Analytical Methods for the Detection and Quantification of ADCs in Biological Matrices
Abstract
1. Introduction
2. Label-Free Bioanalytical Methods
2.1. Ligand-Binding Assays (LBAs)
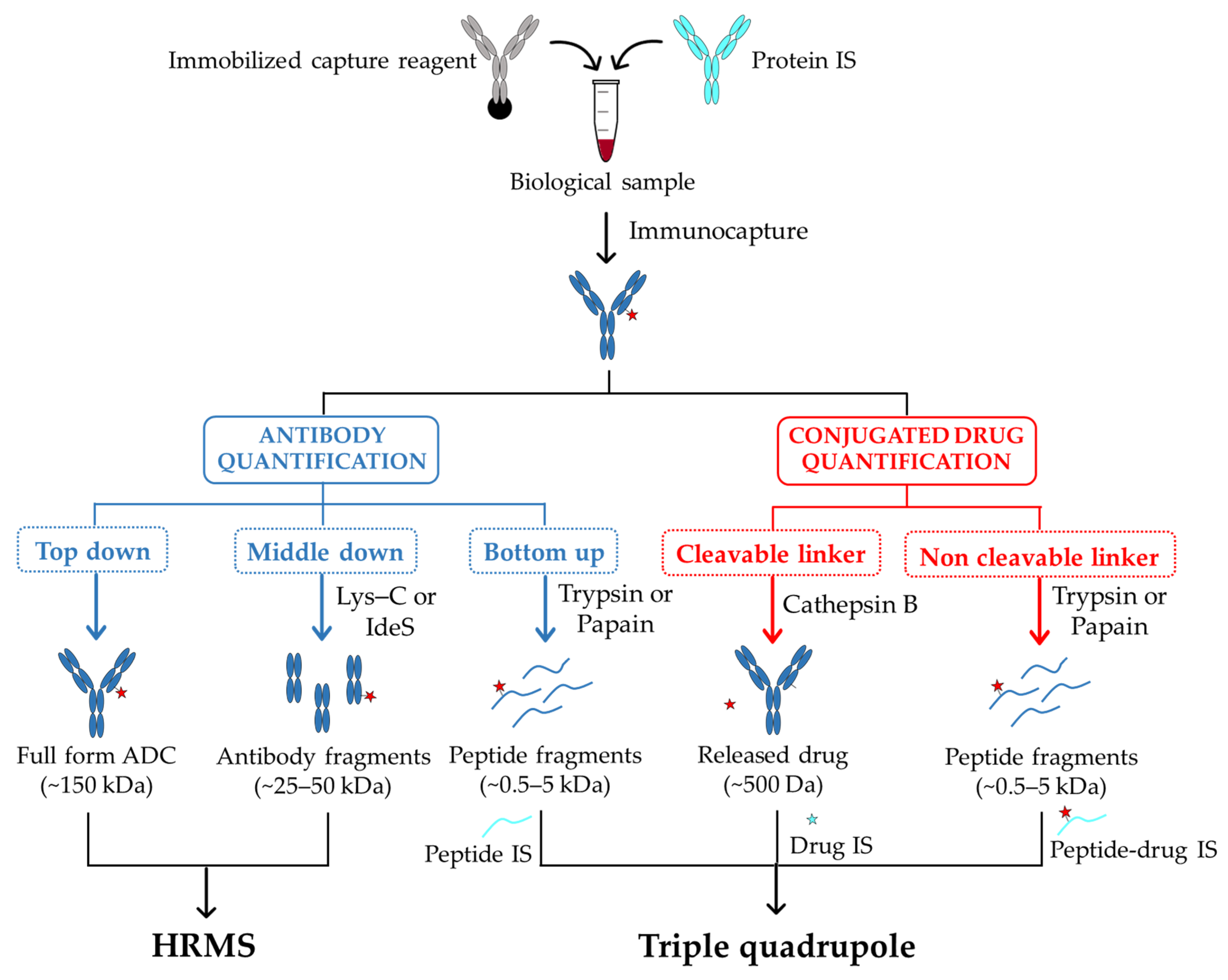
2.2. Hybrid Ligand-Binding Immuno-Affinity Capture Followed by Liquid Chromatography Coupled to Mass Spectrometry (LB-LC–MS)
2.3. Nano-Surface and Molecular-Orientation Limited (nSMOL) Proteolysis
2.4. MS Methods to Assess Pharmacokinetics and Biodistribution of Payload
3. Analytical Methods Requiring a Prelabeling of ADCs
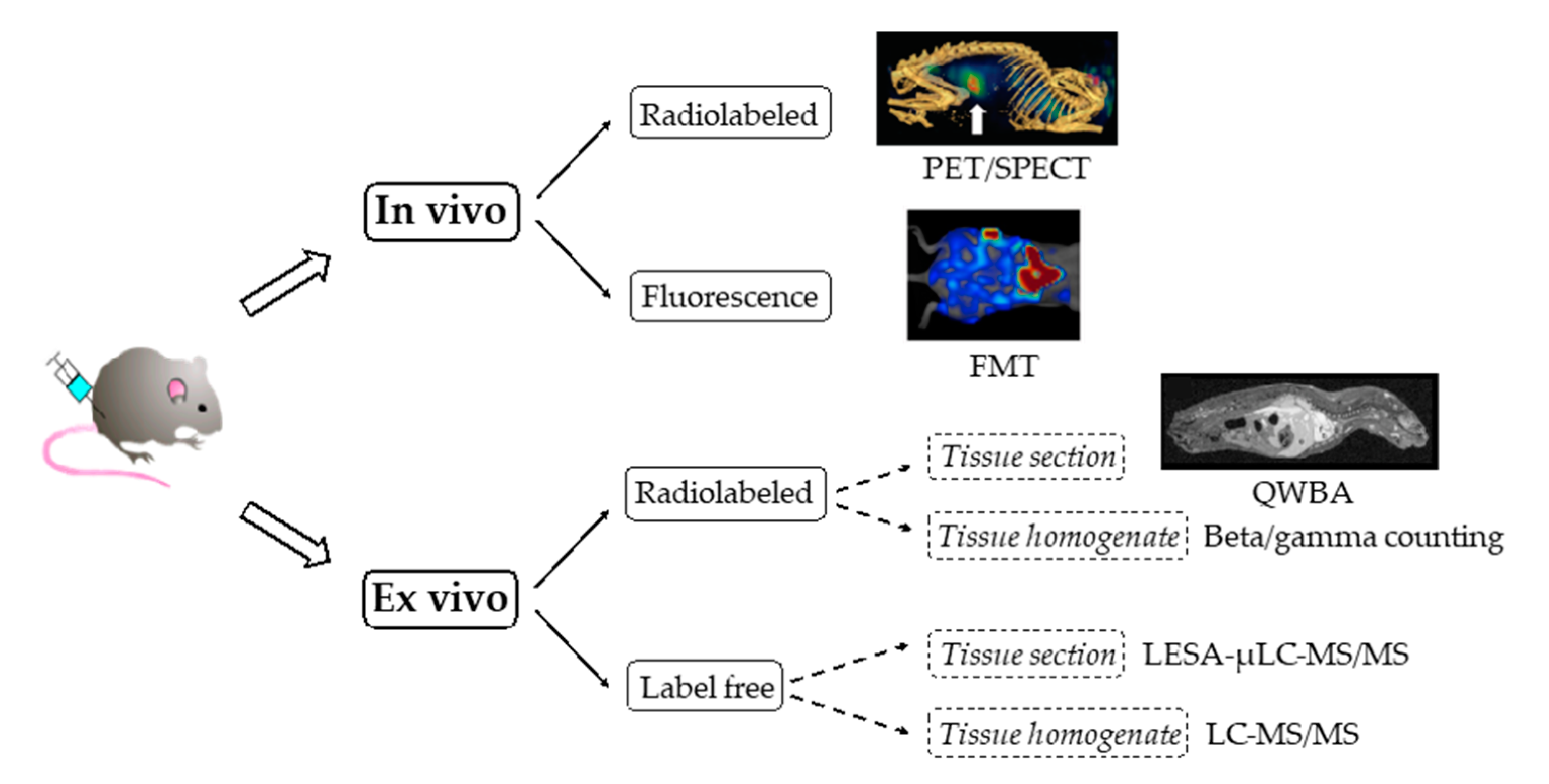
3.1. Noninvasive Molecular Imaging Approaches
3.2. Ex Vivo “Cut and Count” Techniques
3.3. Dual Radiolabeling
3.4. Limitations of Analytical Methods Involving Radiolabeling
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADA | Antidrug antibody |
| ADC | Antibody–drug conjugate |
| ADME | Absorption, distribution, metabolism and excretion |
| CIQA | Cryo-imaging quantitative autoradiography |
| DAR | Drug-to-antibody ratio |
| DM1 | Mertansine |
| DM4 | Ravtansine |
| DOTA | Tetraxetan |
| DTPA | Pentetic acid |
| EIC | Extracted-ion chromatogram |
| ELISA | Enzyme-linked immunosorbent assay |
| EPR | Enhanced permeability and retention |
| Fab | Fragment antigen binding |
| Fc | Fragment crystallizable |
| FcRn | Neonatal Fc receptor |
| FDA | Food and Drug Administration |
| FMT | Fluorescence molecular tomography |
| HER-2 | Human epidermal growth factor receptor 2 |
| HR/AM | High resolution accurate-mass |
| HRMS | High resolution mass spectrometry |
| HRP | Horseradish peroxidase |
| IgG | Immunoglobulin G |
| IS | Internal standard |
| LBA | Ligand binding assay |
| LB-LC–MS | Ligand-binding immuno-affinity capture followed by LC–MS analysis |
| LC–MS | Liquid chromatography tandem mass spectrometry |
| LESA–µLC–MS | Liquid extraction surface analysis coupled to µLC–MS |
| LLOQ | Lower limit of quantification |
| MMAE | Monomethyl auristatin E |
| MMAF | Monomethyl auristatin F |
| nSMOL | Nano-surface and molecular-orientation limited proteolysis |
| PET | Positron emission tomography |
| QWBA | Quantitative whole-body autoradiography |
| SEC | Size exclusion chromatography |
| SHA | Semihomogeneous assay |
| SPE | Solid phase extraction |
| SPECT | Single-photon emission computed tomography |
| T-DM1 | Trastuzumab emtansine/Kadcyla® |
References
- Joubert, N.; Beck, A.; Dumontet, C.; Denevault-Sabourin, C. Antibody–Drug Conjugates: The Last Decade. Pharmaceuticals 2020, 13, 245. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Goetsch, L.; Dumontet, C.; Corvaïa, N. Strategies and challenges for the next generation of antibody-drug conjugates. Nat. Rev. Drug Discov. 2017, 16, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liu, D. Antibody-drug conjugates in clinical trials for lymphoid malignancies and multiple myeloma. J. Hematol. Oncol. 2019, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Coats, S.; Williams, M.; Kebble, B.; Dixit, R.; Tseng, L.; Yao, N.; Tice, D.A.; Soria, J. Antibody–Drug Conjugates: Future Directions in Clinical and Translational Strategies to Improve the Therapeutic Index. Clin. Cancer Res. 2019, 25, 5441–5449. [Google Scholar] [CrossRef]
- Chau, C.H.; Steeg, P.S.; Figg, W.D. Therapeutics Antibody–Drug conjugates for cancer. Therapeutics 2019, 394, 793–804. [Google Scholar] [CrossRef]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Hiroshi Morisaki, J.; et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef]
- Wang, R.E.; Liu, T.; Wang, Y.; Cao, Y.; Du, J.; Luo, X.; Deshmukh, V.; Kim, C.H.; Lawson, B.R.; Tremblay, M.S.; et al. An immunosuppressive antibody-drug conjugate. J. Am. Chem. Soc. 2015, 137, 3229–3232. [Google Scholar] [CrossRef]
- Lim, R.K.V.; Yu, S.; Cheng, B.; Li, S.; Kim, N.J.; Cao, Y.; Chi, V.; Kim, J.Y.; Chatterjee, A.K.; Schultz, P.G.; et al. Targeted Delivery of LXR Agonist Using a Site-Specific Antibody-Drug Conjugate. Bioconjug. Chem. 2015, 26, 2216–2222. [Google Scholar] [CrossRef]
- Svendsen, P.; Graversen, J.H.; Etzerodt, A.; Hager, H.; Røge, R.; Grønbæk, H.; Christensen, E.I.; Møller, H.J.; Vilstrup, H.; Moestrup, S.K. Antibody-Directed Glucocorticoid Targeting to CD163 in M2-type Macrophages Attenuates Fructose-Induced Liver Inflammatory Changes. Mol. Ther. Methods Clin. Dev. 2017, 4, 50–61. [Google Scholar] [CrossRef]
- Pahl, A.; Lutz, C.; Hechler, T. Amanitins and their development as a payload for antibody-drug conjugates. Drug Discov. Today Technol. 2018, 30, 85–89. [Google Scholar] [CrossRef]
- Masters, J.C.; Nickens, D.J.; Xuan, D.; Shazer, R.L.; Amantea, M. Clinical toxicity of antibody drug conjugates: A meta-analysis of payloads. Invest. New Drugs 2017, 36, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q. Site-specific antibody conjugation for ADC and beyond. Biomedicines 2017, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.M.; Coumbe, B.G.T.; Josephs, D.H.; Mele, S.; Ilieva, K.M.; Cheung, A. Antibody structure and engineering considerations for the design and function of Antibody Drug Conjugates (ADCs). Oncoimmunology 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Junutula, J.R.; Raab, H.; Clark, S.; Bhakta, S.; Leipold, D.D.; Weir, S.; Chen, Y.; Simpson, M.; Tsai, S.P.; Dennis, M.S.; et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008, 26, 925–932. [Google Scholar] [CrossRef]
- Hallam, T.J.; Smider, V.V. Unnatural amino acids in novel antibody conjugates. Future Med. Chem. 2014, 6, 1309–1324. [Google Scholar] [CrossRef]
- Hofer, T.; Skeffington, L.R.; Chapman, C.M.; Rader, C. Molecularly defined antibody conjugation through a selenocysteine interface. Biochemistry 2009, 48, 12047–12057. [Google Scholar] [CrossRef]
- Dennler, P.; Chiotellis, A.; Fischer, E.; Brégeon, D.; Belmant, C.; Gauthier, L.; Lhospice, F.; Romagne, F.; Schibli, R. Transglutaminase-based chemo-enzymatic conjugation approach yields homogeneous antibody-drug conjugates. Bioconjug. Chem. 2014, 25, 569–578. [Google Scholar] [CrossRef]
- Pishesha, N.; Ingram, J.R.; Ploegh, H.L. Sortase A: A Model for Transpeptidation and Its Biological Applications. Annu. Rev. Cell Dev. Biol. 2018, 34, 163–188. [Google Scholar] [CrossRef]
- Rabuka, D.; Rush, J.S.; Dehart, G.W.; Wu, P.; Bertozzi, C.R. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc. 2012, 7, 1052–1067. [Google Scholar] [CrossRef]
- Badescu, G.; Bryant, P.; Bird, M.; Henseleit, K.; Swierkosz, J.; Parekh, V.; Tommasi, R.; Pawlisz, E.; Jurlewicz, K.; Farys, M.; et al. Bridging disulfides for stable and defined antibody drug conjugates. Bioconjug. Chem. 2014, 25, 1124–1136. [Google Scholar] [CrossRef]
- Van Geel, R.; Wijdeven, M.A.; Heesbeen, R.; Verkade, J.M.M.; Wasiel, A.A.; Van Berkel, S.S.; Van Delft, F.L. Chemoenzymatic Conjugation of Toxic Payloads to the Globally Conserved N-Glycan of Native mAbs Provides Homogeneous and Highly Efficacious Antibody-Drug Conjugates. Bioconjug. Chem. 2015, 26, 2233–2242. [Google Scholar] [CrossRef]
- Bargh, J.D.; Isidro-Llobet, A.; Parker, J.S.; Spring, D.R. Cleavable linkers in antibody-drug conjugates. Chem. Soc. Rev. 2019, 48, 4361–4374. [Google Scholar] [CrossRef] [PubMed]
- Bross, P.F.; Beitz, J.; Chen, G.; Chen, X.H.; Duffy, E.; Kieffer, L.; Roy, S.; Sridhara, R.; Rahman, A.; Williams, G.; et al. Approval summary: Gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin. Cancer Res. 2001, 7, 1490–1496. [Google Scholar] [PubMed]
- Lamb, Y.N. Inotuzumab Ozogamicin: First Global Approval. Drugs 2017, 77, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Pillow, T.H.; Schutten, M.; Yu, S.F.; Ohri, R.; Sadowsky, J.; Poon, K.A.; Solis, W.; Zhong, F.; Del Rosario, G.; Go, M.A.T.; et al. Modulating therapeutic activity and toxicity of pyrrolobenzodiazepine antibody-drug conjugates with self-immolative disulfide linkers. Mol. Cancer Ther. 2017, 16, 871–878. [Google Scholar] [CrossRef] [PubMed]
- De Claro, R.A.; McGinn, K.; Kwitkowski, V.; Bullock, J.; Khandelwal, A.; Habtemariam, B.; Ouyang, Y.; Saber, H.; Lee, K.; Koti, K.; et al. U.S. Food and Drug Administration approval summary: Brentuximab vedotin for the treatment of relapsed Hodgkin lymphoma or relapsed systemic anaplastic large-cell lymphoma. Clin. Cancer Res. 2012, 18, 5845–5849. [Google Scholar] [CrossRef]
- Deeks, E.D. Polatuzumab Vedotin: First Global Approval. Drugs 2019, 79, 1467–1475. [Google Scholar] [CrossRef]
- Zhong, Y.J.; Shao, L.H.; Li, Y. Cathepsin B-cleavable doxorubicin prodrugs for targeted cancer therapy (Review). Int. J. Oncol. 2013, 42, 373–383. [Google Scholar] [CrossRef]
- Jeffrey, S.C.; Andreyka, J.B.; Bernhardt, S.X.; Kissler, K.M.; Kline, T.; Lenox, J.S.; Moser, R.F.; Nguyen, M.T.; Okeley, N.M.; Stone, I.J.; et al. Development and properties of β-glucuronide linkers for monoclonal antibody-drug conjugates. Bioconjug. Chem. 2006, 17, 831–840. [Google Scholar] [CrossRef]
- Kern, J.C.; Cancilla, M.; Dooney, D.; Kwasnjuk, K.; Zhang, R.; Beaumont, M.; Figueroa, I.; Hsieh, S.C.; Liang, L.; Tomazela, D.; et al. Discovery of Pyrophosphate Diesters as Tunable, Soluble, and Bioorthogonal Linkers for Site-Specific Antibody-Drug Conjugates. J. Am. Chem. Soc. 2016, 138, 1430–1445. [Google Scholar] [CrossRef]
- Erickson, H.K.; Widdison, W.C.; Mayo, M.F.; Whiteman, K.; Audette, C.; Wilhelm, S.D.; Singh, R. Tumor delivery and in vivo processing of disulfide-linked and thioether-linked antibody-maytansinoid conjugates. Bioconjug. Chem. 2010, 21, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Doronina, S.O.; Mendelsohn, B.A.; Bovee, T.D.; Cerveny, C.G.; Alley, S.C.; Meyer, D.L.; Oflazoglu, E.; Toki, B.E.; Sanderson, R.J.; Zabinski, R.F.; et al. Enhanced activity of monomethylauristatin F through monoclonal antibody delivery: Effects of linker technology on efficacy and toxicity. Bioconjug. Chem. 2006, 17, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.; Price, L.; Schorzman, A.; Storrie, M.; Piscitelli, J.; Razo, J.; Zamboni, W. Factors Affecting the Pharmacology of Antibody–Drug Conjugates. Antibodies 2018, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.T.; Robinson, R.; Schorzman, A.N.; Piscitelli, J.A.; Razo, J.F.; Zamboni, W.C. Pharmacologic Considerations in the Disposition of Antibodies and Antibody-Drug Conjugates in Preclinical Models and in Patients. Antibodies 2019, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.Q.; Xu, K.; Liu, L.; Raab, H.; Bhakta, S.; Kenrick, M.; Parsons-Reponte, K.L.; Tien, J.; Yu, S.F.; Mai, E.; et al. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat. Biotechnol. 2012, 30, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Gorovits, B.; Alley, S.C.; Bilic, S.; Booth, B.; Kaur, S.; Oldfield, P.; Purushothama, S.; Rao, C.; Shord, S.; Siguenza, P. Bioanalysis of antibody-drug conjugates: American Association of Pharmaceutical Scientists Antibody-Drug Conjugate Working Group position paper. Bioanalysis 2013, 5, 997–1006. [Google Scholar] [CrossRef]
- Sun, X.; Ponte, J.F.; Yoder, N.C.; Laleau, R.; Coccia, J.; Lanieri, L.; Qiu, Q.; Wu, R.; Hong, E.; Bogalhas, M.; et al. Effects of Drug-Antibody Ratio on Pharmacokinetics, Biodistribution, Efficacy, and Tolerability of Antibody-Maytansinoid Conjugates. Bioconjug. Chem. 2017, 28, 1371–1381. [Google Scholar] [CrossRef]
- Kamath, A.V.; Iyer, S. Preclinical Pharmacokinetic Considerations for the Development of Antibody Drug Conjugates. Pharm. Res. 2015, 32, 3470–3479. [Google Scholar] [CrossRef]
- Sukumaran, S.; Zhang, C.; Leipold, D.D.; Saad, O.M.; Xu, K.; Gadkar, K.; Samineni, D.; Wang, B.; Milojic-blair, M.; Rubinfeld, B.; et al. Development and Translational Application of an Integrated, Mechanistic Model of Antibody-Drug Conjugate Pharmacokinetics. AAPS J. 2016. [Google Scholar] [CrossRef]
- Lin, K.; Tibbitts, J. Pharmacokinetic considerations for antibody drug conjugates. Pharm. Res. 2012, 29, 2354–2366. [Google Scholar] [CrossRef]
- Kumar, S.; King, L.E.; Clark, T.H.; Gorovits, B. Antibody-drug conjugates nonclinical support: From early to late nonclinical bioanalysis using ligand-binding assays. Bioanalysis 2015, 7, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Dowell, J.A.; Korth-Bradley, J. Pharmacokinetics of Gemtuzumab Ozogamicin, an Antibody-Targeted Chemotherapy Agent for the Treatment of Patients with Acute Myeloid Leukemia in First Relapse. J. Clin. Pharmacol. 2001, 41, 1206–1214. [Google Scholar] [CrossRef]
- Lewis Phillips, G.D.; Li, G.; Dugger, D.L.; Crocker, L.M.; Parsons, K.L.; Mai, E.; Blättler, W.A.; Lambert, J.M.; Chari, R.V.J.; Lutz, R.J.; et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008, 68, 9280–9290. [Google Scholar] [CrossRef] [PubMed]
- Pollack, V.A.; Alvarez, E.; Tse, K.F.; Torgov, M.Y.; Xie, S.; Shenoy, S.G.; MacDougall, J.R.; Arrol, S.; Zhong, H.; Gerwien, R.W.; et al. Treatment parameters modulating regression of human melanoma xenografts by an antibody–Drug conjugate (CR011-vcMMAE) targeting GPNMB. Cancer Chemother. Pharmacol. 2007, 60, 423–435. [Google Scholar] [CrossRef]
- Singh, A.P.; Shah, D.K. Measurement and Mathematical Characterization of Cell-Level Pharmacokinetics of Antibody-Drug Conjugates: A Case Study with Trastuzumab-vc-MMAE s. Drug Metab. Dispos. 2017, 45, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Guo, L.; Verma, A.; Wong, G.G.; Shah, D.K. A Cell-Level Systems PK-PD Model to Characterize In Vivo Efficacy of ADCs. Pharmaceutics 2019, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.P.; Chan, P.; Lee, C.; Nelson, C.; Elliott, J.M.; Bechtel, C.; Raab, H.; Xie, D.; Akutagawa, J.; Baudys, J.; et al. Anti-CD22-MCC-DM1 and MC-MMAF conjugates: Impact of assay format on pharmacokinetic parameters determination. Bioconjug. Chem. 2008, 19, 1673–1683. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Audette, C.; Hoffee, M.; Lambert, J.M.; Blättler, W. Erratum: Pharmacokinetics and biodistribution of the antitumor immunoconjugate, cantuzumab mertansine (huC242-DM1), and its two components in mice (Journal of Pharmacology and Experimental Therapeutics (2004) 308 (1073-1082)). J. Pharmacol. Exp. Ther. 2004, 310, 844. [Google Scholar] [CrossRef]
- Kovtun, Y.V.; Audette, C.A.; Ye, Y.; Kovtun, Y.V.; Audette, C.A.; Ye, Y.; Xie, H.; Ruberti, M.F.; Phinney, S.J.; Leece, B.A.; et al. Antibody-Drug Conjugates Designed to Eradicate Tumors with Homogeneous and Heterogeneous Expression of the Target Antigen with Homogeneous and Heterogeneous Expression of the Target Antigen. Cancer Res. 2006, 66, 3214–3221. [Google Scholar] [CrossRef]
- Advani, A.; Coiffier, B.; Czuczman, M.S.; Dreyling, M.; Foran, J.; Gine, E.; Gisselbrecht, C.; Ketterer, N.; Nasta, S.; Rohatiner, A.; et al. Safety, Pharmacokinetics, and Preliminary Clinical Activity of Inotuzumab Ozogamicin, a Novel Immunoconjugate for the Treatment of B-Cell Non-Hodgkin’ s Lymphoma: Results of a Phase I Study. J. Clin. Oncol. 2010, 28, 2085–2093. [Google Scholar] [CrossRef]
- DiJoseph, J.F.; Armellino, D.C.; Boghaert, E.R.; Khandke, K.; Dougher, M.M.; Sridharan, L.; Kunz, A.; Hamann, P.R.; Gorovits, B.; Udata, C.; et al. Antibody-targeted chemotherapy with CMC-544: A CD22-targeted immunoconjugate of calicheamicin for the treatment of B-lymphoid malignancies. Blood 2004, 103, 1807–1814. [Google Scholar] [CrossRef] [PubMed]
- Alley, S.C.; Benjamin, D.R.; Jeffrey, S.C.; Okeley, N.M.; Meyer, D.L.; Sanderson, R.J.; Senter, P.D. Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjug. Chem. 2008, 19, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, B.A.W.; Ochoa, L.; Hammond, L.A.; Patnaik, A.; Edwards, T.; Takimoto, C.; Smith, L.; De Bono, J.; Schwartz, G.; Mays, T.; et al. Directed to the CanAg Antigen: A Phase I, Pharmacokinetic, and Biologic Correlative Study. J. Clin. Oncol. 2003, 21, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, R.J.; Hering, M.A.; James, S.F.; Sun, M.M.C.; Doronina, S.O.; Siadak, A.W.; Senter, P.D.; Wahl, A.F. In vivo drug-linker stability of an anti-CD30 dipeptide-linked auristatin immunoconjugate. Clin. Cancer Res. 2005, 11, 843–852. [Google Scholar]
- Kozak, K.R.; Tsai, S.P.; Fourie-O’Donohue, A.; Dela Cruz Chuh, J.; Roth, L.; Cook, R.; Chan, E.; Chan, P.; Darwish, M.; Ohri, R.; et al. Total antibody quantification for MMAE-conjugated antibody-drug conjugates: Impact of assay format and reagents. Bioconjug. Chem. 2013, 24, 772–779. [Google Scholar] [CrossRef]
- Chang, H.-P.; Shah, D.K. Determination of ADC concentration by Ligand-Binding Assays. In Methods in Molecular Biology 2078; Springer: Berlin, Germany, 2020; pp. 361–369. ISBN 9781493999286. [Google Scholar]
- Ramagiri, S.; Moore, I. Hybridizing LBA with LC-MS/MS: The new norm for biologics quantification. Bioanalysis 2016, 8, 483–486. [Google Scholar] [CrossRef]
- Zhu, X.; Huo, S.; Xue, C.; An, B.; Qu, J. Current LC-MS-based strategies for characterization and quanti fi cation of antibody-drug conjugates. J. Pharm. Anal. 2020, 10, 209–220. [Google Scholar] [CrossRef]
- Botzanowski, T.; Wagner-Rousset, E.; Rabuka, D.; Beck, A.; Drake, P.M.; Cianf, S. Insights from native mass spectrometry approaches for top- and middle- level characterization of site-specific antibody-drug conjugates. MAbs 2017, 9, 801–811. [Google Scholar] [CrossRef]
- Fung, E.N.; Bryan, P.; Kozhich, A. Techniques for quantitative LC-MS/MS analysis of protein therapeutics: Advances in enzyme digestion and immunocapture. Bioanalysis 2016, 8, 847–856. [Google Scholar] [CrossRef]
- Van Den Broek, I.; Van Dongen, W.D. LC-MS-based quantification of intact proteins: Perspective for clinical and bioanalytical applications. Bioanalysis 2015, 7, 1943–1958. [Google Scholar] [CrossRef]
- Wang, J.; Gu, H.; Liu, A.; Kozhich, A.; Rangan, V.; Myler, H.; Luo, L.; Wong, R.; Sun, H.; Wang, B.; et al. Antibody-drug conjugate bioanalysis using LB-LC-MS/MS hybrid assays: Strategies, methodology and correlation to ligand-binding assays. Bioanalysis 2016, 8, 1383–1401. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ortiz, R.; Tran, L.; Hall, M.; Spahr, C.; Walker, K.; Laudemann, J.; Miller, S.; Salimi-Moosavi, H.; Lee, J.W. General LC-MS/MS method approach to quantify therapeutic monoclonal antibodies using a common whole antibody internal standard with application to preclinical studies. Anal. Chem. 2012, 84, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Su, D.; Ng, C.; Liu, L.; Yu, S.F.; Pillow, T.H.; Del Rosario, G.; Darwish, M.; Lee, B.C.; Ohri, R.; et al. High-Resolution Accurate-Mass Mass Spectrometry Enabling In-Depth Characterization of in Vivo Biotransformations for Intact Antibody-Drug Conjugates. Anal. Chem. 2017, 89, 5476–5483. [Google Scholar] [CrossRef]
- Jian, W.; Kang, L.; Burton, L.; Weng, N. A workflow for absolute quantitation of large therapeutic proteins in biological samples at intact level using LC-HRMS. Bioanalysis 2016, 8, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Lanshoeft, C.; Cianférani, S.; Heudi, O. Generic Hybrid Ligand Binding Assay Liquid Chromatography High-Resolution Mass Spectrometry-Based Workflow for Multiplexed Human Immunoglobulin G1 Quantification at the Intact Protein Level: Application to Preclinical Pharmacokinetic Studies. Anal. Chem. 2017, 89, 2628–2635. [Google Scholar] [CrossRef]
- Jin, W.; Burton, L.; Moore, I. LC–HRMS quantitation of intact antibody drug conjugate trastuzumab emtansine from rat plasma. Bioanalysis 2018, 10, 851–862. [Google Scholar] [CrossRef]
- He, J.; Yu, S.F.; Yee, S.; Kaur, S.; Xu, K. Characterization of in vivo biotransformations for trastuzumab emtansine by high-resolution accurate-mass mass spectrometry. MAbs 2018, 10, 960–967. [Google Scholar] [CrossRef]
- Liu, H.; Manuilov, A.V.; Chumsae, C.; Babineau, M.L.; Tarcsa, E. Quantitation of a recombinant monoclonal antibody in monkey serum by liquid chromatography-mass spectrometry. Anal. Biochem. 2011, 414, 147–153. [Google Scholar] [CrossRef]
- Wagner-Rousset, E.; Janin-Bussat, M.C.; Colas, O.; Excoffier, M.; Ayoub, D.; Haeuw, J.F.; Rilatt, I.; Perez, M.; Corvaïa, N.; Beck, A. Antibody-drug conjugate model fast characterization by LC-MS following IdeS proteolytic digestion. MAbs 2014, 6, 173–184. [Google Scholar] [CrossRef]
- Kotapati, S.; Passmore, D.; Yamazoe, S.; Kishore, R.; Sanku, K.; Cardarelli, P.M.; Deshpande, S.; Strop, P.; Dollinger, G.; Rajpal, A. Article A Universal Affinity Capture LC-MS Assay for Evaluation of Biotransformation of Site-Specific Antibody Drug Conjugates in Preclinical Studies A Universal Affinity Capture LC-MS Assay for Evaluation of Biotransformation of Site-Specific Antibody Dr. Anal. Chem. 2019, 92, 2065–2073. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Kozhich, A.; Passmore, D.; Gu, H.; Wong, R.; Zambito, F.; Rangan, V.S.; Myler, H.; Aubry, A.F.; Arnold, M.E.; et al. Quantitative bioanalysis of antibody-conjugated payload in monkey plasma using a hybrid immuno-capture LC-MS/MS approach: Assay development, validation, and a case study. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 1002, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Rago, B.; Tumey, L.N.; Wei, C.; Barletta, F.; Clark, T.; Hansel, S.; Han, X. Quantitative Conjugated Payload Measurement Using Enzymatic Release of Antibody−Drug Conjugate with Cleavable Linker. Bioconjug. Chem. 2017, 28, 620–626. [Google Scholar] [CrossRef]
- Sanderson, R.J.; Nicholas, N.D.; Baker Lee, C.; Hengel, S.M.; Lyon, R.P.; Benjamin, D.R.; Alley, S.C. Antibody-conjugated drug assay for protease-cleavable antibody-drug conjugates. Bioanalysis 2016, 8, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gu, C.; Gruenhagen, J.; Yehl, P.; Chetwyn, N.P.; Medley, C.D. An enzymatic deconjugation method for the analysis of small molecule active drugs on antibody-drug conjugates. MAbs 2016, 8, 698–705. [Google Scholar] [CrossRef]
- Shi, C.; Goldberg, S.; Lin, T.; Dudkin, V.; Widdison, W.; Harris, L.; Wilhelm, S.; Jmeian, Y.; Davis, D.; O’Neil, K.; et al. LC/MS/MS Bioanalysis of Protein-Drug Conjugates—The Importance of Incorporating Succinimide Hydrolysis Products. Anal. Chem. 2018, 90, 5314–5321. [Google Scholar] [CrossRef]
- Wei, C.; Zhang, G.; Clark, T.; Barletta, F.; Tumey, L.N.; Rago, B.; Hansel, S.; Han, X. Where Did the Linker-Payload Go? A Quantitative Investigation on the Destination of the Released Linker-Payload from an Antibody-Drug Conjugate with a Maleimide Linker in Plasma. Anal. Chem. 2016, 88, 4979–4986. [Google Scholar] [CrossRef]
- Dong, L.; Li, C.; Locuson, C.; Chen, S.; Qian, M.G. A Two-Step Immunocapture LC/MS/MS Assay for Plasma Stability and Payload Migration Assessment of Cysteine-Maleimide-Based Antibody Drug Conjugates. Anal. Chem. 2018, 90, 5989–5994. [Google Scholar] [CrossRef]
- Iwamoto, N.; Shimada, T.; Umino, Y.; Aoki, C.; Aoki, Y.; Sato, T.-A.; Hamada, A.; Nakagama, H. Selective detection of complementarity-determining regions of monoclonal antibody by limiting protease access to the substrate: Nano-. Analyst 2014, 139, 576–580. [Google Scholar] [CrossRef]
- Iwamoto, N.; Shimomura, A.; Tamura, K.; Hamada, A. LC–MS bioanalysis of Trastuzumab and released emtansine using nano-surface and molecular-orientation limited (nSMOL) proteolysis and liquid–liquid partition in plasma of Trastuzumab emtansine-treated breast cancer patients. J. Pharm. Biomed. Anal. 2017, 145, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, N.; Hamada, A.; Shimada, T. Antibody drug quantitation in coexistence with anti-drug antibodies on nSMOL bioanalysis. Anal. Biochem. 2018, 540–541, 30–37. [Google Scholar] [CrossRef]
- Heudi, O.; Barteau, S.; Picard, F.; Kretz, O. Quantitative analysis of maytansinoid (DM1) in human serum by on-line solid phase extraction coupled with liquid chromatography tandem mass spectrometry—Method validation and its application to clinical samples. J. Pharm. Biomed. Anal. 2016, 120, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Sullivan, M.; Espinosa, O.; Yang, L. A sensitive LC-MS/MS method for the determination of free maytansinoid DM4 concentrations—Method development, validation, and application to the nonclinical studies of antitumor agent DM4 conjugated hu-anti-Cripto MAb B3F6 (B3F6-DM4) in rats and monkeys. Int. J. Mass Spectrom. 2012, 312, 53–60. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Furuta, M.; Manabe, S.; Koga, Y.; Yasunaga, M.; Matsumura, Y. Imaging mass spectrometry for the precise design of antibody-drug conjugates. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Leal, M.; Wentland, J.A.; Han, X.; Zhang, Y.; Rago, B.; Duriga, N.; Spriggs, F.; Kadar, E.; Song, W.; McNally, J.; et al. Preclinical Development of an anti-5T4 Antibody-Drug Conjugate: Pharmacokinetics in Mice, Rats, and NHP and Tumor/Tissue Distribution in Mice. Bioconjug. Chem. 2015, 26, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Walles, M.; Rudolph, B.; Wolf, T.; Bourgailh, J.; Suetterlin, M.; Moenius, T.; Peraus, G.; Heudi, O.; Elbast, W.; Lanshoeft, C.; et al. New insights in tissue distribution, metabolism, and excretion of [3H]-labeled antibody maytansinoid conjugates in female tumor-bearing nude rats. Drug Metab. Dispos. 2016, 44, 897–910. [Google Scholar] [CrossRef] [PubMed]
- Carmon, K.S.; Azhdarinia, A. Application of Immuno-PET in Antibody—Drug Conjugate Development. Mol. Imaging 2018, 17, 1–10. [Google Scholar] [CrossRef]
- Sijbrandi, N.J.; Merkul, E.; Muns, J.A.; Waalboer, D.C.J.; Adamzek, K.; Bolijn, M.; Somsen, G.W.; Haselberg, R.; Steverink, P.J.G.M.; Houthoff, H.; et al. A Novel Platinum (II)—Based Bifunctional ADC Linker Benchmarked Using 89 Zr-Desferal and Auristatin F—Conjugated Trastuzumab. Cancer Res. 2017, 77, 257–268. [Google Scholar] [CrossRef]
- Chia, P.; Parakh, S.; Tsao, M.; Pham, N.; Gan, H.K.; Cao, D.; Burvenich, I.J.G.; Rigopoulos, A.; Reilly, E.B.; John, T.; et al. Targeting and Efficacy of Novel mAb806-Antibody-Drug Conjugates in Malignant Mesothelioma. Pharmaceuticals 2020, 13, 289. [Google Scholar] [CrossRef]
- Lütje, S.; Gerrits, D.; Molkenboer-kuenen, J.D.; Herrmann, K.; Fracasso, G.; Colombatti, M.; Boerman, O.C.; Heskamp, S. Characterization of Site-Specifically Conjugated Monomethyl Auristatin E–And Duocarmycin-Based Anti-PSMA Antibody–Drug Conjugates for Treatment of PSMA-Expressing Tumors. J. Nucl. Med. 2018, 59, 494–502. [Google Scholar] [CrossRef]
- Boswell, C.A.; Yadav, D.B.; Mundo, E.E.; Yu, S.F.; Lacap, J.A.; Fourie-O’Donohue, A.; Kozak, K.R.; Ferl, G.Z.; Zhang, C.; Ho, J.; et al. Biodistribution and efficacy of an anti-TENB2 antibody-drug conjugate in a patient-derived model of prostate cancer. Oncotarget 2019, 10, 6234–6244. [Google Scholar] [CrossRef][Green Version]
- Boswell, C.A.; Mundo, E.E.; Zhang, C.; Stainton, S.L.; Yu, S.F.; Lacap, J.A.; Mao, W.; Kozak, K.R.; Fourie, A.; Polakis, P.; et al. Differential effects of predosing on tumor and tissue uptake of an 111in-labeled anti-TENB2 antibody-drug conjugate. J. Nucl. Med. 2012, 53, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- ter Weele, E.J.; van Scheltinga, A.G.T.T.; Kosterink, J.G.W.; Pot, L.; Vedelaar, S.R.; Lamberts, L.E.; Williams, S.P.; Lub-de Hooge, M.N.; de Vries, E.G.E. Imaging the distribution of an antibody-drug conjugate constituent targeting mesothelin with 89Zr and IRDye 800CW in mice bearing human pancreatic tumor xenografts. Oncotarget 2015, 6, 42081–42090. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Giddabasappa, A.; Gupta, V.R.; Norberg, R.; Gupta, P.; Spilker, M.E.; Wentland, J.; Rago, B.; Eswaraka, J.; Leal, M.; Sapra, P. Biodistribution and targeting of anti-5t4 antibody-drug conjugate using fluorescence molecular tomography. Mol. Cancer Ther. 2016, 15, 2530–2540. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhao, X.; Sun, X.; Li, J.; Wang, W.; Zhang, L.; Gou, S. Preclinical pharmacokinetics of a novel anti-c-Met antibody–drug conjugate, SHR-A1403, in rodents and non-human primates. Xenobiotica 2019, 49, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Saad, O.M.; Shen, B.Q.; Xu, K.; Khojasteh, S.C.; Girish, S.; Kaur, S. Bioanalytical approaches for characterizing catabolism of antibody-drug conjugates. Bioanalysis 2015, 7, 1583–1604. [Google Scholar] [CrossRef] [PubMed]
- Mandler, R.; Kobayashi, H.; Hinson, E.R.; Brechbiel, M.W.; Waldmann, T.A. Herceptin-Geldanamycin Immunoconjugates: Pharmacokinetics, Biodistribution, and Enhanced Antitumor Activity. Cancer Res. 2004, 64, 1460–1467. [Google Scholar] [CrossRef]
- Journal, A.I.; Gong, J.; Guo, F.; Cheng, W.; Fan, H.; Miao, Q. Preliminary biological evaluation of 123I-labelled anti-CD30-LDM in CD30-positive lymphomas murine models. Artif. Cells Nanomed. Biotechnol. 2020, 48, 408–414. [Google Scholar] [CrossRef]
- Boswell, C.A.; Mundo, E.E.; Zhang, C.; Bumbaca, D.; Valle, N.R.; Kozak, K.R.; Fourie, A.; Chuh, J.; Koppada, N.; Saad, O.; et al. Impact of drug conjugation on pharmacokinetics and tissue distribution of Anti-STEAP1 antibody-drug conjugates in rats. Bioconjug. Chem. 2011, 22, 1994–2004. [Google Scholar] [CrossRef]
- Herbertson, R.A.; Tebbutt, N.C.; Lee, F.T.; MacFarlane, D.J.; Chappell, B.; Micallef, N.; Lee, S.T.; Saunder, T.; Hopkins, W.; Smyth, F.E.; et al. Phase I biodistribution and pharmacokinetic study of Lewis Y-targeting immunoconjugate CMD-193 in patients with advanced epithelial cancers. Clin. Cancer Res. 2009, 15, 6709–6715. [Google Scholar] [CrossRef]
- Shen, B.-Q.; Bumbaca, D.; Saad, O.; Yue, Q.; V Pastuskovas, C.; Cyrus Khojasteh, S.; Tibbitts, J.; Kaur, S.; Wang, B.; Chu, Y.-W.; et al. Catabolic Fate and Pharmacokinetic Characterization of Trastuzumab Emtansine (T-DM1): An Emphasis on Preclinical and Clinical Catabolism. Curr. Drug Metab. 2012, 13, 901–910. [Google Scholar] [CrossRef]
- Alley, S.C.; Zhang, X.; Okeley, N.M.; Anderson, M.; Law, C.L.; Senter, P.D.; Benjamin, D.R. The pharmacologic basis for antibody-auristatin conjugate activity. J. Pharmacol. Exp. Ther. 2009, 330, 932–938. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Vugts, D.J.; Visser, G.W.M.; Stigter-Van Walsum, M.; Bolijn, M.; Spiga, M.; Lazzari, P.; Shankar, E.; Sani, M.; Zanda, M.; et al. Development of novel ADCs: Conjugation of tubulysin analogues to trastuzumab monitored by dual radiolabeling. Cancer Res. 2014, 74, 5700–5710. [Google Scholar] [CrossRef] [PubMed]
- Ilovich, O.; Qutaish, M.; Hesterman, J.Y.; Orcutt, K.; Hoppin, J.; Polyak, I.; Seaman, M.; Abu-Yousif, A.O.; Cvet, D.; Bradley, D.P. Dual-isotope cryoimaging quantitative autoradiography: Investigating antibody-drug conjugate distribution and payload delivery through imaging. J. Nucl. Med. 2018, 59, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Muns, J.A.; Montserrat, V.; Houthoff, H.; Codée-van der Schilden, K.; Zwaagstra, O.; Sijbrandi, N.J.; Merkul, E.; Van Dogen, G.A.M.S. In vivo Characterization of Platinum(II)-base linker Technology for the Development of Antibody—Drug Conjugates: Taking Advantage of Dual Labeling with. J. Nucl. Med. 2018, 59, 1146–1152. [Google Scholar] [CrossRef]
- An, B.; Zhang, M.; Pu, J.; Shen, S.; Qu, Y.; Chen, Y.; Huo, S.; Wang, X.; Polli, J.R.; Balthasar, J.P.; et al. High-throughput, sensitive LC-MS quantification of biotherapeutics and biomarkers using an antibody-free, peptide-level multiple-mechanism enrichment via strategically regulated pH, ionic and solvent strengths The Department of Pharmaceutical Sciences. Anal. Chem. 2019, 91, 3475–3483. [Google Scholar] [CrossRef]

| ADC | FDA Approval | Indication | Target Antigen | Antibody | Linker | Payload | Conjugation | Average DAR |
|---|---|---|---|---|---|---|---|---|
| Mylotarg® | 2000, withdrawn in 2010 and reapproved in 2017 | Relapsed or refractory acute myeloid leukemia | CD33 | IgG4 humanized | Hydrazone (cleavable) | Calicheamicine | Lysine | 2.5 |
| Adcetris® | 2011 | Cutaneous anaplastic large cell lymphoma | CD30 | IgG1 chimeric | Val-Cit (cleavable) | MMAE | Cysteine | 4 |
| Kadcyla® | 2013 | HER2-positive metastatic breast cancer | HER2 | IgG1 humanized | SMCC * (noncleavable) | DM1 | Lysine | 3.5 |
| Besponsa® | 2017 | Acute lymphoblastic leukemia | CD22 | IgG4 humanized | Hydrazone (cleavable) | Calicheamicine | Lysine | 6 |
| Polivy® | 2019 | Diffuse large B-cell lymphoma | CD79b | IgG1 humanized | Val-Cit (cleavable) | MMAE | Cysteine | 4 |
| Enhertu® | 2019 | HER2-positive metastatic breast cancer | HER2 | IgG1 humanized | Gly-Gly-Phe-Gly (cleavable) | DXd | Cysteine | 7.5 |
| Padcev® | 2019 | Locally advanced or metastatic urothelial cancer | Nectin-4 | IgG1 fully human | Val-Cit (cleavable) | MMAE | Cysteine | 3.8 |
| Trodelvy® | 2020 | Triple negative breast cancer | Trop-2 | IgG1 humanized | Carbonate (cleavable) | SN-38 | Cysteine | 7.6 |
| Blenrep® | 2020 | Relapsed or refractory multiple myeloma | BCMA | IgG1 humanized | MC * (noncleavable) | MMAF | Cysteine | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cahuzac, H.; Devel, L. Analytical Methods for the Detection and Quantification of ADCs in Biological Matrices. Pharmaceuticals 2020, 13, 462. https://doi.org/10.3390/ph13120462
Cahuzac H, Devel L. Analytical Methods for the Detection and Quantification of ADCs in Biological Matrices. Pharmaceuticals. 2020; 13(12):462. https://doi.org/10.3390/ph13120462
Chicago/Turabian StyleCahuzac, Héloïse, and Laurent Devel. 2020. "Analytical Methods for the Detection and Quantification of ADCs in Biological Matrices" Pharmaceuticals 13, no. 12: 462. https://doi.org/10.3390/ph13120462
APA StyleCahuzac, H., & Devel, L. (2020). Analytical Methods for the Detection and Quantification of ADCs in Biological Matrices. Pharmaceuticals, 13(12), 462. https://doi.org/10.3390/ph13120462





