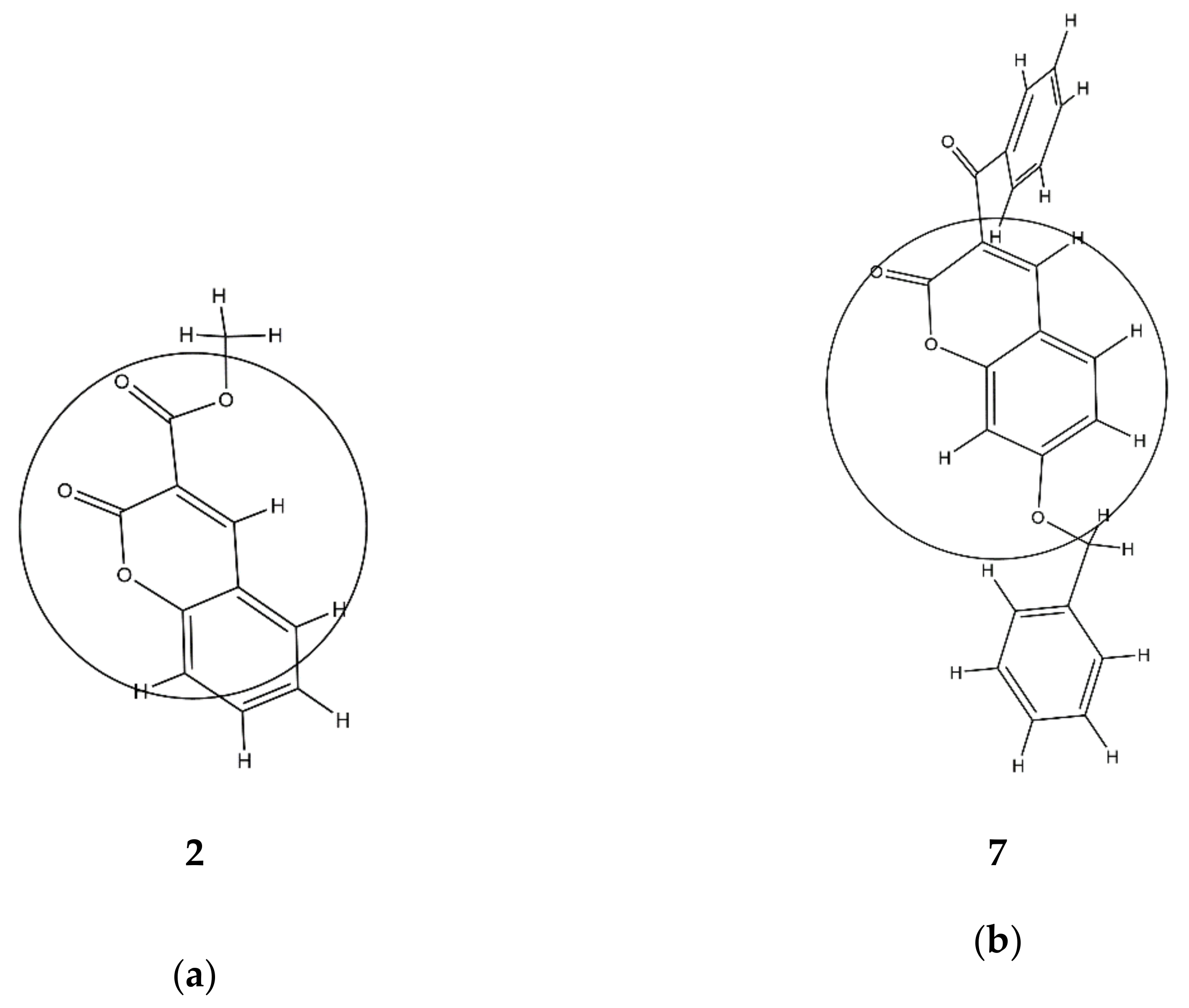
RDF035p in Equation (2) suggests the occurrence of some linear dependence between the inhibition power and the 3D molecular distribution of polarizabilities, calculated for the radius of 3.5 Å from the geometrical center of each molecule. Molecules with more atoms with higher polarizabilities in this area, such as carbon (
p = 1.76) [
42], have higher values of
RDF035p. Thus, compound
7 has a higher value of
RDF035p and therefore better inhibition ability than compound
2 since it has more carbon compounds and fewer oxygen atoms (
p = 0.802) in an area of 3.5 Å from the geometrical center of each molecule (
Table S1,
Figure 2). This supports previous findings that the replacement of a phenyl group attached on the coumarin ring by a 2-pyridyl group or by a morpholinyl group decreased the inhibitory activity of 6- and 7-substituted coumarins against lipoxygenase [
43]. Additionally, in the study of lipoxygenase inhibition by
O-prenylated 3-carboxycoumarins,
O-isopentenyl derivatives demonstrated no considerable lipoxygenase inhibition, while
O-geranyl and
O-farnesyl derivatives demonstrated potent inhibitory activity [
44].
The third variable, descriptor HATS8p, is the leverage-weighted autocorrelation of lag 8/weighted by atomic polarizabilities, which belongs to the GETAWAY (GEometry, Topology, and Atom-Weights AssemblY) descriptors [
42]. The values of this descriptor depend on the 3D relative atom location in the molecule. Therefore, compounds with a larger number of atoms with enhanced atomic polarizabilities, especially Cl and Br at the topological distance of 2, possess higher HATS8p values, such as
15 (0.282), which has a lower inhibitory effect.