1. Introduction
Lettuce, a leafy vegetable crop cultivated globally, is often found to experience nutrient deficiencies during its growth cycle due to various environmental and management factors, commonly known as deficiency phenomena. Such deficiencies can lead to stunted growth, discoloration, and leaf deformation, subsequently reducing yield and nutritional value [
1]. Traditionally, deficiency is diagnosed by experimentally analyzing soil and plant tissues, involving methods such as morphological diagnosis, chemical diagnosis, fertilization diagnosis, and enzymatic diagnosis methods [
2], which, however, have not yet been extensively adopted in agricultural practices for their inability to balance accuracy, cost, detection speed, and universality [
1]. Recent years have seen the gradual application of computer vision technology to diagnosing the nutritional status of crops. This technology is primarily categorized into methods based on chlorophyll content, multispectral and hyperspectral imaging technologies, and digital image analysis of abnormal features in plant parts [
3]. These methods possess their own limitations despite their respective advantages. For instance, methods measuring chlorophyll content are challenged in the early stages of plant growth, where chlorophyll changes are not significant enough for timely deficiency detection. Multispectral and hyperspectral imaging techniques struggle with achieving high energy efficiency, resolution, signal-to-noise ratio, and stability. On the other hand, methods like digital image analysis of abnormal plant part features are known for their ability to rapidly and accurately identify deficiency signs based on changes in color and morphological abnormalities in leaf images. For example, Kang Xiaoyan predicted the nitrogen content of lettuce through principal component and multiple regression analyses [
4], and Wei et al. rapidly graded the quality of external spinach through feature extraction [
5]. Despite advances in image processing technology for crop nutritional diagnosis, the effectiveness of existing models and algorithms varies significantly across different crops, lacking universality. Existing studies mostly focus on mature leafy vegetables and tend to rely on single-leaf image analysis, typically involving destructive sampling and leading to resource wastage, and there is relatively scarce research on the identification of deficiency symptoms in lettuce crops [
6].
Moreover, there was numerous noise interference in the process of transmitting lettuce images, which not only reduced the image quality but also affected the recognition results obtained based on these images. To reduce the impact of noise on image quality, a variety of denoising algorithms were developed [
7]. Traditional adaptive median filtering algorithms and some improved median filtering methods achieved good denoising effects on software platforms, but these algorithms mostly employed serial sorting methods [
8], which resulted in a massive computational amount and highly complex algorithms in the imaging process and thus extended image processing time [
9].
However, appropriate image enhancement techniques were required to further enhance image quality after denoising, especially in cases where characteristic colors are lacking. Adaptive Histogram Equalization (AHE), developed by Stark and his colleagues, was aimed at effectively enhancing local contrast [
10]. Yet, it was criticized for neglecting the overall image attributes, thereby affecting the quality and rendering effects of the entire image. The Average Weighted Histogram Equalization (AvHeq) method, proposed by Lin and his team [
11], effectively preserved the brightness information of images but failed to take into account the color information. The Brightness Preserving Bi-Histogram Equalization (BBHE), developed by Kim et al., selected average brightness as the separation threshold while maintaining the overall brightness before and after enhancement [
12]. This method may partially enhance images with a large dynamic range or uneven brightness. The Contrast Limited Adaptive Histogram Equalization (CLAHE) method, put forward by Lee J et al., was designed to limit the amplification of noise and excessive contrast enhancement in local areas caused by histogram equalization [
13]. Subsequently, an improved CLAHE algorithm, featuring adaptive parameters T1 and T2, was introduced by Fang Danyang et al. [
14]. This algorithm was effective in preventing the creation of artifacts yet exhibited deficiencies in handling global contrast issues.
Ultimately, image segmentation, as a key step in the processing flow, was utilized to divide the image into distinct regions with marked differences based on characteristics such as grayscale, color, texture, and shape, which facilitated subsequent feature extraction. While there were a variety of segmentation methods, there was a lack of a universal image segmentation algorithm. Methods still needed to be selected based on specific image features and application environments. Standard techniques, including region growing, area segmentation, and merging, were employed when images and their adjacent pixels were similar or even the same [
15]. Edge tracking and detection were typically employed for image segmentation based on edge textures or sudden changes in grayscale values. The traditional edge detection techniques include the Canny algorithm, Sobel algorithm, Prewitt algorithm, and Log algorithm [
16]. Among them, the Canny algorithm was widely recognized for its balanced performance, whose effectiveness in complex scenarios with a significant amount of noise, however, was limited. On this basis, the Canny algorithm underwent extensive research and adjustments by many experts, aiming to improve its efficacy in noisy environments. Yuanfeng Liu et al. focused on the target area and then employed an improved Canny operator for noise reduction, thereby enhancing the target detection accuracy [
17], but accompanied by blurring image edges, potentially the omission of weak edges in the image. Wenbo Fu and his team combined CNN and Canny into the C-Canny algorithm, which improved the threshold positioning accuracy [
18].
Despite progress in computer vision-based approaches for crop nutritional diagnosis, there remains a gap in the universality and robustness of segmentation algorithms tailored to practical applications [
19,
20]. Existing studies [
21,
22] primarily concentrate on isolated experimental conditions and lack comprehensive evaluations of real-world agricultural scenarios where variations in lighting, background complexity, and noise interference significantly impact image quality and recognition accuracy. Notably, while traditional image segmentation techniques such as region growing and edge detection have been widely used [
23], their adaptability to varying lettuce deficiency symptoms under field conditions remains underexplored. Furthermore, recent deep learning-based segmentation models have demonstrated improved accuracy in plant phenotyping and disease detection; however, their computational complexity and dependence on extensive labeled datasets limit their feasibility for real-time applications in resource-constrained environments. Studies [
24,
25] utilizing convolutional neural networks (CNNs), such as U-Net and Mask R-CNN, have achieved high precision in leaf segmentation, yet they often struggle with processing speed and the generalization required for diverse lettuce cultivars. Additionally, research on integrating advanced edge detection techniques with adaptive segmentation methods remains limited, particularly in addressing the challenges of detecting subtle deficiency symptoms in lettuce canopies. Given these limitations, this study aims to bridge the gap by developing a Field-Programmable Gate Array (FPGA)-based nutrient deficiency detection system that incorporates an enhanced Canny edge detection algorithm and a multi-dimensional image analysis framework. By integrating adaptive thresholding and multi-directional gradient estimation, the proposed approach seeks to improve segmentation accuracy while maintaining computational efficiency.
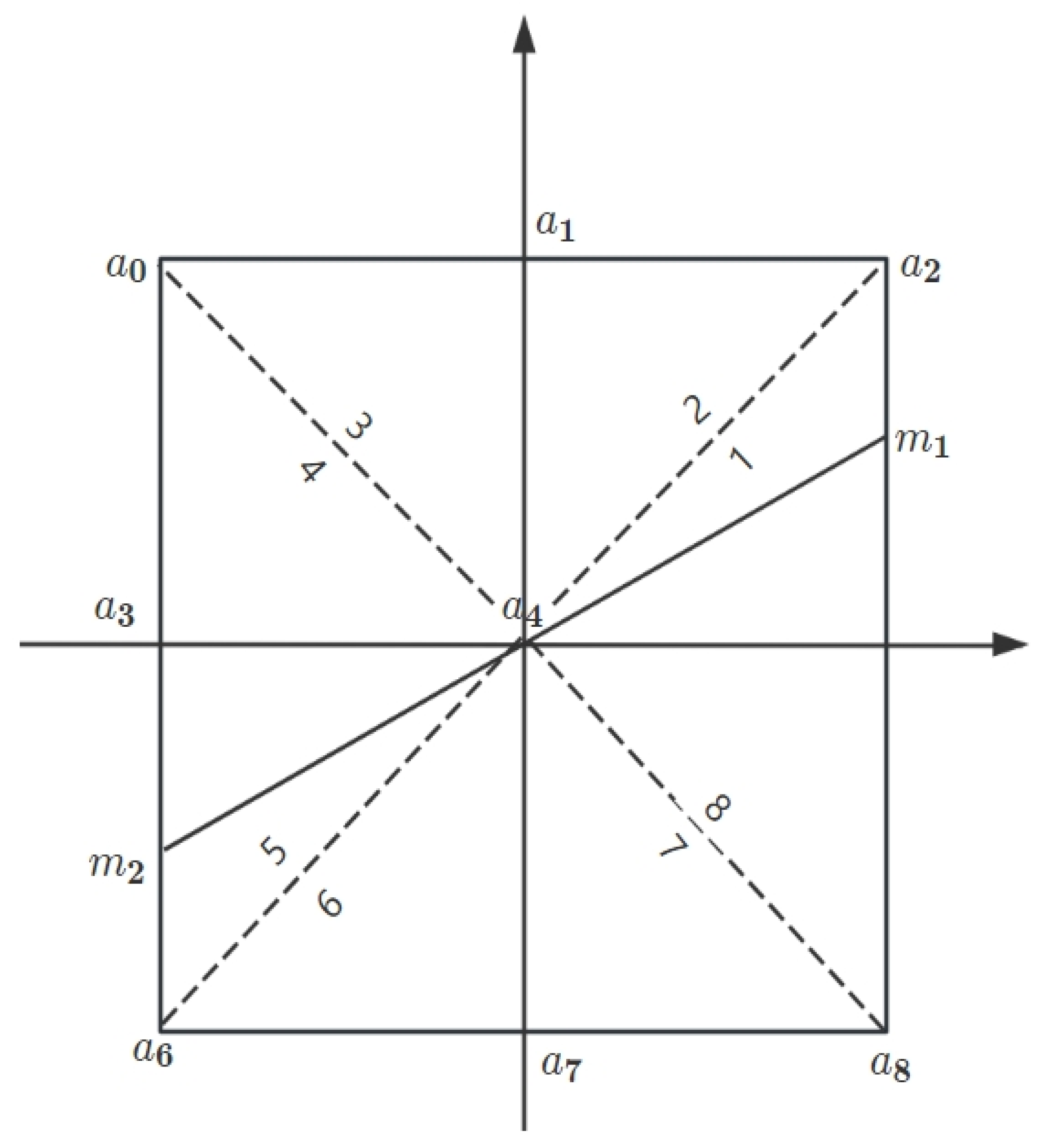
In terms of noise reduction, a median filtering algorithm was proposed in this study based on a dynamic window histogram to effectively balance noise suppression and image detail preservation, significantly reducing the computational burden, and achieving rapid updates by utilizing a sliding window histogram technique or an adaptive. This algorithm was implemented on a hardware platform to compensate for the limitations of traditional PC serial processing in real-time processing. Furthermore, to address the potential issues of texture and detail loss, as well as over-enhancement caused by image enhancement algorithms, an improved CLAHE algorithm was introduced in this study. This improvement was manifested in three aspects: Firstly, an adaptive adjustment mechanism based on the overall brightness of the image was introduced to optimize the precision and effect of contrast enhancement while suppressing noise generated in the process; secondly, an adaptive smoothing distribution mechanism based on local histogram statistics was introduced, which dynamically adjusted contrast enhancement strategies according to local image features to preserve image details; and lastly, considering the need for image reconstruction, a 5 × 5 pixel neighborhood bicubic interpolation technique was employed to improve the detail preservation capability during image enlargement or resolution enhancement and thus enhance the overall visual quality of the image. Additionally, an adaptive global–local contrast enhancement algorithm (CLGCE) was proposed, which combines the advantages of Global Histogram Equalization (GHE) and the improved CLAHE technique. This algorithm achieved a greater balance in processing global brightness and local details. Finally, in order to better segment the characteristic color regions of lettuce, the Canny edge detection algorithm was enhanced to improve its accuracy and adaptability in complex image processing. Firstly, the gradient calculation of the algorithm was expanded by employing Sobel operators in eight directions, namely horizontal and vertical, as well as 45°, 135°, 180°, 225°, 270°, and 315° directions. This multi-directional gradient calculation helped to more comprehensively capture image edge information, especially the details in non-standard directions. Secondly, an improved method was introduced, considering gradients in diagonal directions and combining linear interpolation techniques, allowing image edges to be more accurately processed and estimated. Furthermore, an automatic threshold calculation strategy was proposed to dynamically compute thresholds based on image characteristics, significantly enhancing the algorithm’s adaptability to different image conditions. Based on these improvements, a novel multi-dimensional image analysis algorithm was further developed, specifically designed to accurately segment the defective areas of lettuce in complex environments. The high-precision characteristics of the improved multi-directional gradient adaptive threshold Canny edge detection algorithm were integrated into this algorithm, along with the efficiency of traditional threshold segmentation and the local adaptability of gradient-guided adaptive threshold segmentation. This integration was aimed at significantly enhancing the detection precision and processing efficiency of the defective areas of lettuce, even in variable and complex environments.
A reference for the development of deficiency analysis techniques in intelligent agriculture is provided by this lettuce nutrient deficiency detection method, which is characterized by its real-time, non-destructive, and automated nature. This research contributes to the advancement of intelligent agriculture by providing a real-time, non-destructive, and scalable solution for lettuce nutrient deficiency diagnosis, addressing key challenges in image noise reduction, segmentation accuracy, and adaptability across diverse cultivation environments.
2. Experiments and Materials
The experiments on the nutrient deficiency of hydroponic lettuce were conducted from 10 March to 1 May 2024 in the Plant Factory of the College of Electrical and Information Engineering at Northeast Agricultural University. The experimental environment is shown in
Figure 1.
Lettuce seeds with good germination potential and healthy appearance were selected for the experiment and sown in seedling trays. During this stage, appropriate humidity and temperature conditions were maintained to promote seed germination. After meticulous cultivation for 14 days, the lettuce seedlings were transplanted into a hydroponic system. In the process of cultivating hydroponic lettuce, light intensity and nutrient solution concentration were identified as the primary environmental factors influencing growth, with a growth cycle of 40 days. A plant factory system was utilized during the experiment to uniformly regulate environmental parameters such as carbon dioxide concentration, temperature, airflow, and humidity, ensuring healthy growth under standardized conditions. In the hydroponic system, the pH of the nutrient solution was maintained between 5.5 and 6.5, and the electrical conductivity (EC value) was controlled within the range of 1400 to 1800 μS/cm. Red and blue light supplementation was applied in a timed manner, with the red-to-blue light ratio adjusted according to the growth stage of lettuce to ensure efficient photosynthesis and prevent abnormalities caused by insufficient light. Air circulation was achieved through a scheduled ventilation system that simulated natural conditions, providing an adequate carbon dioxide supply and preventing “tip burn”. The ambient temperature was controlled within the range of 18 to 22 °C, while the nutrient solution temperature was maintained at approximately 18 °C. Humidity level was kept between 40% and 85% to avoid excessive moisture, which potentially leads to rotting.
Figure 2 illustrates the entire cultivation process of hydroponic lettuce, from sowing on 10 March 2024 to harvesting on 1 May 2024.
To study the effect of nutrient solution on the change in the color of the hydroponic lettuce canopy, cultivation groups with varying degrees of nitrogen, phosphorus, and potassium deficiency, along with the control group exhibiting normal growth, were designed. The nutrient solution used in this study was prepared according to the Hoagland formula, a widely used method for providing essential nutrients to plants in hydroponic systems [
26]. This formula provides a balanced concentration of nutrients, including nitrogen, phosphorus, and potassium, which are critical for plant growth. The optimal concentrations of these nutrients for hydroponic lettuce cultivation are generally considered to be 15.000 mmol/L of nitrogen, 1.000 mmol/L of phosphorus, and 6.000 mmol/L of potassium, which were adopted in the control group of our experiment (
Table 1). For the experimental groups, varying levels of these nutrients were systematically reduced to simulate deficiencies, with concentrations adjusted based on the severity of the deficiency: mild, moderate, or severe. The nutrient solution composition for each deficiency group was deliberately altered to reflect the targeted deficiency while maintaining a constant concentration of other essential nutrients to ensure plant growth under stress conditions. This approach allows a comparison of nutrient deficiency impacts across different plant growth stages and nutrient levels.
To ensure the accuracy and applicability of the experimental results, the experiment was conducted by subdividing the deficiency levels into mild, moderate, and severe so that subtle growth changes in plants could be captured under varying degrees of nutritional stress, thereby enabling the more precise diagnosis of deficiencies. Additionally, to enhance the reliability and statistical significance of the experimental data, a large number of samples were prepared for each deficiency level. The hydroponic lettuce sample data collected in this experiment will provide key sample evidence for establishing a real-time diagnosis system for nutrient deficiencies in hydroponic lettuce. The detailed data are listed in
Table 2.
4. Lettuce Real-Time Diagnostic System
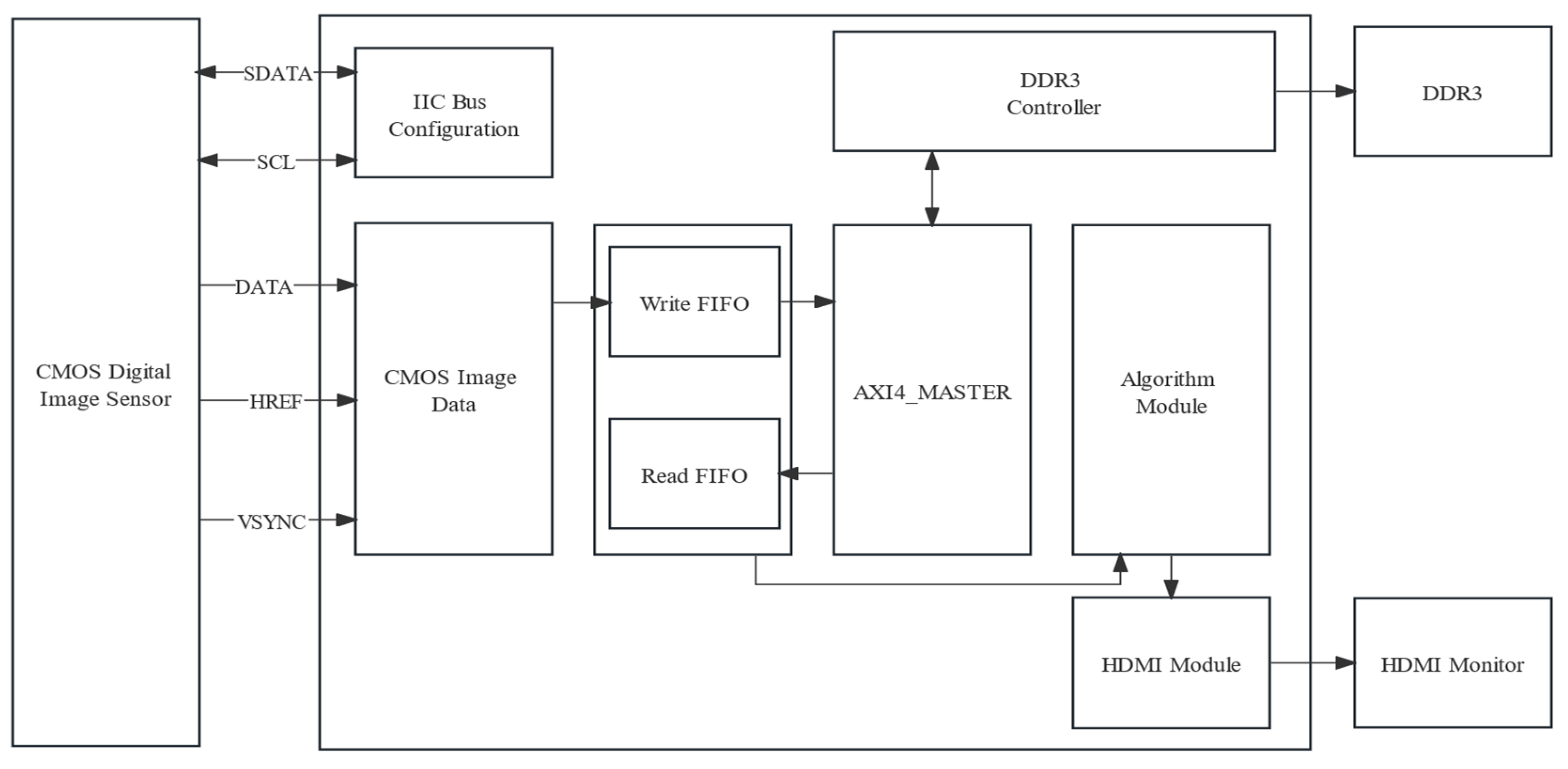
The hardware architecture constructed in this study was divided into four modules, namely a real-time image capture module, a storage module, an algorithm processing module, and an HDMI display module. The overall framework of the system is depicted in
Figure 9. Each module had its own specific role: the image capture module, the storage module, the algorithm processing module, and the HDMI display module were, respectively, dedicated to real-time image acquisition, image data storage, nutritional deficiency analysis on the captured images, and displaying processed images in real-time on an HDMI screen.
The ARTIX-7 series 200T XC7A200T-2FBG484I FPGA device from XILINX (San Jose, CA, USA) was used by the master controller to coordinate the data conversion of the OV5640 image sensor and SDRAM memory, drive the HDMI display screen, and implement algorithmic processing of image data, as shown in
Figure 10.
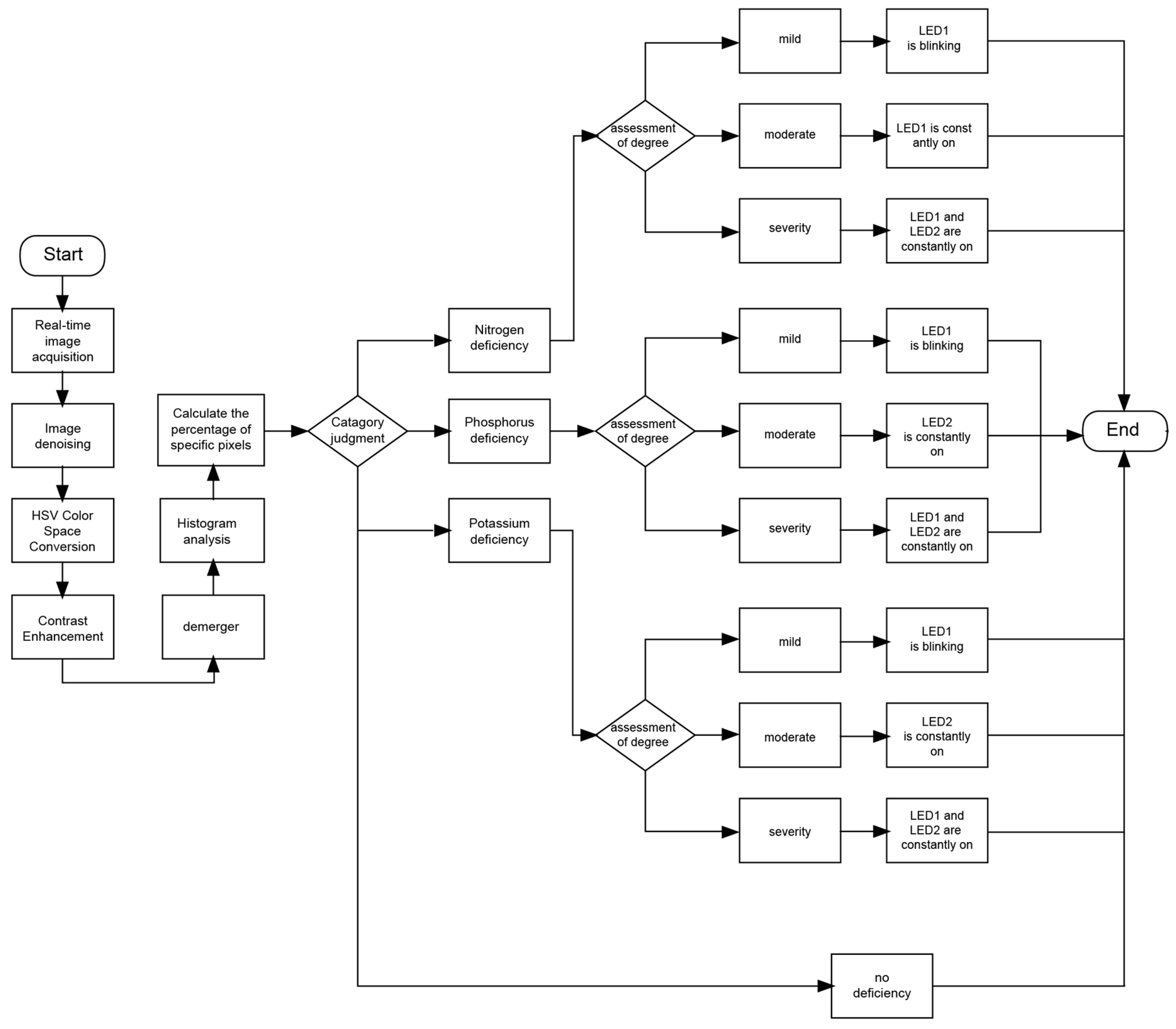
To enhance the reliability of classification results, the system integrated a multi-stage classification process. The classification procedure did not involve traditional machine learning training, validation, or testing stages but instead relied on predefined decision rules to optimize nutrient deficiency detection. Image data collected from the real-time capture module were processed to extract characteristic color features and construct a feature space. A decision-tree-based classification model was then applied to categorize lettuce nutrient deficiency levels based on these extracted features. The classification decisions were determined using predefined criteria derived from labeled lettuce images with verified nutrient deficiency conditions, ensuring consistent and interpretable outcomes.
The classification performance of the system was quantitatively evaluated using precision, recall, and F1-score metrics. These metrics were computed based on the confusion matrices, which captured the system’s ability to correctly identify nutrient deficiencies at different growth stages. The confusion matrices were constructed by comparing system predictions with ground truth labels obtained from expert assessments, and classification errors were analyzed based on the non-diagonal elements of the matrices. Statistical validation of classification performance was conducted using a chi-square test to assess the significance of misclassifications, ensuring that the conclusions drawn from the results were robust and reliable. The classification errors mentioned in line 658 were calculated by determining the proportion of misclassified instances in relation to the total number of classified samples. Misclassification trends were analyzed to identify potential sources of error, such as variations in lighting conditions and leaf texture inconsistencies. These insights contributed to the refinement of the algorithm and guided the future development of the system toward incorporating additional feature extraction techniques, including texture and shape analysis, to enhance classification accuracy.
The workflow is described as follows. After a power reset, the system was initiated, and each module entered the initialization phase. This phase was managed by the central control FPGA chip, which not only drove the camera module, storage module, algorithm module, and HDMI module but also provided the necessary clock signals and configured the internal registers of each module. After initialization, the CMOS sensor collaborated with the FPGA and captured RGB video data using line and field signals. And these data were transferred to the FPGA and buffered in DDR3 memory and then further processed in the algorithm module. The primary task was to denoise the captured images of the lettuce canopy, convert the image data format from RGB565 to HSV mode, and enhance contrast for better detail information. Then, the characteristic colors and background of the lettuce canopy were precisely segmented, and the segmented images were displayed in real-time on an HDMI monitor. To classify the nutrient deficiencies, a histogram-based classification model was implemented. This model analyzed the distribution of pixel values within each characteristic color region, applying a threshold-based decision rule to distinguish among different nutrient deficiency levels. The classification process involved constructing feature vectors based on the pixel frequency distribution and mapping them to predefined deficiency categories. The system subsequently conducted histogram analysis on the segmented pixel values, classified them based on the frequency of the predominant pixel values in each characteristic color, and determined the nutritional deficiencies of the lettuce by lighting up different LED lights. The workflow diagram of this study demonstrated each step from data collection to the display of final results, as shown in
Figure 11.
5. Experimental Results and Analysis
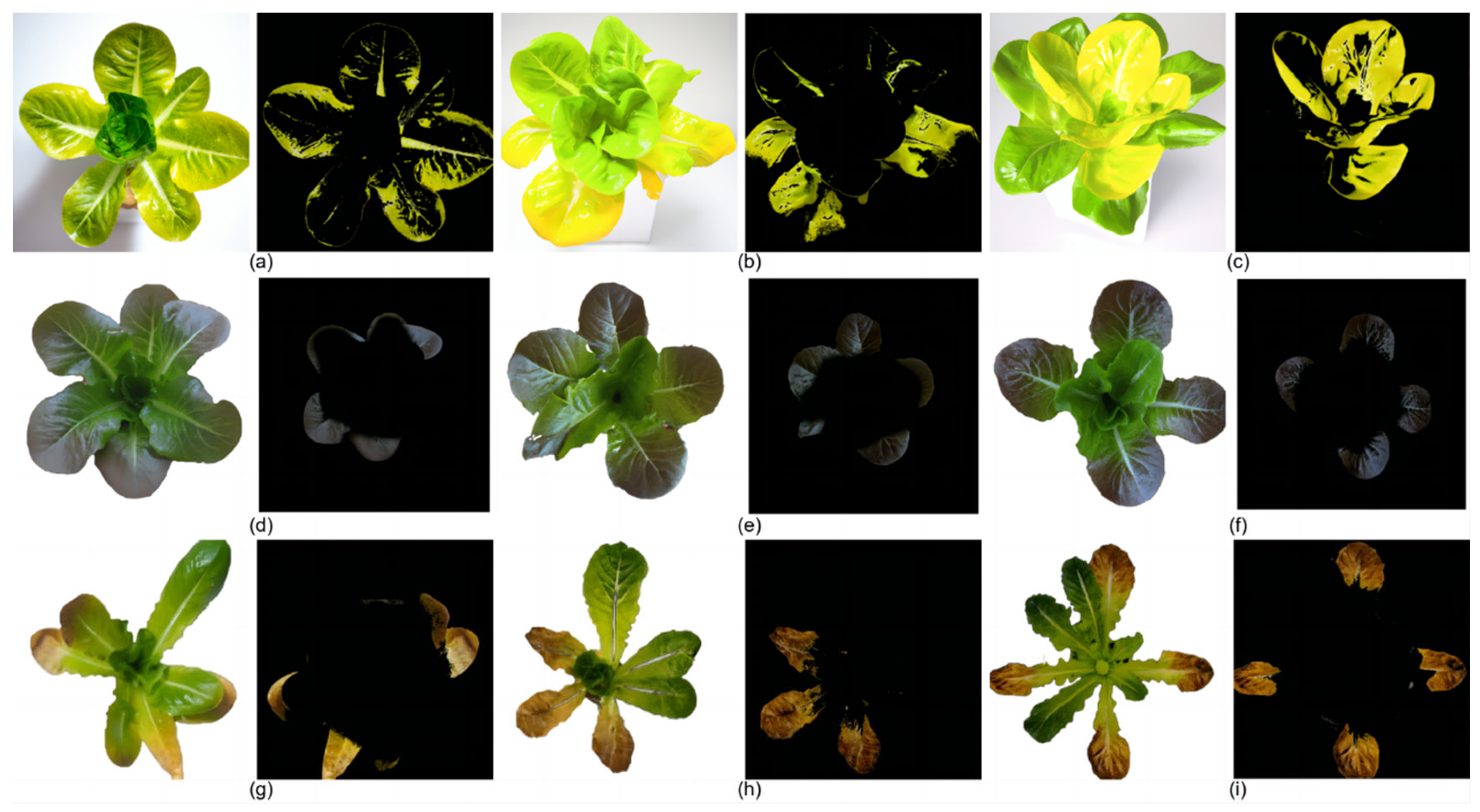
In this study, in the real-time nutrient deficiency detection process in mature lettuce leaves, the characteristic colors of nutrient-deficient areas were accurately segmented from the background. The nutrient deficiency segmentation results for mature lettuce in this study are depicted as follows:
Figure 12a–c for the N-deficient group,
Figure 12d–f for the P-deficient group, and
Figure 12g–i for the K-deficient group.
To ensure the generalization capability of the proposed system, the dataset was collected from multiple hydroponic lettuce cultivation setups under varying environmental conditions, including different lighting intensities and background variations. Additionally, data augmentation techniques, such as brightness normalization and contrast enhancement, were applied to mitigate biases associated with specific acquisition conditions. However, future studies will concentrate on validating the system on an independent dataset obtained from external sources to further assess its robustness in diverse practical scenarios. All reported accuracy, precision, recall, and F1-score values represent the mean of multiple trials conducted across different lettuce growth stages. Standard deviations were calculated and are provided in
Table 3 to illustrate the variability in system performance. A K-fold cross-validation approach (K = 5) was employed to estimate the mean and standard deviations of all reported metrics, ensuring a more reliable performance evaluation. In cases where K-fold cross-validation was not applicable, alternative resampling methods were used to assess the stability of the diagnostic outcomes. To further validate the reproducibility of the results, each experiment was conducted multiple times, with the number of trials per growth stage and nutrient deficiency level recorded. Confidence intervals were calculated for key performance metrics to provide statistical insights into the system’s reliability. These results confirm that the system maintains high accuracy and consistency in diagnosing nutrient deficiencies in lettuce across different growth stages.
To ensure the reliability of the results, statistical measures including mean, standard deviation, and confidence intervals were calculated for the accuracy values across multiple trials. These statistical parameters provide a more robust assessment of the system’s performance by quantifying variations in detection accuracy under different conditions. Each experiment was conducted across multiple trials to mitigate the effects of variability in image acquisition and processing. A K-fold cross-validation approach was employed to assess the system’s generalizability and stability, ensuring that the reported performance metrics are not overly optimistic due to a specific set of test samples. To further validate the effectiveness of the proposed diagnostic system, a comparative analysis was conducted against existing lettuce nutrient deficiency detection methods. Performance metrics such as precision, recall, and F1-score were benchmarked against alternative image-based diagnostic techniques, and the observed differences were statistically analyzed using paired t-tests (or ANOVA, if applicable) to determine the significance of performance improvements.
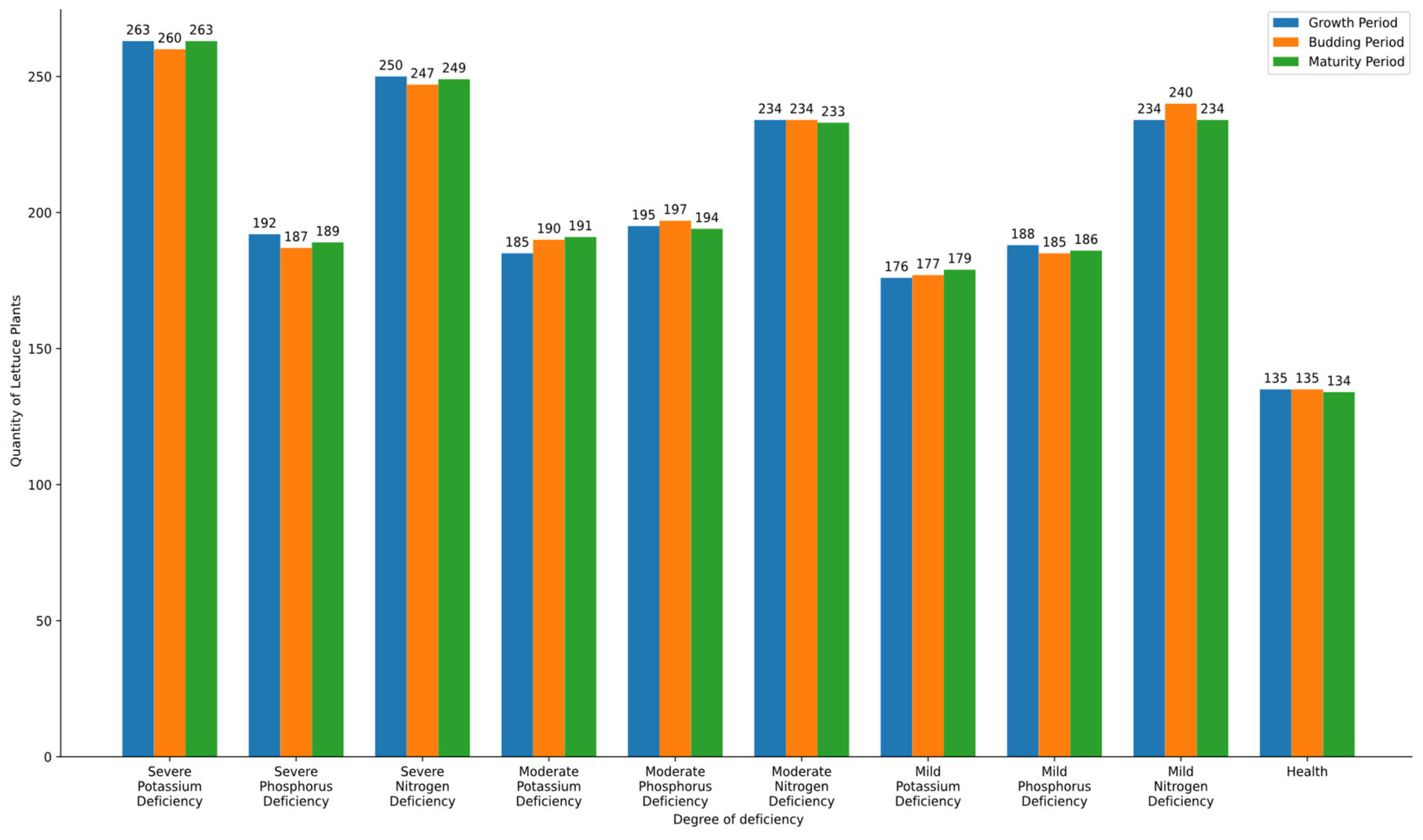
After multiple experiments, the number of lettuce plants identified by the system at different growth stages and varying levels of nutrient deficiencies was collected and organized. To evaluate the effectiveness of the proposed model, a comparison with existing classification techniques, such as machine learning-based classifiers (e.g., SVM, CNN), should be conducted. However, in this study, a hardware-optimized histogram analysis approach was prioritized to ensure real-time processing on FPGA. Future research will explore the integration of alternative classification techniques to assess performance differences and further improve diagnostic accuracy. To clearly illustrate the identification of nutrient deficiencies in lettuce at different time stages, a bar graph was created, as shown in
Figure 13.
Table 3 further presents the accuracy of the system in identifying lettuce plants with different levels of nutrient deficiencies across various growth stages.
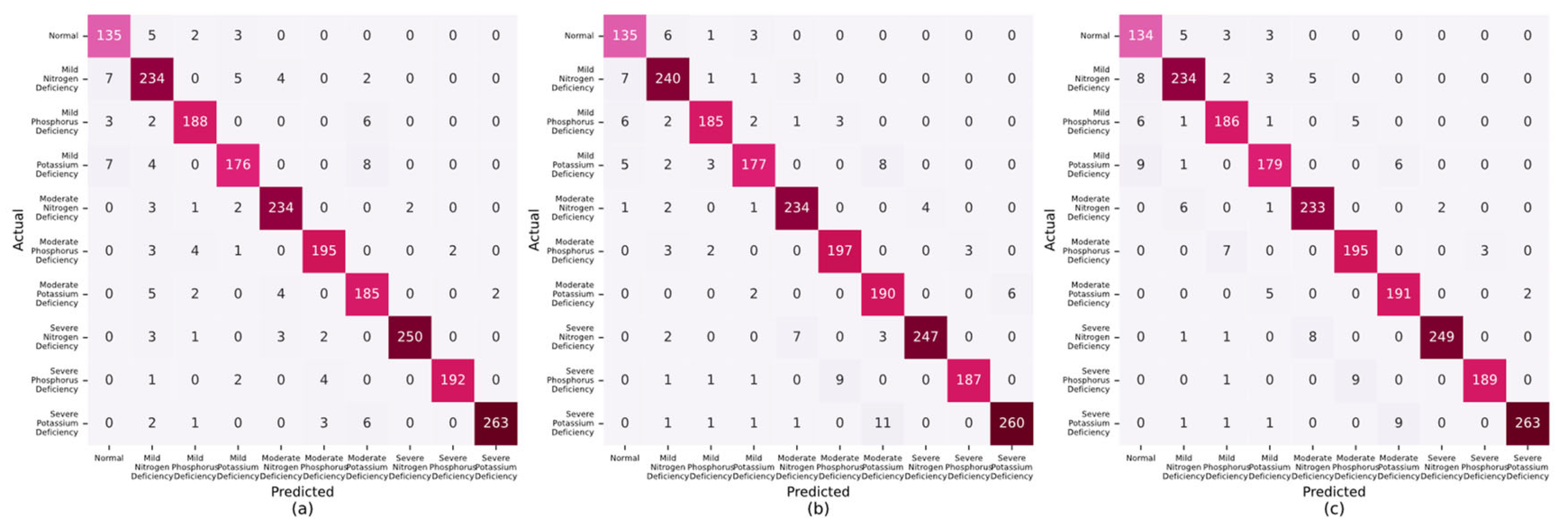
A comparative analytical approach was adopted to assess the efficacy of the proposed lettuce nutrient deficiency detection system. This process entailed aligning the nutrient deficiencies that were recognized by the system with the genuine deficiencies noted in lettuce at different growth stages. The confusion matrices, which were used to illustrate the accuracy of lettuce identification in the growing period, budding period, and maturity period, are presented in
Figure 14a–c. Elevated values along the principal diagonal of these matrices underscored the system’s proficiency in accurately identifying instances. In contrast, the values in the non-diagonal elements highlighted discrepancies between the nutrient-deficient states predicted by the system and the actual nutrient-deficient states, clearly indicating the categories of misclassification. These findings corroborated the system’s robust capability to discern three distinct levels of nutrient deficiencies in lettuce across diverse growth stages.
The real-time diagnostic system developed in this study demonstrated significant improvements in efficiency, accuracy, and real-time processing compared to existing nutrient deficiency detection methods [
24,
25]. Traditional approaches, such as manual inspection or non-automated image processing techniques, often suffer from subjectivity, time inefficiency, and susceptibility to environmental factors, such as lighting variations. Some recent studies have explored machine learning-based classification methods for nutrient deficiency detection; however, these typically rely on offline processing, require extensive labeled datasets, and may not provide real-time diagnostic capabilities.
In contrast to conventional methods, this study employed an FPGA-based hardware-accelerated architecture, enabling parallel processing and significantly reducing computational latency. The system integrated advanced image processing techniques, including dynamic window algorithms and contrast enhancement, ensuring robustness in diverse environmental conditions. Additionally, while deep learning-based approaches provide promising classification accuracy, they are often computationally intensive and require substantial training data. In contrast, the proposed system efficiently processes real-time image data without necessitating extensive model training, making it more suitable for practical agricultural applications. Although direct comparison with existing methods is challenging due to variations in experimental conditions, dataset characteristics, and evaluation metrics, the proposed system demonstrates high reliability in detecting nutrient deficiencies across different growth stages, achieving an average precision of 0.944, recall of 0.943, and F1 score of 0.943. As shown in
Figure 15, the nutrient deficiency diagnosis performance in lettuce at various growth stages is illustrated:
Figure 15a radar chart of precision for lettuce nutrient deficiency detection,
Figure 15b radar chart of recall for lettuce nutrient deficiency detection, and
Figure 15c radar chart of F1 score for lettuce nutrient deficiency detection. These performance metrics suggest that the system provides a viable alternative to both traditional visual assessment and data-intensive machine learning approaches, striking a balance between accuracy, real-time processing, and practical applicability in hydroponic farming. Furthermore, the effect of the plant’s growth stage on the accuracy and quality of nutrient deficiency detection was considered. It was observed that the system’s performance varied slightly across the different growth stages, with the highest accuracy typically achieved during the budding and maturity periods. The precision of deficiency detection in the growing period was slightly lower, possibly due to the morphological and physiological changes in the plant at this stage. This suggests that the growth stage could influence the system’s ability to detect nutrient deficiencies, especially in the early stages of lettuce development. Future research could explore the impact of specific growth stages in greater detail to optimize the detection process for various developmental phases.
6. Conclusions
This study fully utilized the parallel processing capabilities of FPGA (Field-Programmable Gate Array), combined with multidimensional image analysis algorithms, optimized image enhancement algorithms, and image denoising techniques, to propose a novel real-time diagnostic system for nutrient deficiencies in hydroponically cultivated lettuce. The system was able to precisely identify nutrient deficiencies of lettuce across different growth stages, with an average precision of 0.944, a recall of 0.943, and an F1 score of 0.943, underscoring its high diagnostic efficiency and accuracy, which satisfied the requirements for automation, real-time processing, and non-destructive detection.
The main innovations of this study are reflected in the following aspects: First, the system employed real-time image processing methods to significantly enhance image processing efficiency and reduce computational latency through hardware-accelerated algorithms, thus providing an efficient solution for the real-time diagnostic needs of agricultural applications. Second, by leveraging the FPGA-based parallel processing architecture, the system enabled efficient analysis of multidimensional image features, which not only extracted color features but also focused on edge details and local variations in the image, making it capable of precisely detecting subtle nutrient-deficient regions under various environmental conditions. Furthermore, the proposed dynamic window algorithm effectively handled noise and lighting variation, ensuring the robustness of the system in complex real-world scenarios.
Nevertheless, this study has certain limitations. Specifically, the system still heavily relies on color features in some cases, which are inherently susceptible to the influence of lighting conditions and leaf aging, potentially leading to classification errors. Future research will focus on further expanding the range of features, such as incorporating information on leaf texture, shape, and light reflection properties, while integrating multi-angle machine learning techniques to develop a more comprehensive and accurate diagnostic model for nutrient deficiencies.
Although this study demonstrated the feasibility and effectiveness of the proposed system in identifying nutrient deficiencies in lettuce, there remains room for further exploration in future research. In particular, quantitative assessments of image quality, such as peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM), will be considered to evaluate the impact of image enhancement, noise reduction, and edge detection techniques on overall image clarity and diagnostic accuracy. Moreover, further experimental comparisons with other existing systems will provide a more comprehensive understanding of the proposed system’s performance in various real-world conditions. Future research will also concentrate on extending the analysis to include texture-based features and multi-angle imaging techniques, which may enhance classification robustness and reduce dependency on color features that are sensitive to environmental factors, such as lighting and leaf aging.
By proposing an innovative, hardware-accelerated real-time diagnostic system, this study provides an efficient and practical solution for detecting nutrient deficiencies in hydroponically cultivated lettuce and offers valuable insights and directions for advancing precision agricultural technologies, paving the way for more intelligent and sustainable agricultural practices.