Cyclin-Dependent Kinase and Antioxidant Gene Expression in Cancers with Poor Therapeutic Response
Abstract
1. Introduction
1.1. Cell Cycle Regulation in Cancer
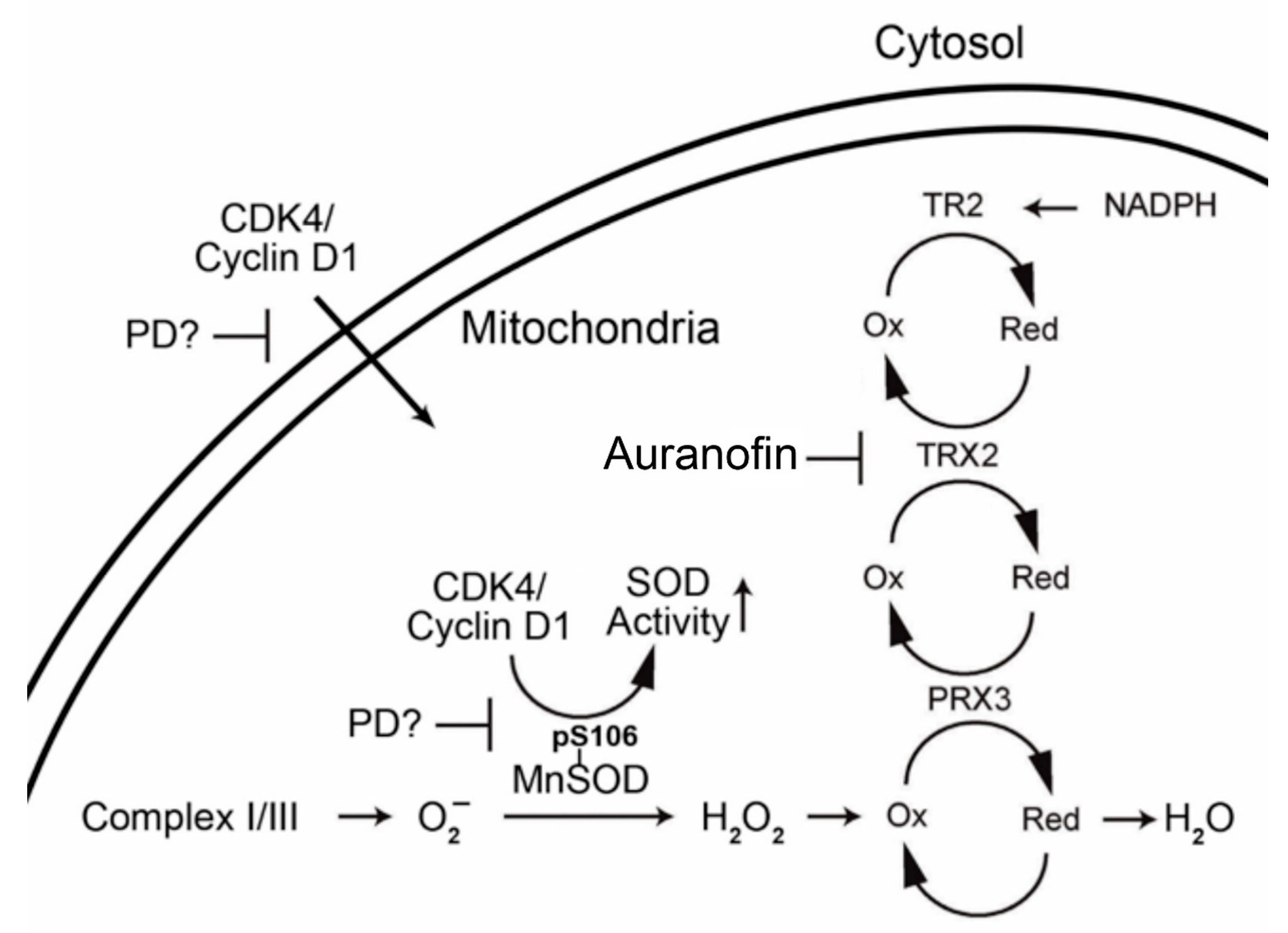
1.2. Mitochondrial Antioxidant Defense
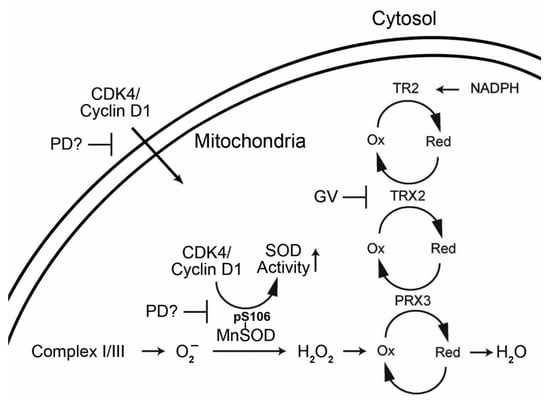
1.3. Interrelationship of Cell Cycle-Related Genes and Mitochondrial Antioxidant Defense Genes
1.4. The Cancer Genome Atlas (TCGA)
2. Results
2.1. mRNA Expression
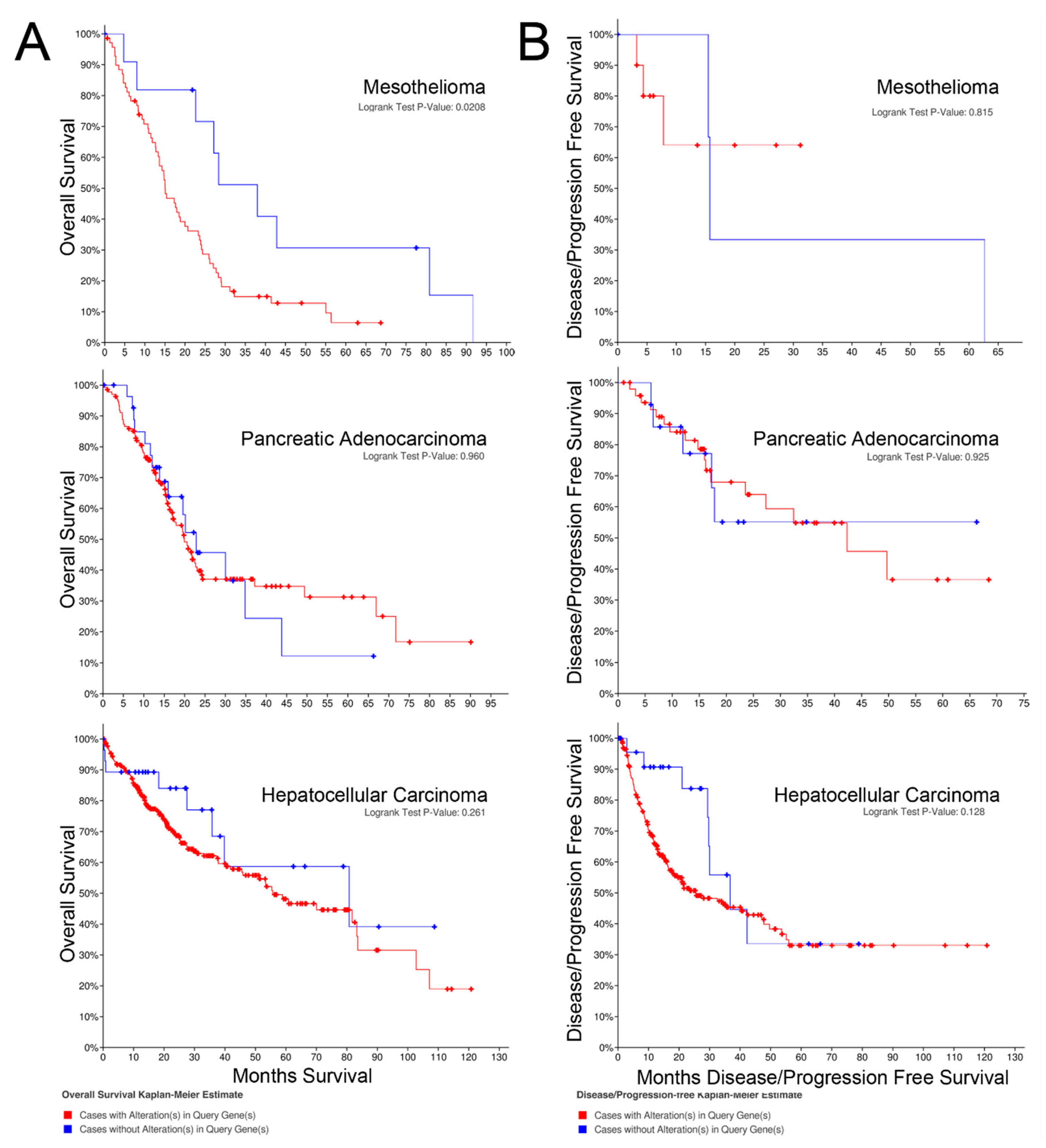
2.2. Overall Survival
2.3. Progression-free Survival
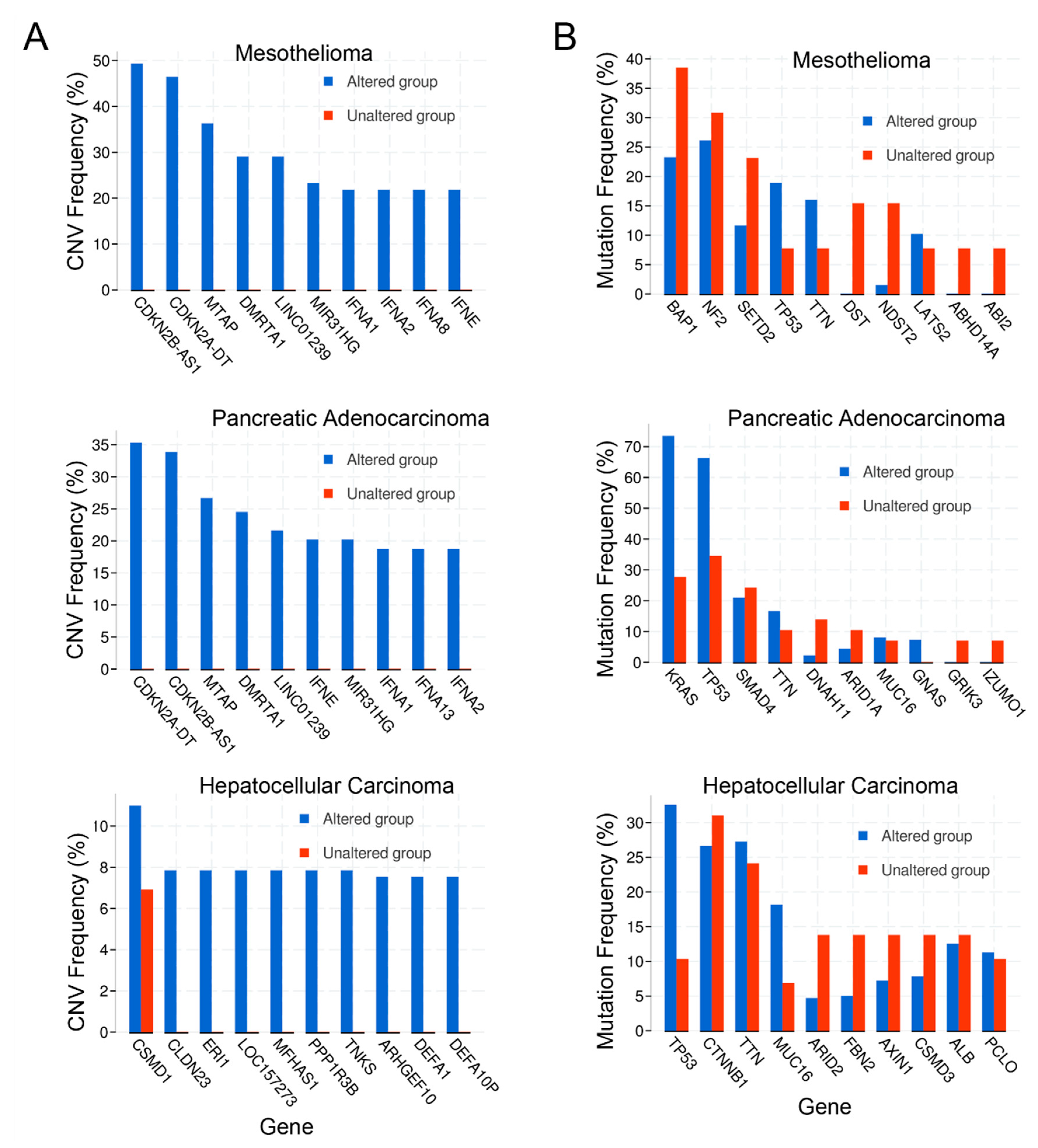
2.4. Copy Number Alterations
2.5. Mutations
3. Discussion
4. Conclusions
5. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Vaccaro, V.; Sperduti, I.; Milella, M. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 365, 768–769. [Google Scholar]
- Vogelzang, N.J.; Rusthoven, J.J.; Symanowski, J.; Denham, C.; Kaukel, E.; Ruffie, P.; Gatzemeier, U.; Boyer, M.; Emri, S.; Manegold, C.; et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2003, 21, 2636–2644. [Google Scholar] [CrossRef]
- De Gooijer, C.J.; Baas, P.; Burgers, J.A. Current chemotherapy strategies in malignant pleural mesothelioma. Transl. Lung Cancer Res. 2018, 7, 574–583. [Google Scholar] [CrossRef]
- Ikeda, M.; Morizane, C.; Ueno, M.; Okusaka, T.; Ishii, H.; Furuse, J. Chemotherapy for hepatocellular carcinoma: Current status and future perspectives. Jpn. J. Clin. Oncol. 2018, 48, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Springfeld, C.; Jager, D.; Buchler, M.W.; Strobel, O.; Hackert, T.; Palmer, D.H.; Neoptolemos, J.P. Chemotherapy for pancreatic cancer. Presse Med. 2019, 48, e159–e174. [Google Scholar] [CrossRef] [PubMed]
- Zalcman, G.; Mazieres, J.; Margery, J.; Greillier, L.; Audigier-Valette, C.; Moro-Sibilot, D.; Molinier, O.; Corre, R.; Monnet, I.; Gounant, V.; et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): A randomised, controlled, open-label, phase 3 trial. Lancet 2016, 387, 1405–1414. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Alley, E.W.; Lopez, J.; Santoro, A.; Morosky, A.; Saraf, S.; Piperdi, B.; van Brummelen, E. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): Preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2017, 18, 623–630. [Google Scholar] [CrossRef]
- Zhu, A.X.; Finn, R.S.; Edeline, J.; Cattan, S.; Ogasawara, S.; Palmer, D.; Verslype, C.; Zagonel, V.; Fartoux, L.; Vogel, A.; et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): A non-randomised, open-label phase 2 trial. Lancet Oncol. 2018, 19, 940–952. [Google Scholar] [CrossRef]
- Benson, C.; Kaye, S.; Workman, P.; Garrett, M.; Walton, M.; de Bono, J. Clinical anticancer drug development: Targeting the cyclin-dependent kinases. Br. J. Cancer 2005, 92, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Frizelle, S.P.; Grim, J.; Zhou, J.; Gupta, P.; Curiel, D.T.; Geradts, J.; Kratzke, R.A. Re-expression of p16INK4a in mesothelioma cells results in cell cycle arrest, cell death, tumor suppression and tumor regression. Oncogene 1998, 16, 3087–3095. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Diehl, J.A. Nuclear cyclin D1: An oncogenic driver in human cancer. J. Cell. Physiol. 2009, 220, 292–296. [Google Scholar] [CrossRef]
- Choi, Y.J.; Anders, L. Signaling through cyclin D-dependent kinases. Oncogene 2014, 33, 1890–1903. [Google Scholar] [CrossRef]
- Cadoo, K.A.; Gucalp, A.; Traina, T.A. Palbociclib: An evidence-based review of its potential in the treatment of breast cancer. Breast Cancer 2014, 6, 123–133. [Google Scholar]
- De Assis, L.V.; Locatelli, J.; Isoldi, M.C. The role of key genes and pathways involved in the tumorigenesis of Malignant Mesothelioma. Biochim. Biophys. Acta 2014, 1845, 232–247. [Google Scholar] [CrossRef]
- Cen, L.; Carlson, B.L.; Schroeder, M.A.; Ostrem, J.L.; Kitange, G.J.; Mladek, A.C.; Fink, S.R.; Decker, P.A.; Wu, W.; Kim, J.S.; et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol. 2012, 14, 870–881. [Google Scholar] [CrossRef]
- Brautigam, L.; Schutte, L.D.; Godoy, J.R.; Prozorovski, T.; Gellert, M.; Hauptmann, G.; Holmgren, A.; Lillig, C.H.; Berndt, C. Vertebrate-specific glutaredoxin is essential for brain development. Proc. Natl. Acad. Sci. USA 2011, 108, 20532–20537. [Google Scholar] [CrossRef]
- Nonn, L.; Williams, R.R.; Erickson, R.P.; Powis, G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 2003, 23, 916–922. [Google Scholar] [CrossRef]
- Perez, V.I.; Lew, C.M.; Cortez, L.A.; Webb, C.R.; Rodriguez, M.; Liu, Y.; Qi, W.; Li, Y.; Chaudhuri, A.; Van Remmen, H.; et al. Thioredoxin 2 haploinsufficiency in mice results in impaired mitochondrial function and increased oxidative stress. Free Radic. Biol. Med. 2008, 44, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.M.; Zhang, H.; Jones, D.P. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol. Sci. 2006, 91, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Benhar, M.; Forrester, M.T.; Hess, D.T.; Stamler, J.S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 2008, 320, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Hosoi, F.; Yamaguchi-Iwai, Y.; Nakamura, H.; Masutani, H.; Ueda, S.; Nishiyama, A.; Takeda, S.; Wada, H.; Spyrou, G.; et al. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 2002, 21, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- Damdimopoulos, A.E.; Miranda-Vizuete, A.; Pelto-Huikko, M.; Gustafsson, J.A.; Spyrou, G. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J. Biol. Chem. 2002, 277, 33249–33257. [Google Scholar] [CrossRef] [PubMed]
- Rigobello, M.P.; Folda, A.; Baldoin, M.C.; Scutari, G.; Bindoli, A. Effect of auranofin on the mitochondrial generation of hydrogen peroxide. Role of thioredoxin reductase. Free Radic. Res. 2005, 39, 687–695. [Google Scholar] [CrossRef]
- Cox, A.G.; Brown, K.K.; Arner, E.S.; Hampton, M.B. The thioredoxin reductase inhibitor auranofin triggers apoptosis through a Bax/Bak-dependent process that involves peroxiredoxin 3 oxidation. Biochem. Pharmacol. 2008, 76, 1097–1109. [Google Scholar] [CrossRef]
- Schuh, E.; Pfluger, C.; Citta, A.; Folda, A.; Rigobello, M.P.; Bindoli, A.; Casini, A.; Mohr, F. Gold(I) carbene complexes causing thioredoxin 1 and thioredoxin 2 oxidation as potential anticancer agents. J. Med. Chem. 2012, 55, 5518–5528. [Google Scholar] [CrossRef]
- Okrah, K.; Tarighat, S.; Liu, B.; Koeppen, H.; Wagle, M.C.; Cheng, G.; Sun, C.; Dey, A.; Chang, M.T.; Sumiyoshi, T.; et al. Transcriptomic analysis of hepatocellular carcinoma reveals molecular features of disease progression and tumor immune biology. NPJ Precis. Oncol. 2018, 2, 25. [Google Scholar] [CrossRef]
- Zhang, X.; De Milito, A.; Demiroglu-Zergeroglu, A.; Gullbo, J.; D’Arcy, P.; Linder, S. Eradicating Quiescent Tumor Cells by Targeting Mitochondrial Bioenergetics. Trends Cancer 2016, 2, 657–663. [Google Scholar] [CrossRef]
- Zhang, X.; Fryknas, M.; Hernlund, E.; Fayad, W.; De Milito, A.; Olofsson, M.H.; Gogvadze, V.; Dang, L.; Pahlman, S.; Schughart, L.A.; et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat. Commun. 2014, 5, 3295. [Google Scholar] [CrossRef]
- Bollard, J.; Miguela, V.; Ruiz de Galarreta, M.; Venkatesh, A.; Bian, C.B.; Roberto, M.P.; Tovar, V.; Sia, D.; Molina-Sanchez, P.; Nguyen, C.B.; et al. Palbociclib (PD-0332991), a selective CDK4/6 inhibitor, restricts tumour growth in preclinical models of hepatocellular carcinoma. Gut 2016, 66, 1286–1296. [Google Scholar] [CrossRef]
- Franco, J.; Balaji, U.; Freinkman, E.; Witkiewicz, A.K.; Knudsen, E.S. Metabolic Reprogramming of Pancreatic Cancer Mediated by CDK4/6 Inhibition Elicits Unique Vulnerabilities. Cell Rep. 2016, 14, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Bonelli, M.A.; Digiacomo, G.; Fumarola, C.; Alfieri, R.; Quaini, F.; Falco, A.; Madeddu, D.; La Monica, S.; Cretella, D.; Ravelli, A.; et al. Combined Inhibition of CDK4/6 and PI3K/AKT/mTOR Pathways Induces a Synergistic Anti-Tumor Effect in Malignant Pleural Mesothelioma Cells. Neoplasia 2017, 19, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Rotblat, B.; Grunewald, T.G.; Leprivier, G.; Melino, G.; Knight, R.A. Anti-oxidative stress response genes: Bioinformatic analysis of their expression and relevance in multiple cancers. Oncotarget 2013, 4, 2577–2590. [Google Scholar] [CrossRef] [PubMed]
- Song, I.S.; Jeong, Y.J.; Han, J. Mitochondrial metabolism in cancer stem cells: A therapeutic target for colon cancer. BMB Rep. 2015, 48, 539–540. [Google Scholar] [CrossRef]
- Anders, L.; Ke, N.; Hydbring, P.; Choi, Y.J.; Widlund, H.R.; Chick, J.M.; Zhai, H.; Vidal, M.; Gygi, S.P.; Braun, P.; et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 2011, 20, 620–634. [Google Scholar] [CrossRef]
- Roder, C.; Thomson, M.J. Auranofin: Repurposing an old drug for a golden new age. Drugs R D 2015, 15, 13–20. [Google Scholar] [CrossRef]
- Wang, Z.; Jensen, M.A.; Zenklusen, J.C. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol. Biol. 2016, 1418, 111–141. [Google Scholar]
- The Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research, N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017, 32, 185–203. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research, N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef] [PubMed]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- De Rienzo, A.; Archer, M.A.; Yeap, B.Y.; Dao, N.; Sciaranghella, D.; Sideris, A.C.; Zheng, Y.; Holman, A.G.; Wang, Y.E.; Dal Cin, P.S.; et al. Gender-Specific Molecular and Clinical Features Underlie Malignant Pleural Mesothelioma. Cancer Res. 2016, 76, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, E.; Castelli, G.; Testa, U. Pancreatic Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Biomedicines 2017, 5, 65. [Google Scholar] [CrossRef]
- Weinberg, F.; Hamanaka, R.; Wheaton, W.W.; Weinberg, S.; Joseph, J.; Lopez, M.; Kalyanaraman, B.; Mutlu, G.M.; Budinger, G.R.; Chandel, N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA 2010, 107, 8788–8793. [Google Scholar] [CrossRef]
- Piskounova, E.; Agathocleous, M.; Murphy, M.M.; Hu, Z.; Huddlestun, S.E.; Zhao, Z.; Leitch, A.M.; Johnson, T.M.; DeBerardinis, R.J.; Morrison, S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015, 527, 186–191. [Google Scholar] [CrossRef]
- Sarosiek, K.A.; Ni Chonghaile, T.; Letai, A. Mitochondria: Gatekeepers of response to chemotherapy. Trends Cell Biol. 2013, 23, 612–619. [Google Scholar] [CrossRef]
- Funes, J.M.; Quintero, M.; Henderson, S.; Martinez, D.; Qureshi, U.; Westwood, C.; Clements, M.O.; Bourboulia, D.; Pedley, R.B.; Moncada, S.; et al. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc. Natl. Acad. Sci. USA 2007, 104, 6223–6228. [Google Scholar] [CrossRef]
- Le, A.; Stine, Z.E.; Nguyen, C.; Afzal, J.; Sun, P.; Hamaker, M.; Siegel, N.M.; Gouw, A.M.; Kang, B.H.; Yu, S.H.; et al. Tumorigenicity of hypoxic respiring cancer cells revealed by a hypoxia-cell cycle dual reporter. Proc. Natl. Acad. Sci. USA 2014, 111, 12486–12491. [Google Scholar] [CrossRef] [PubMed]
- Policastro, L.L.; Ibanez, I.L.; Notcovich, C.; Duran, H.A.; Podhajcer, O.L. The tumor microenvironment: Characterization, redox considerations, and novel approaches for reactive oxygen species-targeted gene therapy. Antioxid. Redox Signal. 2013, 19, 854–895. [Google Scholar] [CrossRef] [PubMed]
- Smolkova, K.; Plecita-Hlavata, L.; Bellance, N.; Benard, G.; Rossignol, R.; Jezek, P. Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int. J. Biochem. Cell Biol. 2011, 43, 950–968. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.F.; Yao, K.; Du, X.; Li, Y.; Yang, X.Y.; Yu, M.; Li, M.Z.; Cui, Q.H. PGC-1alpha regulates the cell cycle through ATP and ROS in CH1 cells. J. Zhejiang Univ. Sci. B 2016, 17, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Freije, W.A.; Guptan, P.; Banerjee, U. Metabolic control of G1-S transition: Cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J. Cell Biol. 2010, 188, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Qin, L.; Shi, Y.; Candas, D.; Fan, M.; Lu, C.L.; Vaughan, A.T.; Shen, R.; Wu, L.S.; Liu, R.; et al. CDK4-mediated MnSOD activation and mitochondrial homeostasis in radioadaptive protection. Free Radic. Biol. Med. 2015, 81, 77–87. [Google Scholar] [CrossRef]
- Wolf, D.A. Is reliance on mitochondrial respiration a “chink in the armor” of therapy-resistant cancer? Cancer Cell 2014, 26, 788–795. [Google Scholar] [CrossRef]
- Sarsour, E.H.; Kalen, A.L.; Goswami, P.C. Manganese superoxide dismutase regulates a redox cycle within the cell cycle. Antioxid. Redox Signal. 2014, 20, 1618–1627. [Google Scholar] [CrossRef]
- Woodcock, J.; Woosley, R. The FDA critical path initiative and its influence on new drug development. Annu. Rev. Med. 2008, 59, 1–12. [Google Scholar] [CrossRef]
- Pessetto, Z.Y.; Weir, S.J.; Sethi, G.; Broward, M.A.; Godwin, A.K. Drug repurposing for gastrointestinal stromal tumor. Mol. Cancer Ther. 2013, 12, 1299–1309. [Google Scholar] [CrossRef]
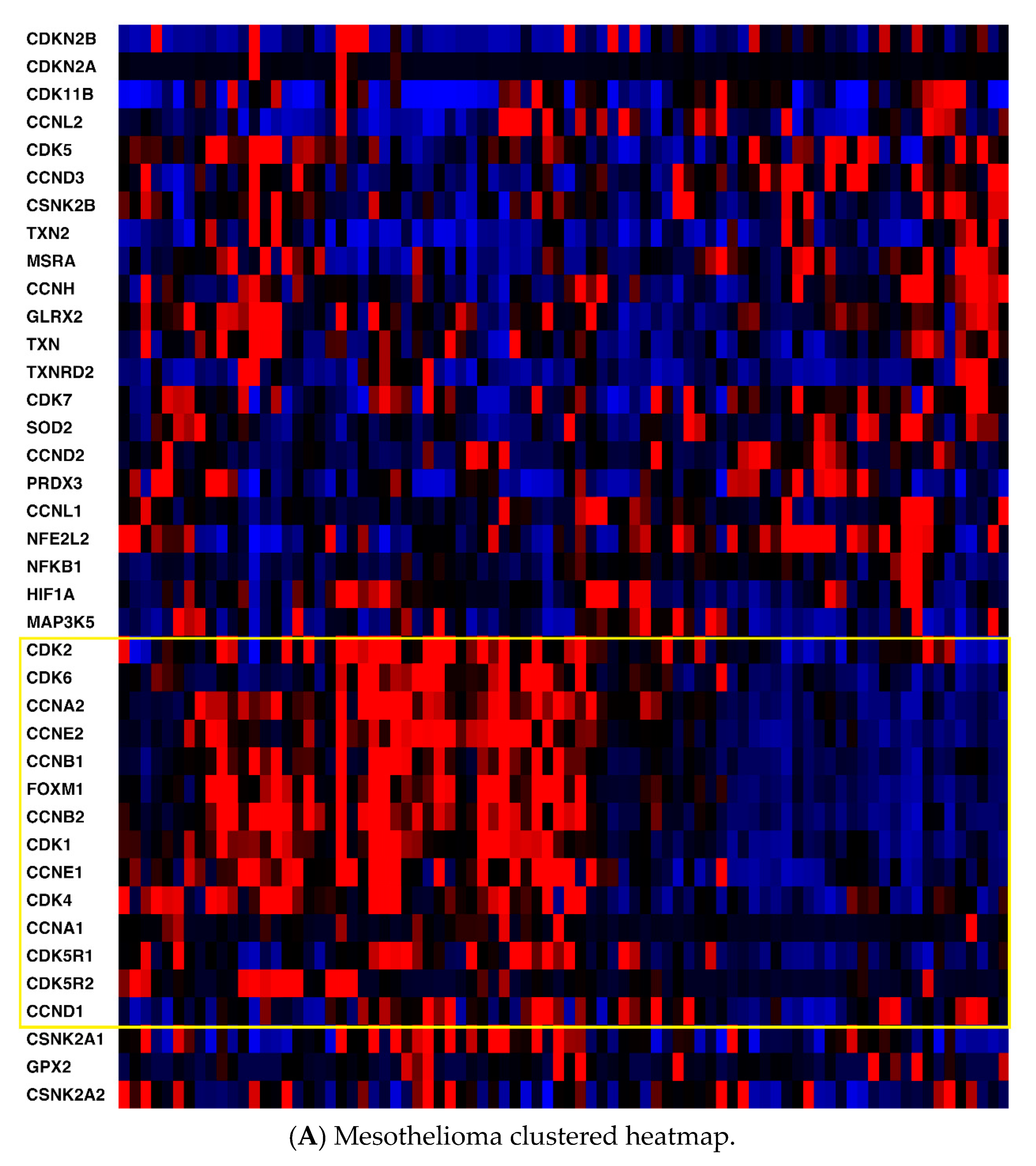
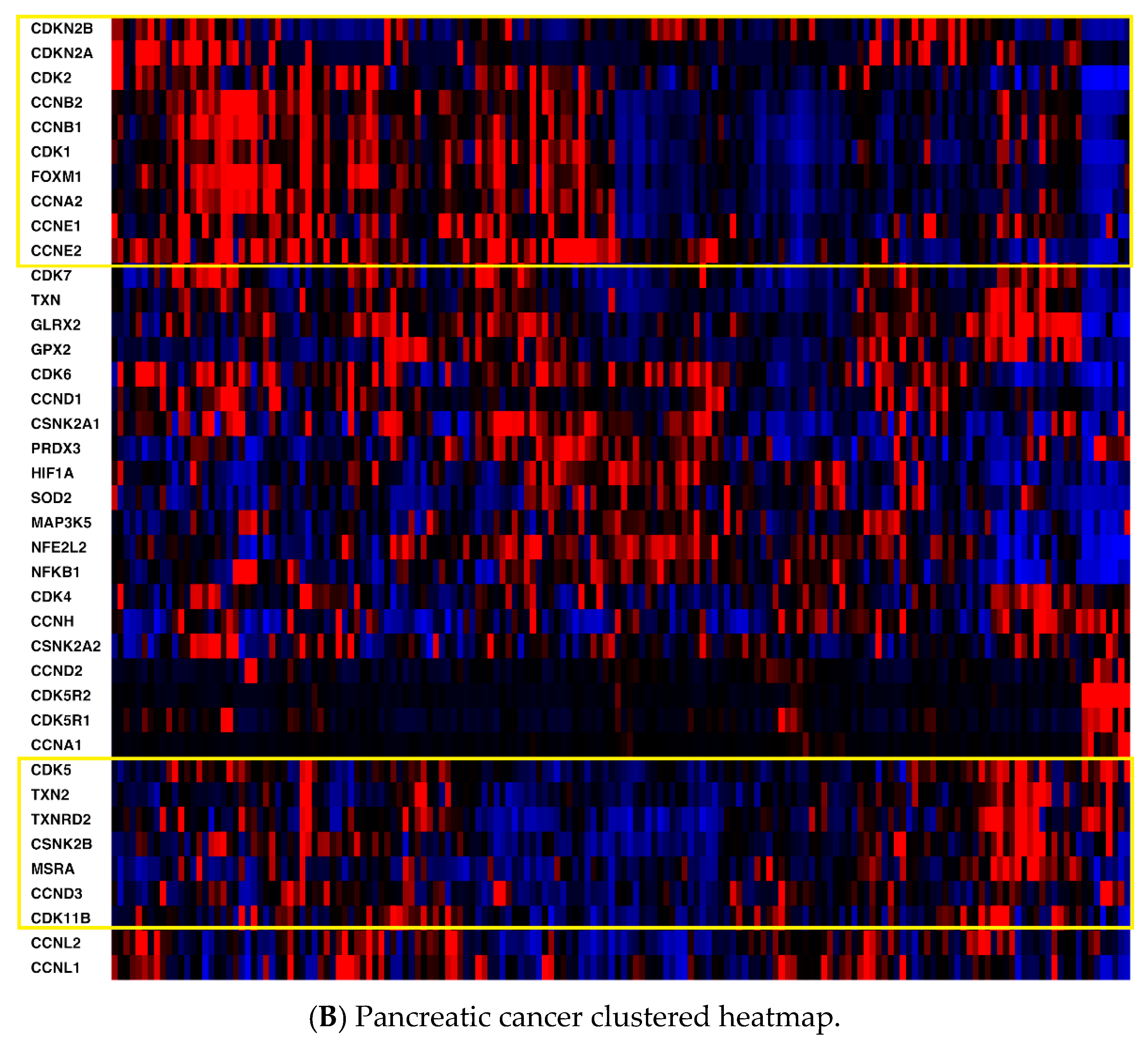
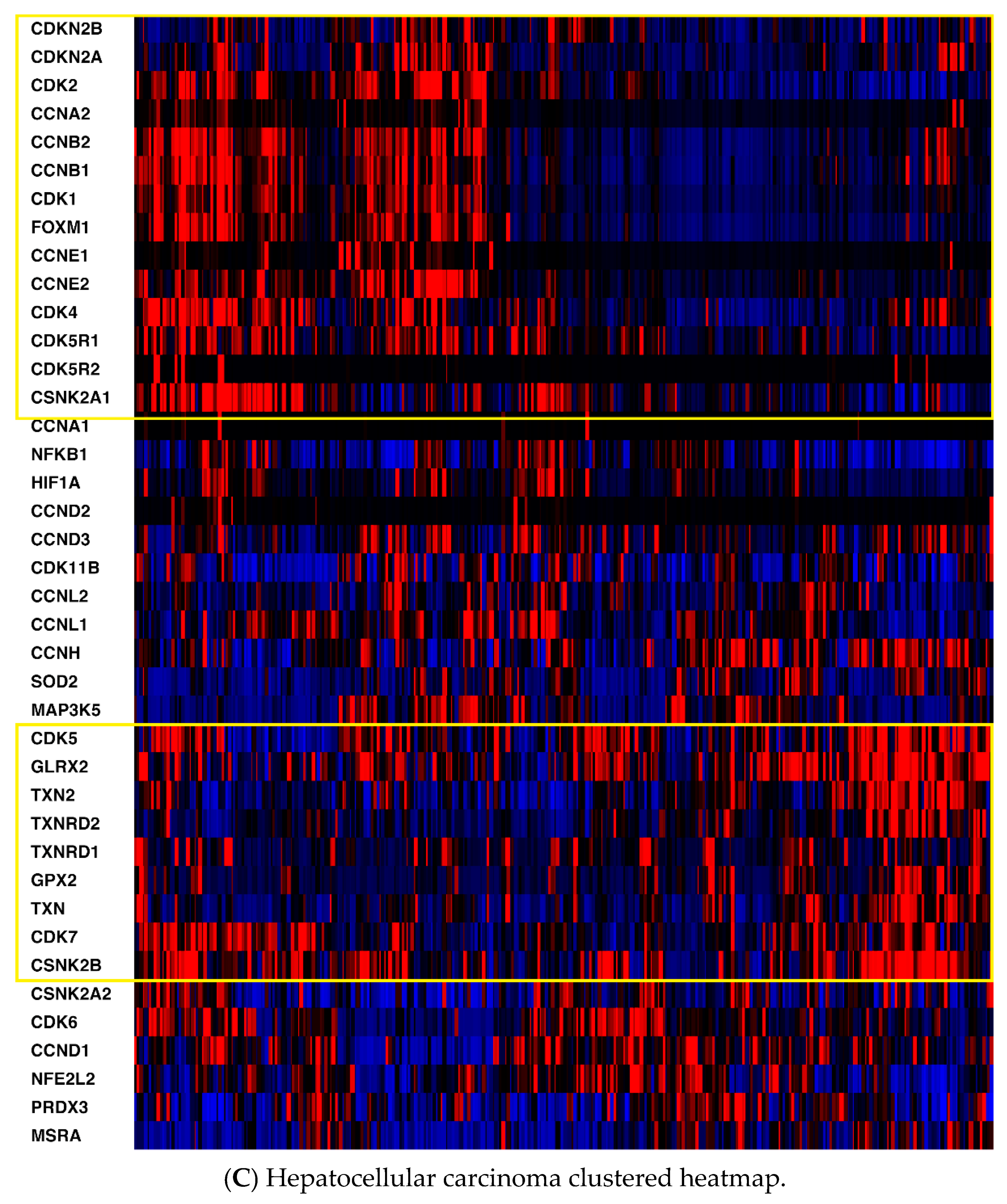
| CDKN2A | CDKN2B | CDK1 | CDK2 | CDK4 | CDK5 | CDK6 | CDK7 |
| CDK11B | CCNA1 | CCNA2 | CCNB1 | CCNB2 | CCND1 | CCND2 | CCND3 |
| CCNE1 | CCNE2 | CDK5R1 | CDK5R2 | CCNH | CCNL1 | CCNL2 | FOXM1 |
| TXN | TXNRD1 | TXN2 | TXNRD2 | PRDX3 | GLRX2 | GPX2 | SOD2 |
| MSRA | HIF1A | NFKB1 | CSNK2A1 | CSNK2A2 | CSNK2B | NFE2L2 | MAP3K5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scaria, G.S.; Kren, B.T.; Klein, M.A. Cyclin-Dependent Kinase and Antioxidant Gene Expression in Cancers with Poor Therapeutic Response. Pharmaceuticals 2020, 13, 26. https://doi.org/10.3390/ph13020026
Scaria GS, Kren BT, Klein MA. Cyclin-Dependent Kinase and Antioxidant Gene Expression in Cancers with Poor Therapeutic Response. Pharmaceuticals. 2020; 13(2):26. https://doi.org/10.3390/ph13020026
Chicago/Turabian StyleScaria, George S., Betsy T. Kren, and Mark A. Klein. 2020. "Cyclin-Dependent Kinase and Antioxidant Gene Expression in Cancers with Poor Therapeutic Response" Pharmaceuticals 13, no. 2: 26. https://doi.org/10.3390/ph13020026
APA StyleScaria, G. S., Kren, B. T., & Klein, M. A. (2020). Cyclin-Dependent Kinase and Antioxidant Gene Expression in Cancers with Poor Therapeutic Response. Pharmaceuticals, 13(2), 26. https://doi.org/10.3390/ph13020026