Deep Ensembling of Multiband Images for Earth Remote Sensing and Foramnifera Data
Abstract
:1. Introduction
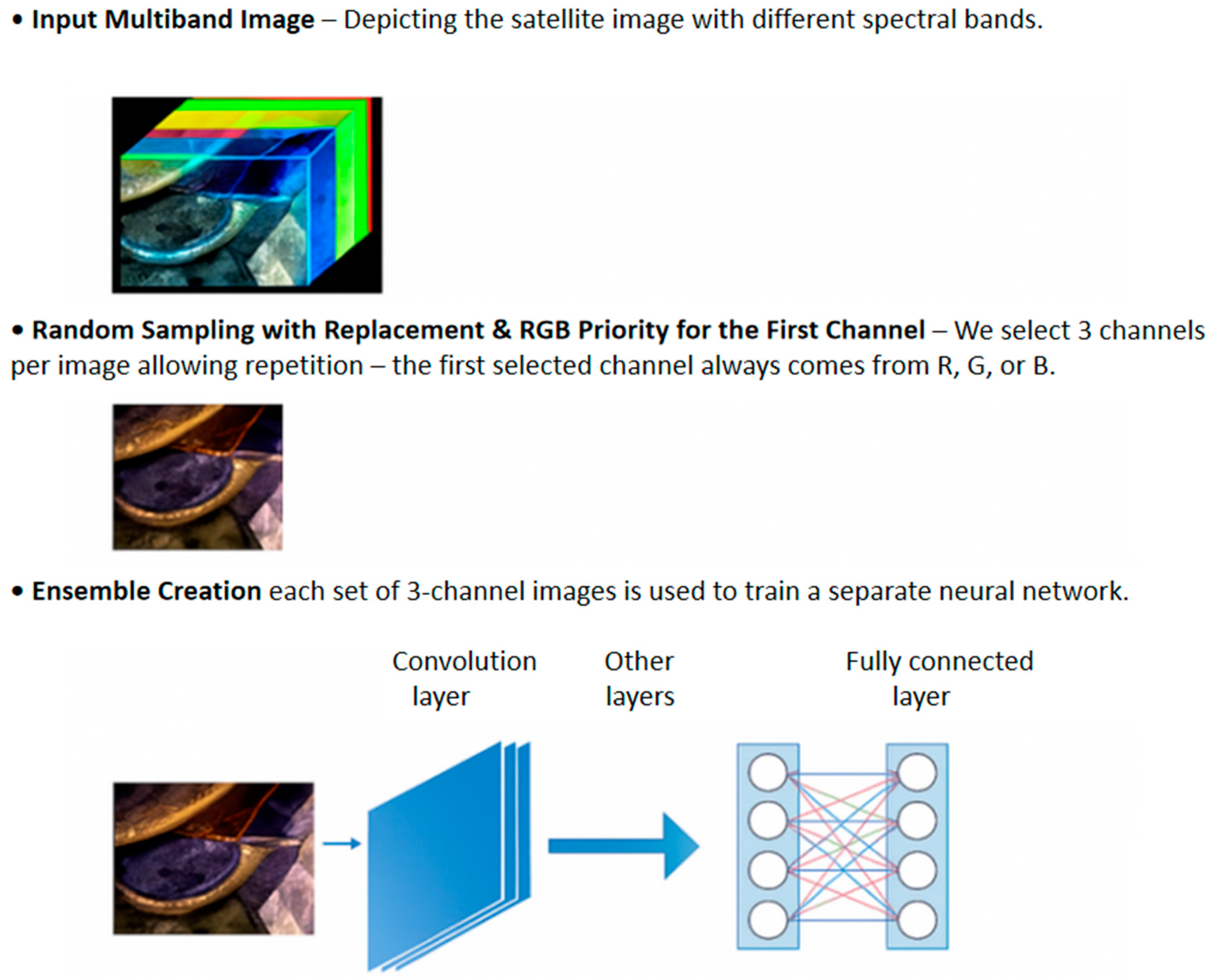
- Presentation of a simple method for randomly creating three-channel images starting from multiband images, useful for CNNs pretrained on very large RGB image datasets, such as ImageNet;
- An example of EL based on DL architectures and the proposed approach for generating three or multichannel images from multiband images, where each network is trained on a given set of generated images (e.g., each network is trained on images created in the same way, where at least one channel is from one of the three RGB channels);
- Although computationally more expensive than other approaches, our proposed method is simple, consisting of only a few lines of code;
- We evaluate our approach using three datasets, following a clear and replicable protocol, thereby ensuring that our work can serve as a strong baseline for future research on multiband classification systems;
- Our approach obtains SOTA with no change in hyperparameters between datasets.
- The code of the proposed approach will be available at https://github.com/LorisNanni/Multi-Band-Image-Analysis-Using-Ensemble-Neural-Networks (accessed on 28 March 2025).
Related Work
2. Materials and Methods
2.1. Foramnifera Dataset
- 178 images of G. bulloides;
- 182 images of G. ruber;
- 150 images of G. sacculifer;
- 174 images of N. incompta;
- 152 images of N. pachyderma;
- 151 images of N. dutertrei;
- 450 images labeled as “rest of the world”, representing other species of planktic foraminifera.
2.2. EuroSAT Dataset
2.3. So2Sat LCZ42
- Band B2—10 m GSD;
- Band B3—10 m GSD;
- Band B4—10 m GSD;
- Band B5—upsampled to 10 m from 20 m GSD;
- Band B6—upsampled to 10 m from 20 m GSD;
- Band B7—upsampled to 10 m from 20 m GSD;
- Band B8—10 m GSD;
- Band B8a—upsampled to 10 m from 20 m GSD;
- Band B11—upsampled to 10 m from 20 m GSD;
- Band B12—upsampled to 10 m from 20 m GSD.
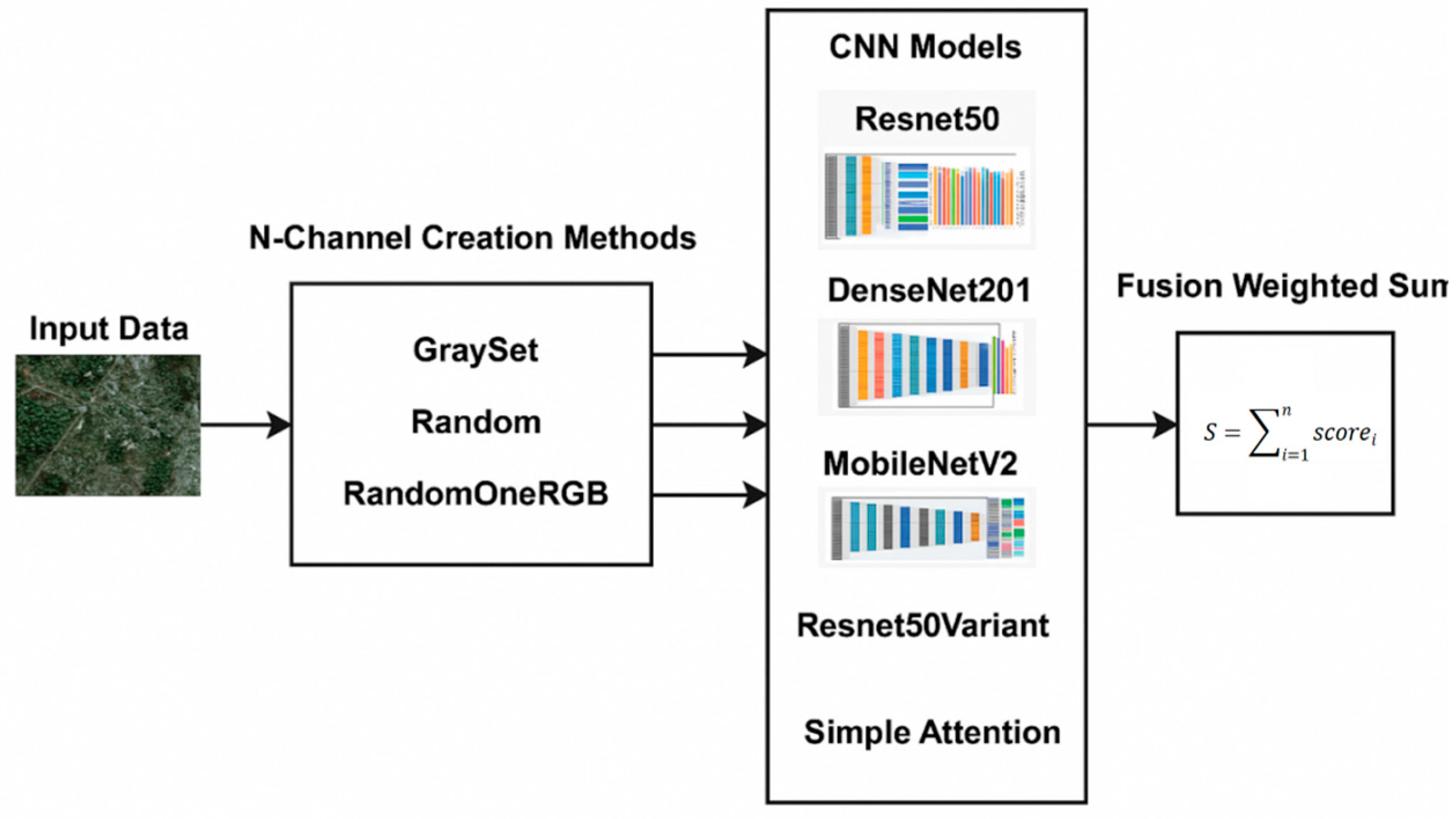
2.4. CNN Ensemble Learning (EL)
- The three-channel images are then used to train a set of three classic CNN networks, Resnet50, DenseNet201, and MobileNetV2, pretrained on ImageNet;
- The new multichannel images are then used to train a set of custom networks, one based on ResNet50 and the other on an attention model, both trained from scratch.
2.4.1. Ensemble Classifiers
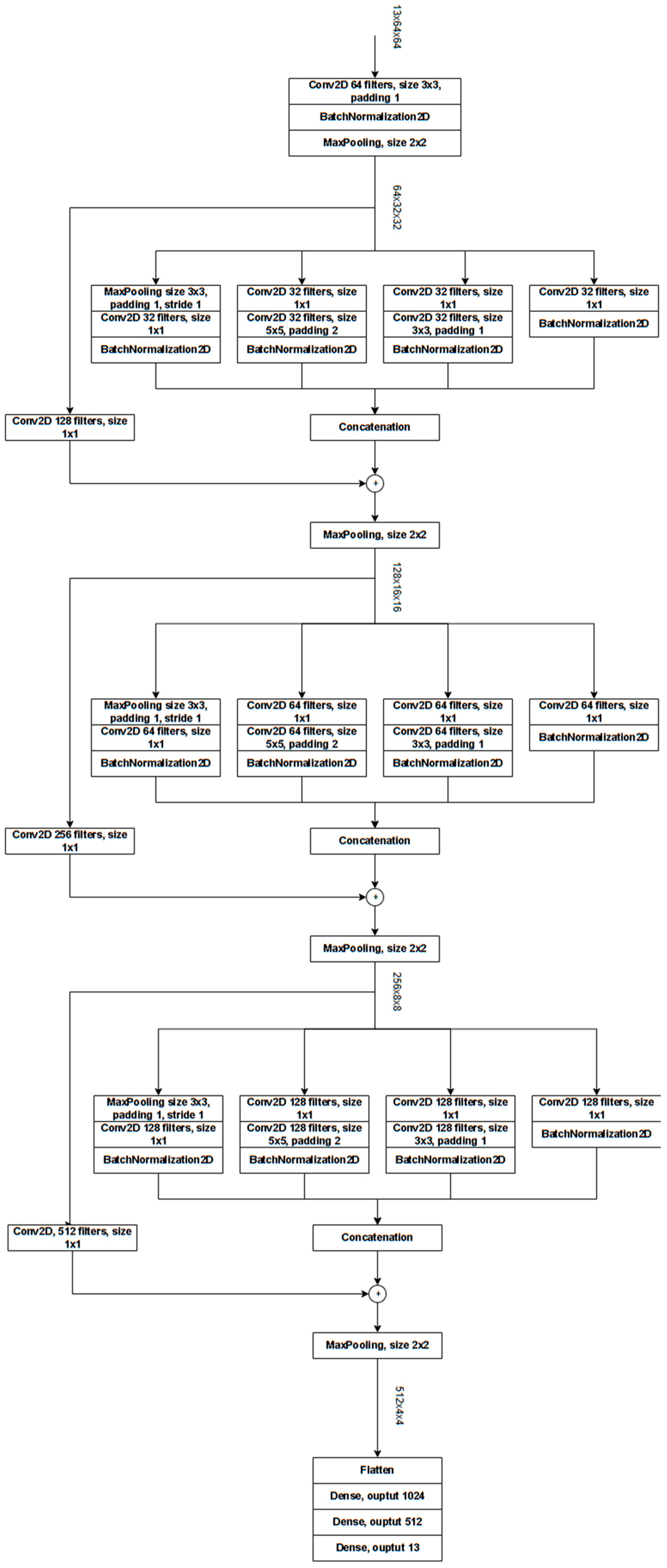
ResNet-Based Custom Architectures (Cres)
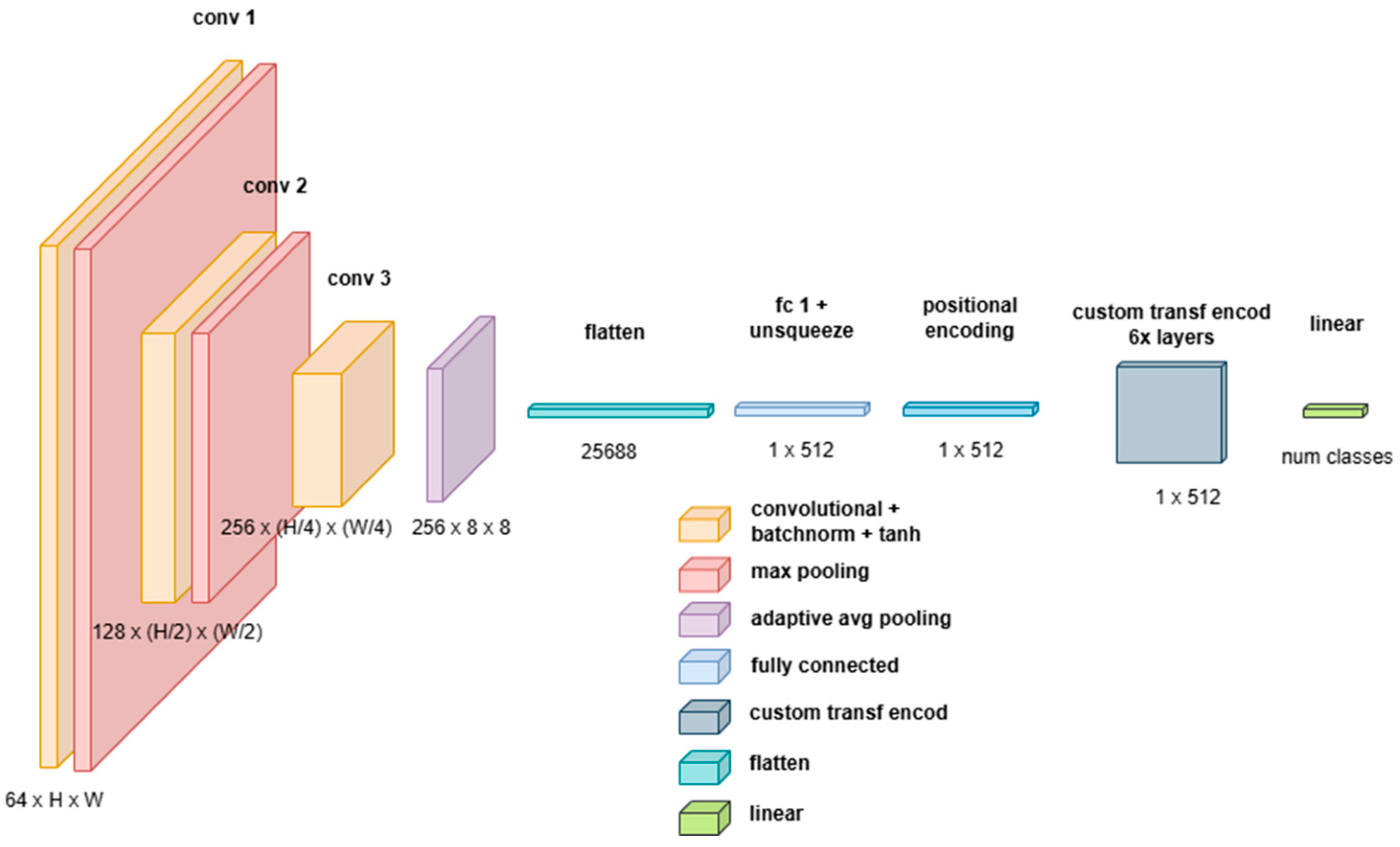
Custom Attention-Based Network (Catt)
- A 3D CNN Backbone for hierarchical feature extraction and dimensionality reduction;
- A Transformer Encoder for long-range dependency modeling and final classification.
- A 3D convolutional layer (Conv3D) with a kernel size of , enabling local feature extraction across spatial and spectral dimensions;
- Batch normalization (BatchNorm3D) to normalize activations and improve stability during training;
- A non-linearity (Tanh activation function), which normalizes values in the range .
- The first two pooling layers use a kernel of , reducing only the height and width but keeping the spectral depth unchanged;
- The final layer employs Adaptive Average Pooling, ensuring a fixed-size feature representation of , regardless of input image size.
- Preserves multispectral and volumetric information; unlike 2D CNNs, which process each spectral channel independently or in stacked formats, 3D CNNs treat spectral and spatial dimensions jointly, maintaining inter-band relationships;
- Reduces computational complexity for the Transformer; the 3D CNN acts as a preprocessing mechanism, reducing the input dimensions passed to the Transformer, thereby helping to mitigate the computational overhead of self-attention mechanisms, which scales quadratically with sequence length;
- Results in robust Spatial Feature Extraction: CNNs provide strong inductive biases for grid-structured data, capturing local correlations in a way that self-attention mechanisms alone struggle to replicate;
- Provide adaptive representation learning; the use of adaptive average pooling guarantees that the Transformer receives a consistent input size, preventing issues with variable input resolutions.
2.4.2. Fusion Method
2.5. Three/Multichannel Image Creation
2.5.1. Three-Channel Image Creation
- becomes the first RGB image;
- becomes the second RGB image;
- becomes the last RGB image.
2.5.2. Multichannel Image Creation
2.6. Performance Metrics
3. Results
- Y(t)_X means that we coupled the X architecture with ensemble Y, where the ensemble has t networks;
- X + Z means that we combine by sum rule the X and Z architectures with networks coupled with Random(20);
- X + Y + Z_z means that we combine by sum rule the X, Y, and Z architectures with networks coupled with Random(z).
- RGB(x) means that x networks are trained using RGB channels and then combined with the sum rule;
- X + Z means that we combine by sum rule the X and Z architectures, both coupled with RandomOneRGB(20);
- X + Y + Z_1 means that we combine by sum rule the X, Y, and Z architectures, coupled with RandomOneRGB(1).
| Approach | Accuracy |
|---|---|
| RGB(1)_Res | 98.54 |
| RGB(5)_Res | 98.80 |
| Random(5)_Res | 98.57 |
| RandomOneRGB(5)_Res | 98.91 |
| RandomOneRGB(20)_Res | 98.94 |
| RandomOneRGB(20)_DN | 99.15 |
| RandomOneRGB(20)_MV2 | 99.22 |
| RandomOneRGB(20)_Cres | 98.87 |
| RandomOneRGB(20)_Catt | 98.24 |
| Res+DN | 99.17 |
| Res+DN+MV2 | 99.22 |
| DN+MV2 | 99.20 |
| Res+DN+MV2+Catt | 99.39 |
| Res+DN+MV2+Cres | 99.41 |
| Res+DN+MV2+Catt+Cres | 99.33 |
| Res+DN+MV2_1 | 99.02 |
| Res+DN+MV2+Catt _1 | 99.20 |
| Res+DN+MV2+Cres_1 | 99.28 |
| Res+DN+MV2+Catt+Cres_1 | 99.22 |
| [58] | 99.24 |
| [59] | 99.22 |
| [60] | 99.20 |
| [61] | 98.96 |
| MobileViTV2 (Apple) [66] | 99.09 |
| ViT-large (Google) [66] | 98.55 |
| SwinTransformer (Microsoft) [66] | 98.83 |
| Approach | Year | Accuracy |
|---|---|---|
| RandomOneRGB(1)_Res | 2025 | 68.67 |
| RandomOneRGB(1)_MV2 | 2025 | 64.78 |
| RandomOneRGB(1)_DN | 2025 | 66.72 |
| RandomOneRGB(1)_Cres | 2025 | 62.54 |
| RandomOneRGB(10)_Res | 2025 | 70.97 |
| RandomOneRGB(10)_MV2 | 2025 | 70.91 |
| RandomOneRGB(3)_DN | 2025 | 70.96 |
| RandomOneRGB(10)_Cres | 2025 | 67.21 |
| Res+MV2 | 2025 | 71.80 |
| Res+MV2+DN | 2025 | 72.42 |
| Res+MV2+Cres | 2025 | 72.37 |
| Res+MV2+Cres+DN | 2025 | 72.79 |
| [62] | 2020 | 61.10 |
| [69] | 2023 | 67.87 |
| [70] | 2023 | 68.51 |
| [71] | 2020 | 69.40 |
| [72] | 2023 | 70.00 |
| [67] without PKC * | 2024 | 71.10 |
| [67] with PKC * | 2024 | 73.80 |
4. Discussions
- ResNet50, 10.86 s;
- DenseNet201, 97.19 s;
- MobileNetV2, 9.42 s.
5. Conclusions
- Bridging model architectures: we combine the strengths of standard CNNs and custom architectures through an ensemble approach, which not only achieves state-of-the-art performance but also offers a more accessible alternative compared to methods that rely solely on highly complex or custom networks;
- Providing ease and availability of implementation: all source code used in this study is freely available on GitHub. In this way, we enhance reproducibility and lower the barrier for researchers and practitioners. This ease of implementation addresses the gap where many SOTA methods are difficult to deploy or replicate.
- Expanding model architectures: while our ensemble incorporates established CNN models and custom architectures, exploring additional deep learning frameworks—such as more transformer-based models or graph neural networks—could further capture the complex spectral and spatial dependencies inherent in multiband data. Thus, we plan on developing more custom networks for managing both multispectral and synthetic aperture radar images;
- Investigating domain adaptation and transfer learning: future work could focus on leveraging domain adaptation strategies to enable the ensemble framework to generalize across different sensor types and imaging conditions. Incorporating transfer learning could also facilitate rapid adaptation to new datasets, particularly in dynamic environmental monitoring scenarios;
- Providing temporal dynamics and multitemporal analysis: extending the current framework to handle time-series multiband images will allow for the analysis of temporal changes in land cover or environmental conditions. This integration would enable monitoring of evolving phenomena and improve the relevance of the approach for real-time applications.
- Exploring scalability and real-time implementation: exploring the scalability of the proposed system in operational environments is essential. Optimizing the framework for real-time processing and deployment—possibly through distributed computing or edge-based implementations—could significantly expand its practical utility in remote sensing applications.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nalepa, J. Recent Advances in Multi- and Hyperspectral Image Analysis. Sensors 2021, 21, 6002. [Google Scholar] [CrossRef]
- Uddin, M.P.; Mamun, M.A.; Hossain, M.A. Feature extraction for hyperspectral image classification. In Proceedings of the 2017 IEEE Region 10 Humanitarian Technology Conference (R10-HTC), Dhaka, Bangladesh, 21–23 December 2017. [Google Scholar]
- Chen, H.; Miao, F.; Chen, Y.; Xiong, Y.; Chen, T. A hyperspectral image classification method using multifeature vectors and optimized KELM. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 2781–2795. [Google Scholar] [CrossRef]
- Shi, G.; Huang, H.; Wang, L. Unsupervised dimensionality reduction for hyperspectral imagery via local geometric structure feature learning. IEEE Geosci. Remote Sens. Lett. 2019, 17, 1425–1429. [Google Scholar] [CrossRef]
- Xu, X.; Li, J.; Li, S.; Plaza, A. Subpixel component analysis for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5564–5579. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, X.; Jiang, J.; Zhang, Y.; Liu, X.; Cai, Z. Spectral–spatial and superpixelwise PCA for unsupervised feature extraction of hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2021, 60, 5502210. [Google Scholar] [CrossRef]
- Champa, A.I.; Rabbi, M.F.; Hasan, S.M.M.; Zaman, A.; Kabir, M.H. Tree-based classifier for hyperspectral image classification via hybrid technique of feature reduction. In Proceedings of the 2021 International Conference on Information and Communication Technology for Sustainable Development (ICICT4SD), Dhaka, Bangladesh, 27–28 February 2021. [Google Scholar]
- Wei, L.; Huang, C.; Wang, Z.; Wang, Z.; Zhou, X.; Cao, L. Monitoring of urban black-odor water based on Nemerow index and gradient boosting decision tree regression using UAV-borne hyperspectral imagery. Remote Sens. 2019, 11, 2402. [Google Scholar] [CrossRef]
- Xu, S.; Liu, S.; Wang, H.; Chen, W.; Zhang, F.; Xiao, Z. A hyperspectral image classification approach based on feature fusion and multi-layered gradient boosting decision trees. Entropy 2020, 23, 20. [Google Scholar] [CrossRef]
- Bazine, R.; Huayi, W.; Boukhechba, K. K-NN similarity measure based on fourier descriptors for hyperspectral images classification. In Proceedings of the 2019 International Conference on Video, Signal and Image Processing, Wuhan, China, 29–31 October 2019. [Google Scholar]
- Bhavatarini, N.; Akash, B.N.; Avinash, A.R.; Akshay, H.M. Object detection and classification of hyperspectral images using K-NN. In Proceedings of the 2023 Second International Conference on Electrical, Electronics, Information and Communication Technologies (ICEEICT), Trichirappalli, India, 5–7 April 2023. [Google Scholar]
- Zhang, L.; Huang, D.; Chen, X.; Zhu, L.; Xie, Z.; Chen, X.; Cui, G.; Zhou, Y.; Huang, G.; Shi, W. Discrimination between normal and necrotic small intestinal tissue using hyperspectral imaging and unsupervised classification. J. Biophotonics 2023, 16, e202300020. [Google Scholar] [CrossRef]
- Chen, G.Y. Multiscale filter-based hyperspectral image classification with PCA and SVM. J. Electr. Eng. 2021, 72, 40–45. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, H.; Huang, Y.; Cheng, D.; Huang, J. A GA and SVM classification model for pine wilt disease detection using UAV-based hyperspectral imagery. Appl. Sci. 2022, 12, 6676. [Google Scholar] [CrossRef]
- Pathak, D.K.; Kalita, S.K.; Bhattacharya, D.K. Hyperspectral image classification using support vector machine: A spectral spatial feature based approach. Evol. Intell. 2022, 15, 1809–1823. [Google Scholar] [CrossRef]
- Jain, V.; Phophalia, A. Exponential weighted random forest for hyperspectral image classification. In Proceedings of the IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019. [Google Scholar]
- Kishore, K.M.S.; Behera, M.K.; Chakravarty, S.; Dash, S. Hyperspectral image classification using minimum noise fraction and random forest. In Proceedings of the 2020 IEEE International Women in Engineering (WIE) Conference on Electrical and Computer Engineering (WIECON-ECE), Bhubaneswar, India, 26–27 December 2020. [Google Scholar]
- Zhao, J.; Yan, H.; Huang, L. A joint method of spatial–spectral features and BP neural network for hyperspectral image classification. Egypt. J. Remote Sens. Space Sci. 2023, 26, 107–115. [Google Scholar]
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A Survey of Convolutional Neural Networks: Analysis, Applications, and Prospects. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 6999–7019. [Google Scholar] [PubMed]
- Khan, A.; Rauf, Z.; Sohail, A.; Khan, A.R.; Asif, H.; Asif, A.; Farooq, U. A survey of the vision transformers and their CNN-transformer based variants. Artif. Intell. Rev. 2023, 56, 2917–2970. [Google Scholar]
- Liu, S.; Chu, R.S.W.; Wang, X.; Luk, W. Optimizing CNN-based hyperspectral image classification on FPGAs. In International Symposium on Applied Reconfigurable Computing; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Butt, M.H.F.; Ayaz, H.; Ahmad, M.; Li, J.P.; Kuleev, R. A fast and compact hybrid CNN for hyperspectral imaging-based bloodstain classification. In Proceedings of the 2022 IEEE Congress on Evolutionary Computation (CEC), Padua, Italy, 18–23 July 2022. [Google Scholar]
- Bhosle, K.; Ahirwadkar, B. Deep learning convolutional neural network (cnn) for cotton, mulberry and sugarcane classification using hyperspectral remote sensing data. J. Integr. Sci. Technol. 2021, 9, 70–74. [Google Scholar]
- Yan, T.; Xu, W.; Lin, J.; Duan, L.; Gao, P.; Zhang, C.; Lv, X. Combining multi-dimensional convolutional neural network (CNN) with visualization method for detection of aphis gossypii glover infection in cotton leaves using hyperspectral imaging. Front. Plant Sci. 2021, 12, 604510. [Google Scholar]
- Yu, C.; Han, R.; Song, M.; Liu, C.; Chang, C.-I. A simplified 2D-3D CNN architecture for hyperspectral image classification based on spatial–spectral fusion. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 2485–2501. [Google Scholar]
- Ghaderizadeh, S.; Abbasi-Moghadam, D.; Sharifi, A.; Zhao, N.; Tariq, A. Hyperspectral image classification using a hybrid 3D-2D convolutional neural networks. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 7570–7588. [Google Scholar]
- Xu, Q.; Yuan, X.; Ouyang, C.; Zeng, Y. Attention-based pyramid network for segmentation and classification of high-resolution and hyperspectral remote sensing images. Remote Sens. 2020, 12, 3501. [Google Scholar] [CrossRef]
- Mdrafi, R.; Du, Q.; Gurbuz, A.C.; Tang, B.; Ma, L.; Younan, N.H. Attention-based domain adaptation using residual network for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6424–6433. [Google Scholar]
- Zhang, M.; Gong, M.; He, H.; Zhu, S. Symmetric all convolutional neural-network-based unsupervised feature extraction for hyperspectral images classification. IEEE Trans. Cybern. 2020, 52, 2981–2993. [Google Scholar]
- Mitra, R.; Marchitto, T.M.; Ge, Q.; Zhong, B.; Kanakiya, B.; Cook, M.S.; Fehrenbacher, J.S.; Ortiz, J.D.; Tripati, A.; Lobaton, E. Automated species-level identification of planktic foraminifera using convolutional neural networks, with comparison to human performance. Mar. Micropaleontol. 2019, 147, 16–24. [Google Scholar]
- Edwards, R.; Wright, A. Foraminifera. In Handbook of Sea-Level Research; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 191–217. [Google Scholar]
- Liu, S.; Thonnat, M.; Berthod, M. Automatic classification of planktonic foraminifera by a knowledge-based system. In Proceedings of the Tenth Conference on Artificial Intelligence for Applications, San Antonio, TX, USA, 1–4 March 1994. [Google Scholar]
- Beaufort, L.; Dollfus, D. Automatic recognition of coccoliths by dynamical neural networks. Mar. Micropaleontol. 2004, 51, 57–73. [Google Scholar]
- Pedraza, L.F.; Hernández, C.A.; López, D.A. A Model to Determine the Propagation Losses Based on the Integration of Hata-Okumura and Wavelet Neural Models. Int. J. Antennas Propag. 2017, 2017, 1034673. [Google Scholar]
- Nanni, L.; Faldani, G.; Brahnam, S.; Bravin, R.; Feltrin, E. Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning. Signals 2023, 4, 524–538. [Google Scholar] [CrossRef]
- Huang, B.; Yang, F.; Yin, M.; Mo, X.; Zhong, C. A Review of Multimodal Medical Image Fusion Techniques. Comput. Math. Methods Med. 2020, 2020, 8279342. [Google Scholar]
- Helber, P.; Bischke, B.; Dengel, A.; Borth, D. EuroSAT: A Novel Dataset and Deep Learning Benchmark for Land Use and Land Cover Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 2217–2226. [Google Scholar]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep Learning in Remote Sensing: A Comprehensive Review and List of Resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar]
- Ma, L.; Liu, Y.; Zhang, X.; Ye, Y.; Yin, G.; Johnson, B.A. Deep learning in remote sensing applications: A meta-analysis and review. ISPRS J. Photogramm. Remote Sens. 2019, 152, 166–177. [Google Scholar]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal Convolutional Neural Network for the Classification of Satellite Image Time Series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef]
- Sellami, A.; Abbes, A.B.; Barra, V.; Farah, I.R. Fused 3-D spectral-spatial deep neural networks and spectral clustering for hyperspectral image classification. Pattern Recognit. Lett. 2020, 138, 594–600. [Google Scholar]
- Zhang, X.; Ye, P.; Xiao, G. VIFB: A visible and infrared image fusion benchmark. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020. [Google Scholar]
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19. [Google Scholar]
- Hermessi, H.; Mourali, O.; Zagrouba, E. Multimodal medical image fusion review: Theoretical background and recent advances. Signal Process. 2021, 183, 108036. [Google Scholar]
- Li, X.; Jing, D.; Li, Y.; Guo, L.; Han, L.; Xu, Q.; Xing, M.; Hu, Y. Multi-Band and Polarization SAR Images Colorization Fusion. Remote Sens. 2022, 14, 4022. [Google Scholar] [CrossRef]
- Moon, W.K.; Lee, Y.-W.; Ke, H.-H.; Lee, S.H.; Huang, C.-S.; Chang, R.-F. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Comput. Methods Programs Biomed. 2020, 190, 105361. [Google Scholar]
- Maqsood, S.; Javed, U. Multi-modal medical image fusion based on two-scale image decomposition and sparse representation. Biomed. Signal Process. Control. 2020, 57, 101810. [Google Scholar]
- Ding, I.-J.; Zheng, N.-W. CNN Deep Learning with Wavelet Image Fusion of CCD RGB-IR and Depth-Grayscale Sensor Data for Hand Gesture Intention Recognition. Sensors 2022, 22, 803. [Google Scholar] [CrossRef] [PubMed]
- Tasci, E.; Uluturk, C.; Ugur, A. A voting-based ensemble deep learning method focusing on image augmentation and preprocessing variations for tuberculosis detection. Neural Comput. Appl. 2021, 33, 15541–15555. [Google Scholar]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New data preprocessing trends based on ensemble of multiple preprocessing techniques. TrAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar]
- Mitra, R.; Marchitto, T.M.; Ge, Q.; Zhong, B.; Lobaton, E. Foraminifera optical microscope images with labelled species and segmentation labels. PANGAEA 2019. [Google Scholar] [CrossRef]
- Kuncheva, L.I. Combining Pattern Classifiers: Methods and Algorithms, 2nd ed.; Wiley: New York, NY, USA, 2014. [Google Scholar]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Las Vegas, NV, USA, 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; Volume 1, p. 3. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, J.; Xu, M.; Liu, L.; Wang, D.; Gao, E.; Han, C.; Guo, H.; Du, B.; Tao, D. MTP: Advancing remote sensing foundation model via multi-task pretraining. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 11632–11654. [Google Scholar] [CrossRef]
- Gesmundo, A. A continual development methodology for large-scale multitask dynamic ML systems. arXiv 2022, arXiv:2209.07326. [Google Scholar]
- Gesmundo, A.; Dean, J. An evolutionary approach to dynamic introduction of tasks in large-scale multitask learning systems. arXiv 2022, arXiv:2205.12755. [Google Scholar]
- Jeevan, P.; Sethi, A. Which Backbone to Use: A Resource-efficient Domain Specific Comparison for Computer Vision. arXiv 2024, arXiv:2406.05612. [Google Scholar]
- Zhu, X.X.; Hu, J.; Qiu, C.; Shi, Y.; Kang, J.; Mou, L.; Bagheri, H.; Haberle, M.; Hua, Y.; Huang, R.; et al. So2Sat LCZ42: A Benchmark Data Set for the Classification of Global Local Climate Zones. IEEE Geosci. Remote. Sens. Mag. 2020, 8, 76–89. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y. Convolutional networks for images, speech, and time series. Handb. Brain Theory Neural Netw. 1995, 3361, 1995. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Shabbir, A.; Ali, N.; Ahmed, J.; Zafar, B.; Rasheed, A.; Sajid, M.; Ahmed, A.; Dar, S.H. Satellite and scene image classification based on transfer learning and fine tuning of ResNet50. Math. Probl. Eng. 2021, 2021, 5843816. [Google Scholar] [CrossRef]
- Le, T.D.; Ha, V.N.; Nguyen, T.T.; Eappen, G.; Thiruvasagam, P.; Garces-Socarras, L.M.; Chatzinotas, S. Onboard Satellite Image Classification for Earth Observation: A Comparative Study of ViT Models. arXiv 2024, arXiv:2409.03901. [Google Scholar]
- Zhong, X.; Li, H.; Shen, H.; Gao, M.; Wang, Z.; He, J. Local Climate Zone Mapping by Coupling Multilevel Features With Prior Knowledge Based on Remote Sensing Images. IEEE Trans. Geosci. Remote. Sens. 2024, 62, 4403014. [Google Scholar] [CrossRef]
- Lin, H.; Wang, H.; Yin, J.; Yang, J. Local Climate Zone Classification via Semi-Supervised Multimodal Multiscale Transformer. IEEE Trans. Geosci. Remote. Sens. 2024, 62, 5212117. [Google Scholar] [CrossRef]
- He, G.; Dong, Z.; Guan, J.; Feng, P.; Jin, S.; Zhang, X. SAR and Multi-Spectral Data Fusion for Local Climate Zone Classification with Multi-Branch Convolutional Neural Network. Remote. Sens. 2023, 15, 434. [Google Scholar] [CrossRef]
- Dimitrovski, I.; Kitanovski, I.; Kocev, D.; Simidjievski, N. Current trends in deep learning for Earth Observation: An open-source benchmark arena for image classification. ISPRS J. Photogramm. Remote. Sens. 2023, 197, 18–35. [Google Scholar] [CrossRef]
- Qiu, C.; Tong, X.; Schmitt, M.; Bechtel, B.; Zhu, X.X. Multilevel Feature Fusion-Based CNN for Local Climate Zone Classification From Sentinel-2 Images: Benchmark Results on the So2Sat LCZ42 Dataset. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2020, 13, 2793–2806. [Google Scholar] [CrossRef]
- Ji, W.; Chen, Y.; Li, K.; Dai, X. Multicascaded Feature Fusion-Based Deep Learning Network for Local Climate Zone Classification Based on the So2Sat LCZ42 Benchmark Dataset. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2023, 16, 449–467. [Google Scholar] [CrossRef]
- Narkhede, M.; Mahajan, S.; Bartakke, P.; Sutaone, M. Towards compressed and efficient CNN architectures via pruning. Discov. Comput. 2024, 27, 29. [Google Scholar] [CrossRef]
- Chou, H.-H.; Chiu, C.-T.; Liao, Y.-P. Cross-layer knowledge distillation with KL divergence and offline ensemble for compressing deep neural network. APSIPA Trans. Signal Inf. Process. 2021, 10, e18. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, M.; Shi, J.Q. A survey on deep neural network pruning: Taxonomy, comparison, analysis, and recommendations. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10558–10578. [Google Scholar] [CrossRef]



| Band | Spatial Central Resolution—Meters | Wavelength—Nanometer |
|---|---|---|
| B01—Aerosols | 60 | 443 |
| B02—Blue | 10 | 490 |
| B03—Green | 10 | 560 |
| B04—Red | 10 | 665 |
| B05—Red edge 1 | 20 | 705 |
| B06—Red edge 2 | 20 | 740 |
| B07—Red edge 3 | 20 | 783 |
| B08—NIR | 10 | 842 |
| B08A—Red edge 4 | 20 | 865 |
| B09—Water vapor | 60 | 945 |
| B10—Cirrus | 60 | 1375 |
| B11—SWIR 1 | 20 | 1610 |
| B12—SWIR 2 | 20 | 2190 |
| Approach | F1-Measure |
|---|---|
| [30] | 85.0 |
| [35] | 90.6 |
| GraySet(10)_Res | 89.4 |
| Random(10)_Res | 91.1 |
| Random(20)_Res | 91.3 |
| Random(20)_DN | 91.5 |
| Random(20)_MV2 | 90.2 |
| Random(10)_Cres | 62.7 |
| Random(20)_Catt | 69.7 |
| Res+DN | 91.8 |
| Res+DN+MV2 | 92.1 |
| DN+MV2 | 92.3 |
| Res+DN+MV2+Catt | 92.5 |
| DN+MV2+Catt | 92.1 |
| Res+DN+MV2_1 | 90.1 |
| Res+DN+MV2_2 | 90.7 |
| Res+DN+MV2_3 | 90.7 |
| Precision (%) | Recall (%) | F1 Score (%) | Accuracy (%) | |
|---|---|---|---|---|
| Human Novices (max) [30] | 65 | 64 | 63 | 63 |
| Human Experts (max) [30] | 83 | 83 | 83 | 83 |
| ResNet50 + Vgg16 [30] | 84 | 86 | 85 | 85 |
| Stand alone Vgg16 [30] | 80 | 82 | 81 | 81 |
| [35] | 90.9 | 90.6 | 90.6 | 90.7 |
| Res+DN | 91.1 | 92.8 | 91.8 | 91.7 |
| Res+DN+MV2 | 91.5 | 93.0 | 92.1 | 91.8 |
| DN+MV2 | 91.6 | 93.4 | 92.3 | 92.0 |
| Res+DN+MV2+Catt | 91.9 | 93.5 | 92.5 | 92.2 |
| Res+DN+MV2_1 | Batch Size = 20 | Batch Size = 30 | Batch Size = 60 |
|---|---|---|---|
| Epochs = 15 | 89.9 | 90.0 | 90.1 |
| Epochs = 20 | 90.1 | 90.1 | 90.2 |
| Epochs = 30 | 90.1 | 90.0 | 90.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanni, L.; Brahnam, S.; Ruta, M.; Fabris, D.; Boscolo Bacheto, M.; Milanello, T. Deep Ensembling of Multiband Images for Earth Remote Sensing and Foramnifera Data. Sensors 2025, 25, 2231. https://doi.org/10.3390/s25072231
Nanni L, Brahnam S, Ruta M, Fabris D, Boscolo Bacheto M, Milanello T. Deep Ensembling of Multiband Images for Earth Remote Sensing and Foramnifera Data. Sensors. 2025; 25(7):2231. https://doi.org/10.3390/s25072231
Chicago/Turabian StyleNanni, Loris, Sheryl Brahnam, Matteo Ruta, Daniele Fabris, Martina Boscolo Bacheto, and Tommaso Milanello. 2025. "Deep Ensembling of Multiband Images for Earth Remote Sensing and Foramnifera Data" Sensors 25, no. 7: 2231. https://doi.org/10.3390/s25072231
APA StyleNanni, L., Brahnam, S., Ruta, M., Fabris, D., Boscolo Bacheto, M., & Milanello, T. (2025). Deep Ensembling of Multiband Images for Earth Remote Sensing and Foramnifera Data. Sensors, 25(7), 2231. https://doi.org/10.3390/s25072231










