An Integrated In Silico and In Vivo Approach to Identify Protective Effects of Palonosetron in Cisplatin-Induced Nephrotoxicity
Abstract
1. Introduction
2. Results
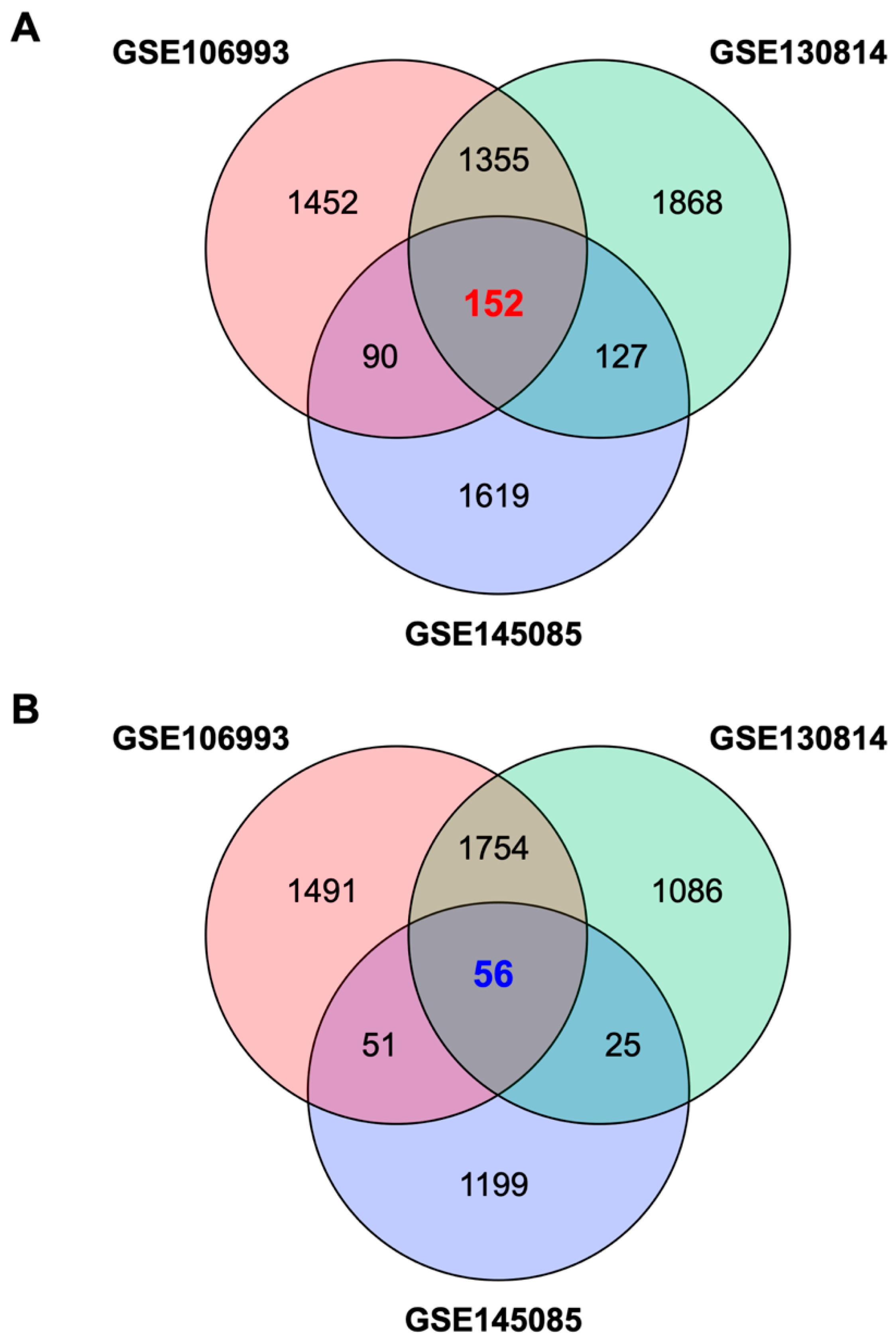
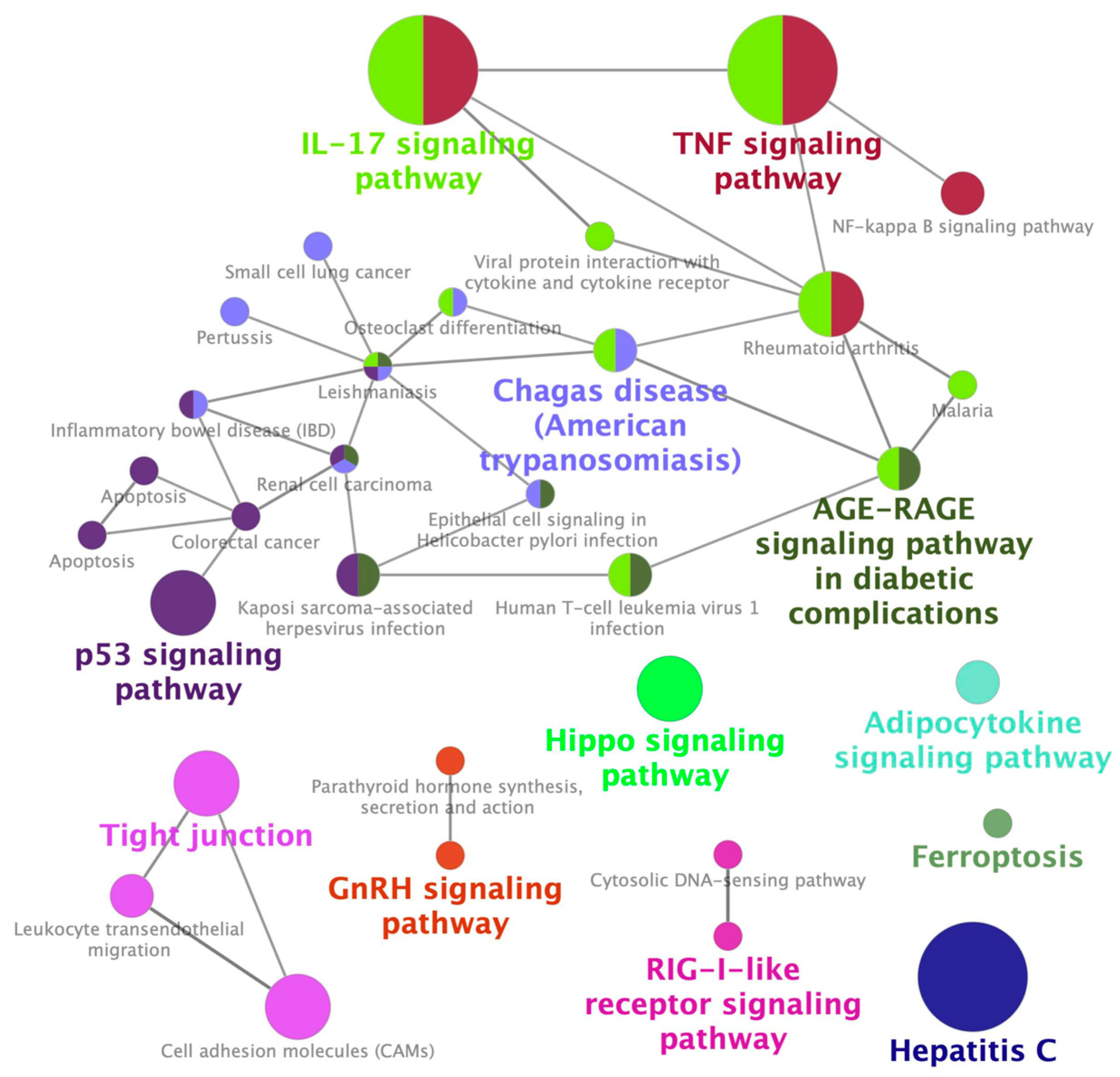
2.1. Identification of a Gene Expression Signature Associated with Cisplatin-Induced Nephrotoxicity
2.2. Identification of Palonosetron as a Potential Therapeutic Drug for CIN
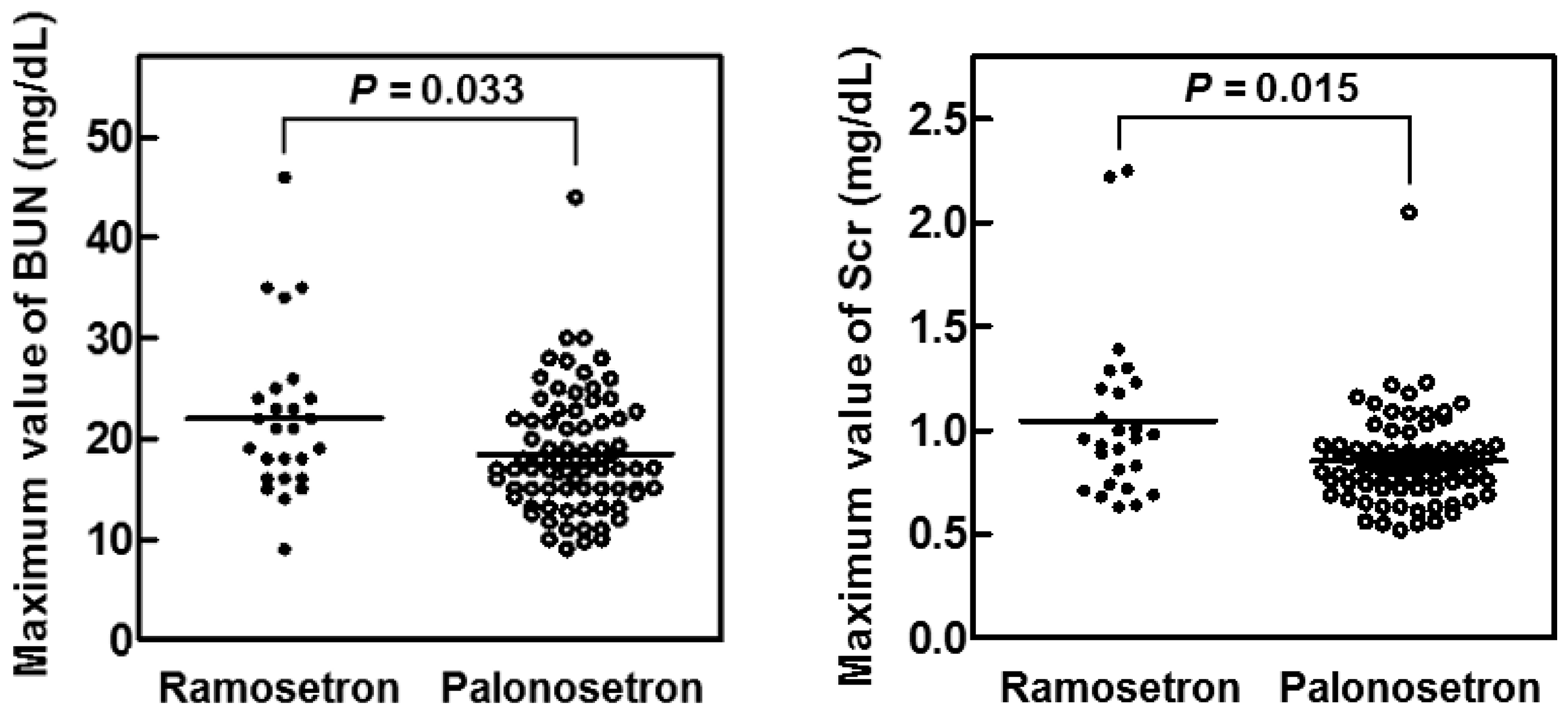
2.3. Suppression of CIN in Patients with Head and Neck Cancer by Palonosetron
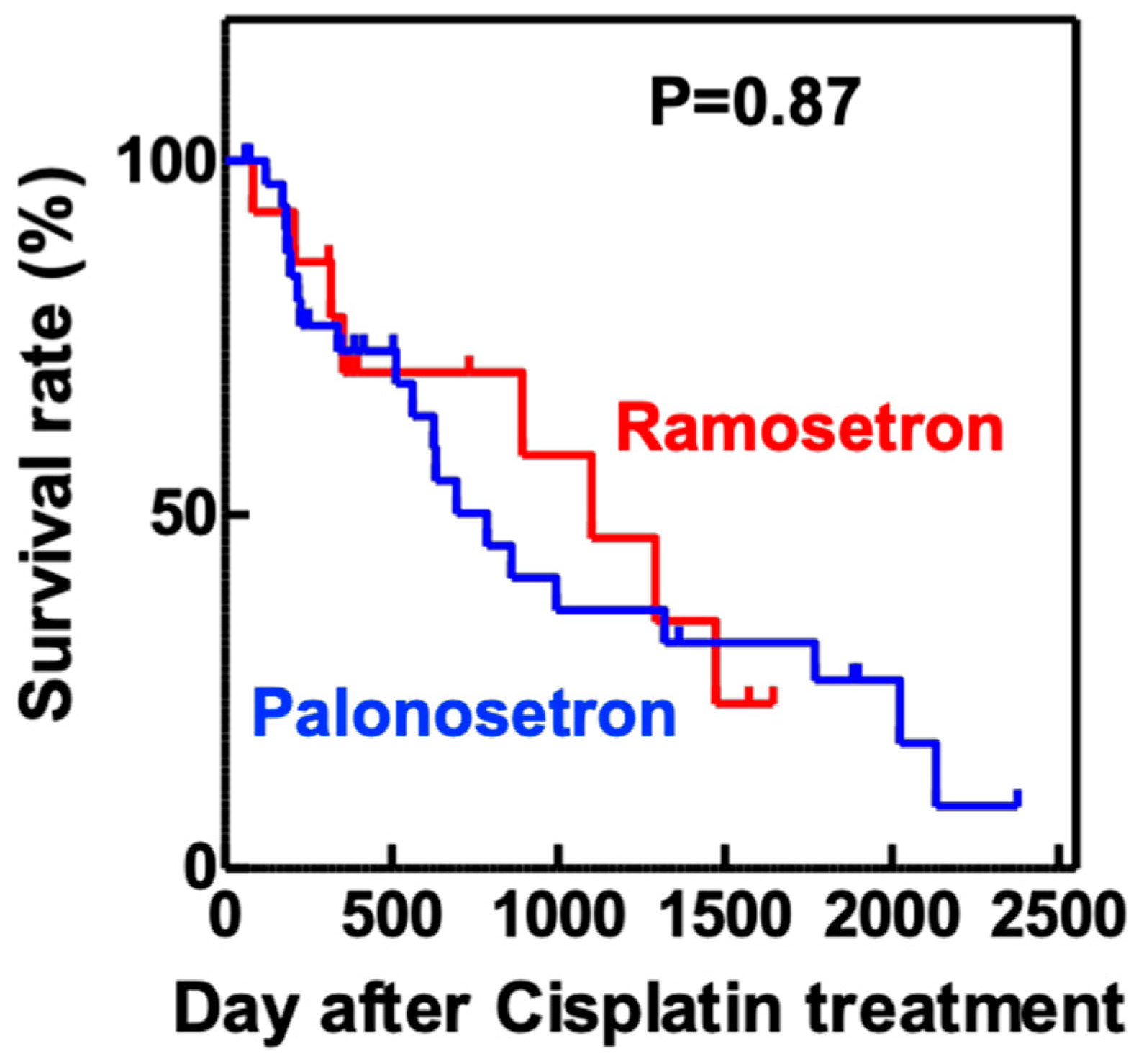
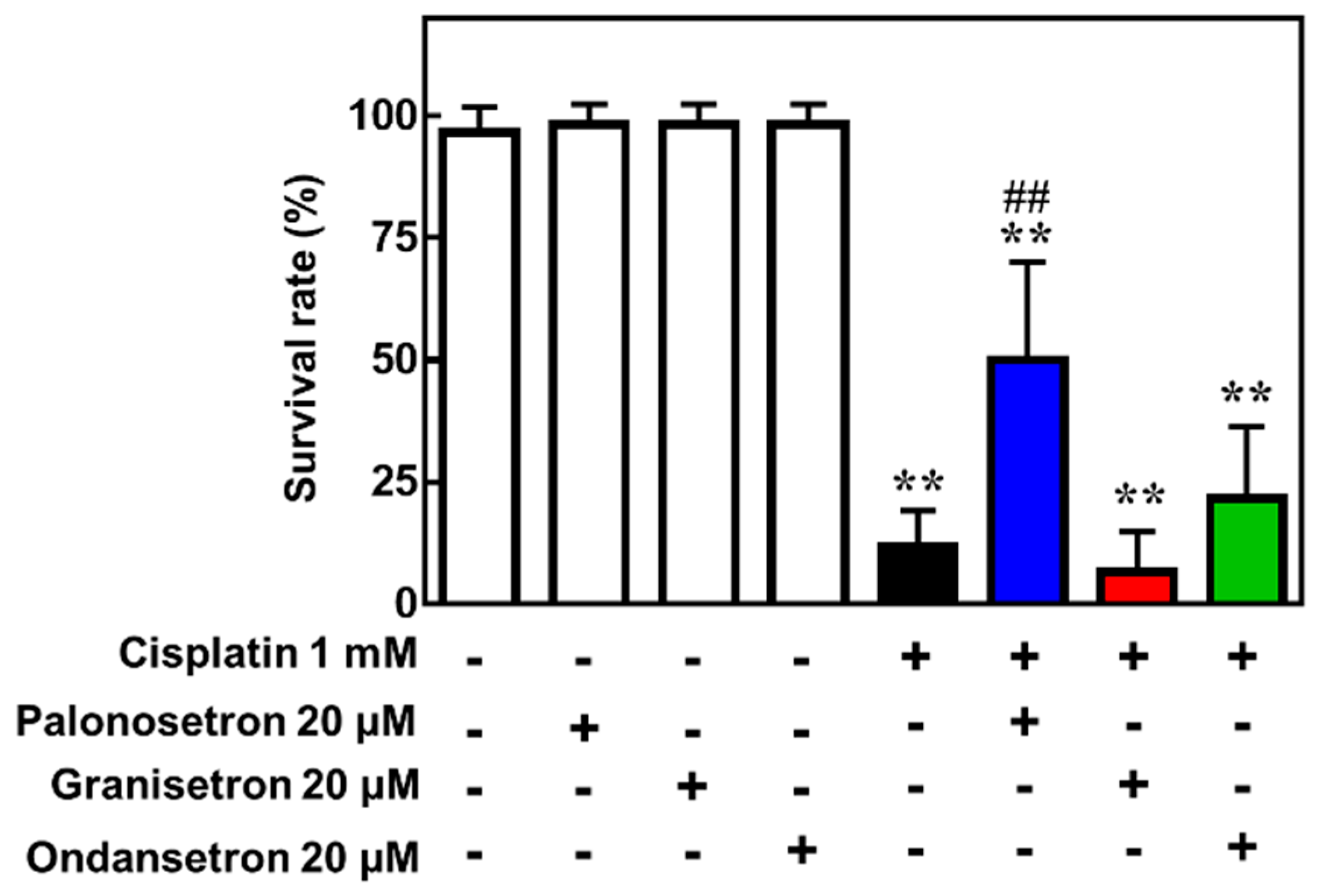
2.4. Palonosetron Treatment Increases the Survival of Cisplatin-Exposed Zebrafish
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Transcriptome Analysis
4.3. Bio/Chemoinformatics Analysis
4.4. Analysis of Electronic Medical Records
4.5. Zebrafish Experiments
4.6. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef] [PubMed]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Perazella, M.A. Pharmacology behind Common Drug Nephrotoxicities. Clin. J. Am. Soc. Nephrol. 2018, 13, 1897–1908. [Google Scholar] [CrossRef]
- Oda, H.; Mizuno, T.; Ikejiri, M.; Nakamura, M.; Tsunoda, A.; Ishihara, M.; Saito, K.; Tamaru, S.; Yamashita, Y.; Nishimura, Y.; et al. Risk factors for cisplatin-induced acute kidney injury: A pilot study on the usefulness of genetic variants for predicting nephrotoxicity in clinical practice. Mol. Clin. Oncol. 2020, 13, 58. [Google Scholar] [CrossRef]
- Hiramatsu, S.I.; Ikemura, K.; Fujisawa, Y.; Iwamoto, T.; Okuda, M. Concomitant lansoprazole ameliorates cisplatin-induced nephrotoxicity by inhibiting renal organic cation transporter 2 in rats. Biopharm. Drug Dispos. 2020, 41, 239–247. [Google Scholar] [CrossRef]
- Holditch, S.J.; Brown, C.N.; Lombardi, A.M.; Nguyen, K.N.; Edelstein, C.L. Recent Advances in Models, Mechanisms, Biomarkers, and Interventions in Cisplatin-Induced Acute Kidney Injury. Int. J. Mol. Sci. 2019, 20, 3011. [Google Scholar] [CrossRef]
- Volarevic, V.; Djokovic, B.; Jankovic, M.G.; Harrell, C.R.; Fellabaum, C.; Djonov, V.; Arsenijevic, N. Molecular mechanisms of cisplatin-induced nephrotoxicity: A balance on the knife edge between renoprotection and tumor toxicity. J. Biomed. Sci. 2019, 26, 25. [Google Scholar] [CrossRef]
- Crona, D.J.; Faso, A.; Nishijima, T.F.; McGraw, K.A.; Galsky, M.D.; Milowsky, M.I. A Systematic Review of Strategies to Prevent Cisplatin-Induced Nephrotoxicity. Oncologist 2017, 22, 609–619. [Google Scholar] [CrossRef]
- Casanova, A.G.; Hernández-Sánchez, M.T.; López-Hernández, F.J.; Martinez-Salgado, C.; Prieto, M.; Vicente-Vicente, L.; Morales, A.I. Systematic review and meta-analysis of the efficacy of clinically tested protectants of cisplatin nephrotoxicity. Eur. J. Clin. Pharmacol. 2020, 76, 23–33. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Hara, H. Editorial: Drug Repositioning: Current Advances and Future Perspectives. Front. Pharmacol. 2018, 9, 1068. [Google Scholar] [CrossRef] [PubMed]
- Ikemura, K.; Hiramatsu, S.; Okuda, M. Drug Repositioning of Proton Pump Inhibitors for Enhanced Efficacy and Safety of Cancer Chemotherapy. Front. Pharmacol. 2017, 8, 911. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Tagawa, M.; Ito, H.; Tsuruma, K.; Hara, H. Overcoming Obstacles to Drug Repositioning in Japan. Front. Pharmacol. 2017, 8, 729. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef]
- Montero-Melendez, T.; Perretti, M. Connections in pharmacology: Innovation serving translational medicine. Drug Discov. Today 2014, 19, 820–823. [Google Scholar] [CrossRef]
- Musa, A.; Ghoraie, L.S.; Zhang, S.D.; Glazko, G.; Yli-Harja, O.; Dehmer, M.; Haibe-Kains, B.; Emmert-Streib, F. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2018, 19, 506–523. [Google Scholar]
- Gns, H.S.; Gr, S.; Murahari, M.; Krishnamurthy, M. An update on Drug Repurposing: Re-written saga of the drug’s fate. Biomed. Pharmacother. 2019, 110, 700–716. [Google Scholar] [CrossRef]
- Späth, M.R.; Bartram, M.P.; Palacio-Escat, N.; Hoyer, K.J.R.; Debes, C.; Demir, F.; Schroeter, C.B.; Mandel, A.M.; Grundmann, F.; Ciarimboli, G.; et al. The proteome microenvironment determines the protective effect of preconditioning in cisplatin-induced acute kidney injury. Kidney Int. 2019, 95, 333–349. [Google Scholar] [CrossRef]
- Cao, Y.; Mi, X.; Zhang, D.; Wang, Z.; Zuo, Y.; Tang, W. Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury. Clin. Sci. 2020, 134, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Digby, J.L.M.; Vanichapol, T.; Przepiorski, A.; Davidson, A.J.; Sander, V. Evaluation of cisplatin-induced injury in human kidney organoids. Am. J. Physiol. Renal. Physiol. 2020, 318, F971–F978. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lachmann, A.; Ma’ayan, A. Mining data and metadata from the gene expression omnibus. Biophys. Rev. 2019, 11, 103–110. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J.; Molina-Jijón, E.; Medina-Campos, O.N.; Rodriguez-Munoz, R.; Reyes, J.L.; Loredo, M.L.; Barrera-Oviedo, D.; Pinzon, E.; Rodriguez-Rangel, D.S.; Pedraza-Chaverri, J. Curcumin prevents cisplatin-induced decrease in the tight and adherens junctions: Relation to oxidative stress. Food Funct. 2016, 7, 279–293. [Google Scholar] [CrossRef] [PubMed]
- FAERS. FDA Adverse Event Reporting System. Available online: https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-latest-quarterly-data-files (accessed on 19 December 2020).
- Nagashima, T.; Shirakawa, H.; Nakagawa, T.; Kaneko, S. Prevention of antipsychotic-induced hyperglycaemia by vitamin D: A data mining prediction followed by experimental exploration of the molecular mechanism. Sci. Rep. 2016, 6, 26375. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Niimura, T.; Zamami, Y.; Hamano, H.; Ishida, S.; Goda, M.; Takechi, K.; Chuma, M.; Imanishi, M.; Ishizawa, K. Pharmacovigilance evaluation of the relationship between impaired glucose metabolism and BCR-ABL inhibitor use by using an adverse drug event reporting database. Cancer Med. 2019, 8, 174–181. [Google Scholar] [CrossRef]
- Ikemura, K.; Hiramatsu, S.I.; Shinogi, Y.; Nakatani, Y.; Tawara, I.; Iwamoto, T.; Katayama, N.; Okuda, M. Concomitant febuxostat enhances methotrexate-induced hepatotoxicity by inhibiting breast cancer resistance protein. Sci. Rep. 2019, 9, 20359. [Google Scholar] [CrossRef]
- Sanger, G.J.; Andrews, P.L.R. A History of Drug Discovery for Treatment of Nausea and Vomiting and the Implications for Future Research. Front. Pharmacol. 2018, 9, 913. [Google Scholar] [CrossRef]
- Darmani, N.A.; Zhong, W.; Chebolu, S.; Mercadante, F. Differential and additive suppressive effects of 5-HT3 (palonosetron)- and NK1 (netupitant)-receptor antagonists on cisplatin-induced vomiting and ERK1/2, PKA and PKC activation. Pharmacol. Biochem. Behav. 2015, 131, 104–111. [Google Scholar] [CrossRef]
- Rojas, C.; Li, Y.; Zhang, J.; Stathis, M.; Alt, J.; Thomas, A.G.; Cantoreggi, S.; Sebastiani, S.; Pietra, C.; Slusher, B.S. The antiemetic 5-HT3 receptor antagonist Palonosetron inhibits substance P-mediated responses in vitro and in vivo. J. Pharmacol. Exp. Ther. 2010, 335, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Un, H.; Ugan, R.A.; Kose, D.; Bayir, Y.; Cadirci, E.; Selli, J.; Halici, Z. A novel effect of Aprepitant: Protection for cisplatin-induced nephrotoxicity and hepatotoxicity. Eur. J. Pharmacol. 2020, 880, 173168. [Google Scholar] [CrossRef] [PubMed]
- Greenwood-Van Meerveld, B.; Mohammadi, E.; Tyler, K.; Pietra, C.; Bee, L.A.; Dickenson, A. Synergistic Effect of 5-Hydroxytryptamine 3 and Neurokinin 1 Receptor Antagonism in Rodent Models of Somatic and Visceral Pain. J. Pharmacol. Exp. Ther. 2014, 351, 146. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhu, W.; Liang, Q.; Liu, J.; Yang, X.; Sun, G. Tropisetron attenuates lipopolysaccharide induced neuroinflammation by inhibiting NF-κB and SP/NK1R signaling pathway. J. Neuroimmunol. 2018, 320, 80–86. [Google Scholar] [CrossRef]
- Zirak, M.R.; Rahimian, R.; Ghazi-Khansari, M.; Abbasi, A.; Razmi, A.; Mehr, S.E.; Mousavizadeh, K.; Dehpour, A.R. Tropisetron attenuates cisplatin-induced nephrotoxicity in mice. Eur. J. Pharmacol. 2014, 738, 222–229. [Google Scholar] [CrossRef]
- Minami, M.; Endo, T.; Yokota, H.; Ogawa, T.; Nemoto, M.; Hamaue, N.; Hirafuji, M.; Yoshioka, M.; Nagahisa, A.; Andrews, P.L.R. Effects of CP-99, 994, a tachykinin NK(1) receptor antagonist, on abdominal afferent vagal activity in ferrets: Evidence for involvement of NK(1) and 5-HT(3) receptors. Eur. J. Pharmacol. 2001, 428, 215–220. [Google Scholar] [CrossRef]
- Hu, W.-P.; You, X.-H.; Guan, B.-C.; Ru, L.-Q.; Chen, J.-G.; Li, Z.-W. Substance P potentiates 5-HT3 receptor-mediated current in rat trigeminal ganglion neurons. Neurosci. Lett. 2004, 365, 147–152. [Google Scholar] [CrossRef]
- Thomas, A.G.; Stathis, M.; Rojas, C.; Slusher, B.S. Netupitant and palonosetron trigger NK1 receptor internalization in NG108-15 cells. Exp. Brain Res. 2014, 232, 2637–2644. [Google Scholar] [CrossRef][Green Version]
- Kaneko, S.; Nagashima, T. Drug Repositioning and Target Finding Based on Clinical Evidence. Biol. Pharm. Bull. 2020, 43, 362–365. [Google Scholar] [CrossRef]
- Banda, J.M.; Evans, L.; Vanguri, R.S.; Tatonetti, N.P.; Ryan, P.B.; Shah, N.H. A curated and standardized adverse drug event resource to accelerate drug safety research. Sci. Data 2016, 3, 160026. [Google Scholar] [CrossRef]
- Nishimura, Y.; Okabe, S.; Sasagawa, S.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Tanaka, T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front. Pharmacol. 2015, 6, 257. [Google Scholar] [CrossRef] [PubMed]
- Uechi, T.; Kenmochi, N. Zebrafish Models of Diamond-Blackfan Anemia: A Tool for Understanding the Disease Pathogenesis and Drug Discovery. Pharmaceuticals 2019, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, J.; Shiromizu, T.; Adachi, Y.; Ikejiri, M.; Nakatani, K.; Tanaka, T.; Nishimura, Y. Generation of a Triple-Transgenic Zebrafish Line for Assessment of Developmental Neurotoxicity during Neuronal Differentiation. Pharmaceuticals 2019, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Kirchberger, S.; Sturtzel, C.; Pascoal, S.; Distel, M. Quo natas, Danio?-Recent Progress in Modeling Cancer in Zebrafish. Front. Oncol. 2017, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Moon, D.; Jung, S.; Lee, J.; Kim, J. Cisplatin nephrotoxicity is induced via poly(ADP-ribose) polymerase activation in adult zebrafish and mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020, 318, R843–R854. [Google Scholar] [CrossRef] [PubMed]
- Wertman, J.N.; Melong, N.; Stoyek, M.R.; Piccolo, O.; Langley, S.; Orr, B.; Steele, S.L.; Razaghi, B.; Berman, J.N. The identification of dual protective agents against cisplatin-induced oto- and nephrotoxicity using the zebrafish model. Elife 2020, 9, e56235. [Google Scholar] [CrossRef]
- Nowicki, M.; Tran, S.; Muraleetharan, A.; Markovic, S.; Gerlai, R. Serotonin antagonists induce anxiolytic and anxiogenic-like behavior in zebrafish in a receptor-subtype dependent manner. Pharmacol. Biochem. Behav. 2014, 126, 170–180. [Google Scholar] [CrossRef]
- Benneh, C.K.; Biney, R.P.; Mante, P.K.; Tandoh, A.; Adongo, D.W.; Woode, E. Maerua angolensis stem bark extract reverses anxiety and related behaviours in zebrafish-Involvement of GABAergic and 5-HT systems. J. Ethnopharmacol. 2017, 207, 129–145. [Google Scholar] [CrossRef]
- López-Bellido, R.; Barreto-Valer, K.; Rodríguez, R.E. Expression of tachykinin receptors (tacr1a and tacr1b) in zebrafish: Influence of cocaine and opioid receptors. J. Mol. Endocrinol. 2013, 50, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van den Beek, M.; Bouvier, D.; Vcech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Da, W.H.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Kelsh, R.N.; Brand, M.; Jiang, Y.J.; Heisenberg, C.; Lin, S.; Haffter, P.; Odenthal, J.; Mullins, M.C.; van Eeden, F.J.; Furutani-Seiki, M.; et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development 1996, 123, 369–389. [Google Scholar] [PubMed]
- Nishimura, Y.; Inoue, A.; Sasagawa, S.; Koiwa, J.; Kawaguchi, K.; Kawase, R.; Maruyama, T.; Kim, S.; Tanaka, T. Using zebrafish in systems toxicology for developmental toxicity testing. Congenit Anom. 2016, 56, 18–27. [Google Scholar] [CrossRef]
| Patients’ Characteristics | All Patients (n = 103) | Ramosetron (n = 26) | Palonosetron (n = 77) | p Value |
|---|---|---|---|---|
| Male | 88 (85) | 22 (85) | 66 (86) | 0.891 |
| Age (years) | 64 (33–78) | 66 (33–78) | 64 (34–76) | 0.111 |
| Body weight (kg) | 52 (34–85) | 52 (34–72) | 51 (34–85) | 0.499 |
| Body surface area (m2) | 1.56 (1.29–2.02) | 1.57 (1.29–1.85) | 1.56 (1.22–2.02) | 0.463 |
| Smoking history | 86 (83) | 21 (81) | 65 (84) | 0.606 |
| Drinking history | 67 (65) | 17 (65) | 50 (65) | 0.995 |
| 5-FU dose (mg/day) | 1260 (900–1640) | 1225 (1000–1500) | 1270 (900–1640) | 0.204 |
| Cisplatin dose (mg/day) | 125 (80–164) | 120 (95–150) | 125 (80–164) | 0.105 |
| Baseline biological parameters | ||||
| BUN (mg/dL) | 11.0 (6.5–19.0) | 12.0 (7.0–19.0) | 11 (6.5–19.0) | 0.445 |
| Scr (mg/dL) | 0.73 (0.44–1.02) | 0.77 (0.45–1.01) | 0.72 (0.44–1.02) | 0.538 |
| Hemoglobin (g/dL) | 12.4 (8.2–16.9) | 12.1 (8.9–15.1) | 12.5 (8.2–16.9) | 0.463 |
| Platelet (×109 L) | 220 (58–417) | 217 (82–407) | 220 (58–417) | 0.566 |
| White blood cells (×109 L) | 5.27 (2.10–14.9) | 6.20 (2.10–14.9) | 5.14 (2.21–14.4) | 0.477 |
| Co-administrated | ||||
| NSAIDs | 12 (12) | 4 (15) | 8 (10) | 0.396 |
| Magnesium oxide | 26 (25) | 7 (27) | 19 (25) | 0.776 |
| Proton pump inhibitors | 14 (14) | 4 (15) | 10 (13) | 0.632 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wakai, E.; Suzumura, Y.; Ikemura, K.; Mizuno, T.; Watanabe, M.; Takeuchi, K.; Nishimura, Y. An Integrated In Silico and In Vivo Approach to Identify Protective Effects of Palonosetron in Cisplatin-Induced Nephrotoxicity. Pharmaceuticals 2020, 13, 480. https://doi.org/10.3390/ph13120480
Wakai E, Suzumura Y, Ikemura K, Mizuno T, Watanabe M, Takeuchi K, Nishimura Y. An Integrated In Silico and In Vivo Approach to Identify Protective Effects of Palonosetron in Cisplatin-Induced Nephrotoxicity. Pharmaceuticals. 2020; 13(12):480. https://doi.org/10.3390/ph13120480
Chicago/Turabian StyleWakai, Eri, Yuya Suzumura, Kenji Ikemura, Toshiro Mizuno, Masatoshi Watanabe, Kazuhiko Takeuchi, and Yuhei Nishimura. 2020. "An Integrated In Silico and In Vivo Approach to Identify Protective Effects of Palonosetron in Cisplatin-Induced Nephrotoxicity" Pharmaceuticals 13, no. 12: 480. https://doi.org/10.3390/ph13120480
APA StyleWakai, E., Suzumura, Y., Ikemura, K., Mizuno, T., Watanabe, M., Takeuchi, K., & Nishimura, Y. (2020). An Integrated In Silico and In Vivo Approach to Identify Protective Effects of Palonosetron in Cisplatin-Induced Nephrotoxicity. Pharmaceuticals, 13(12), 480. https://doi.org/10.3390/ph13120480







