Smart Image-Based Deep Learning System for Automated Quality Grading of Phalaenopsis Seedlings in Outsourced Production
Abstract
1. Introduction
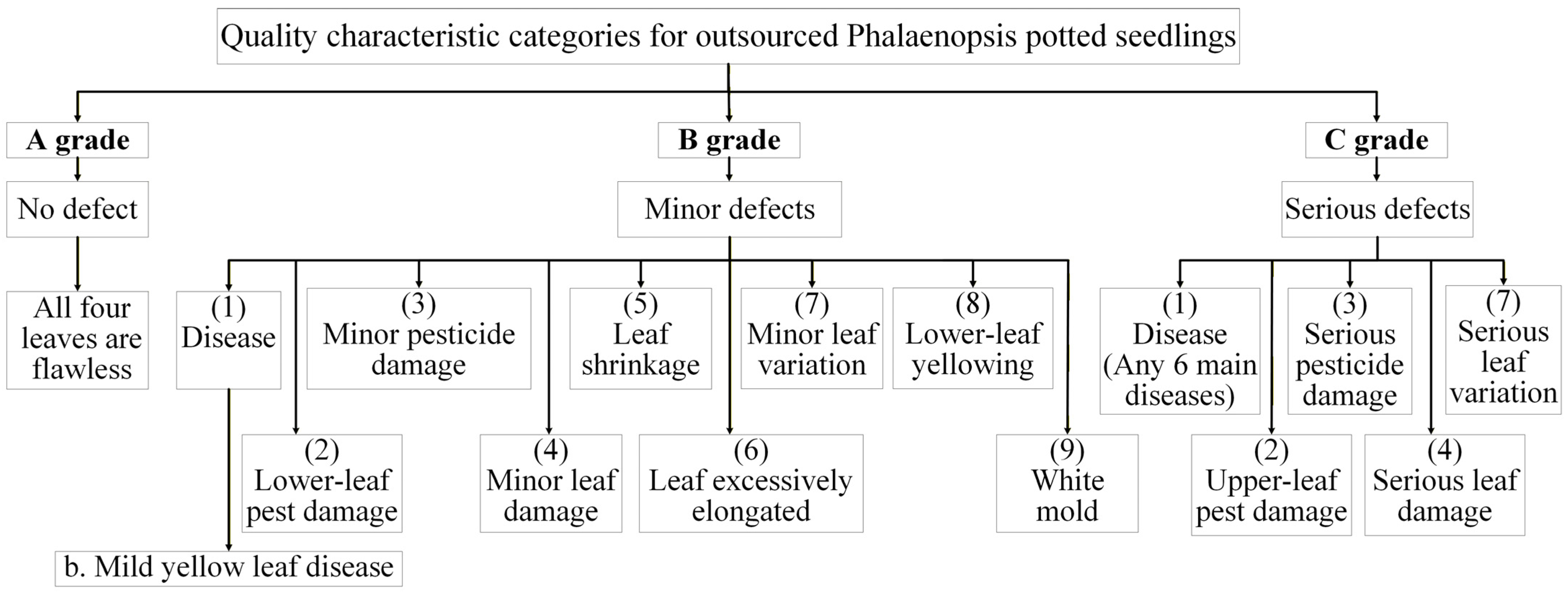
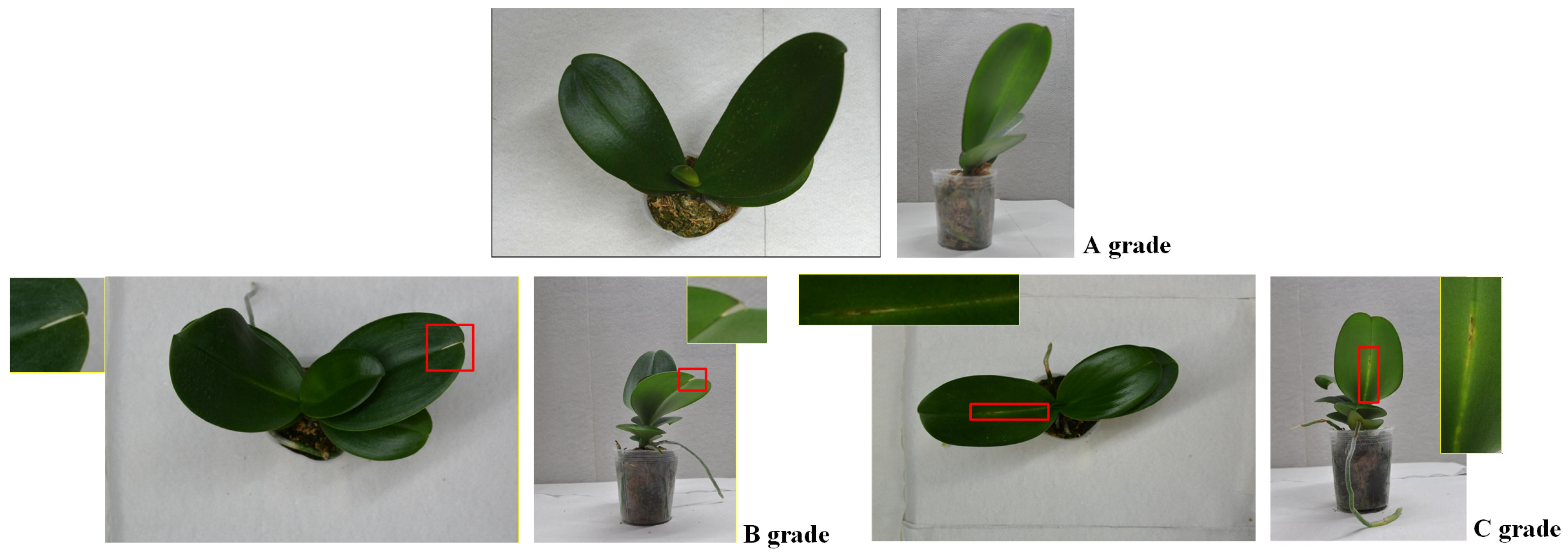
1.1. Defect Types in Phalaenopsis Potted Seedlings
1.2. Quality Grades for Acceptance of Outsourced Phalaenopsis Seedlings
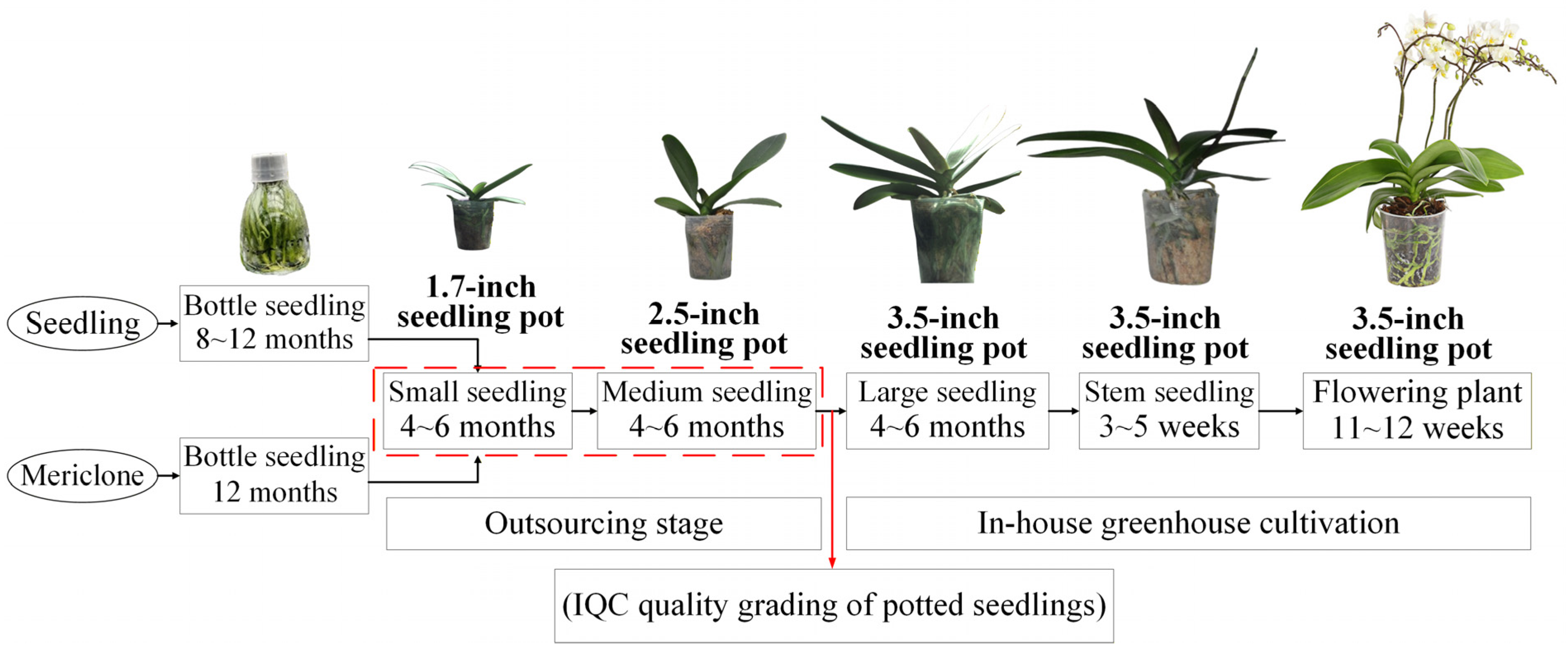
1.3. Current Manual Quality Grading Processes
1.4. Contribution
2. Literature Review
2.1. Machine Vision Applications in Horticultural Quality Assessment
2.2. Deep Learning for Plant Defect Detection and Phenotyping
2.3. Integration of Depth Sensing (RGB-D) for Plant Morphological Analysis
2.4. Multi-View and Multi-Modal Imaging Approaches in Plant Inspection
2.5. Machine Learning for Feature-Based Classification and Grading
2.6. Hybrid Deep Learning and Machine Learning Systems in Agriculture
3. Materials and Methods
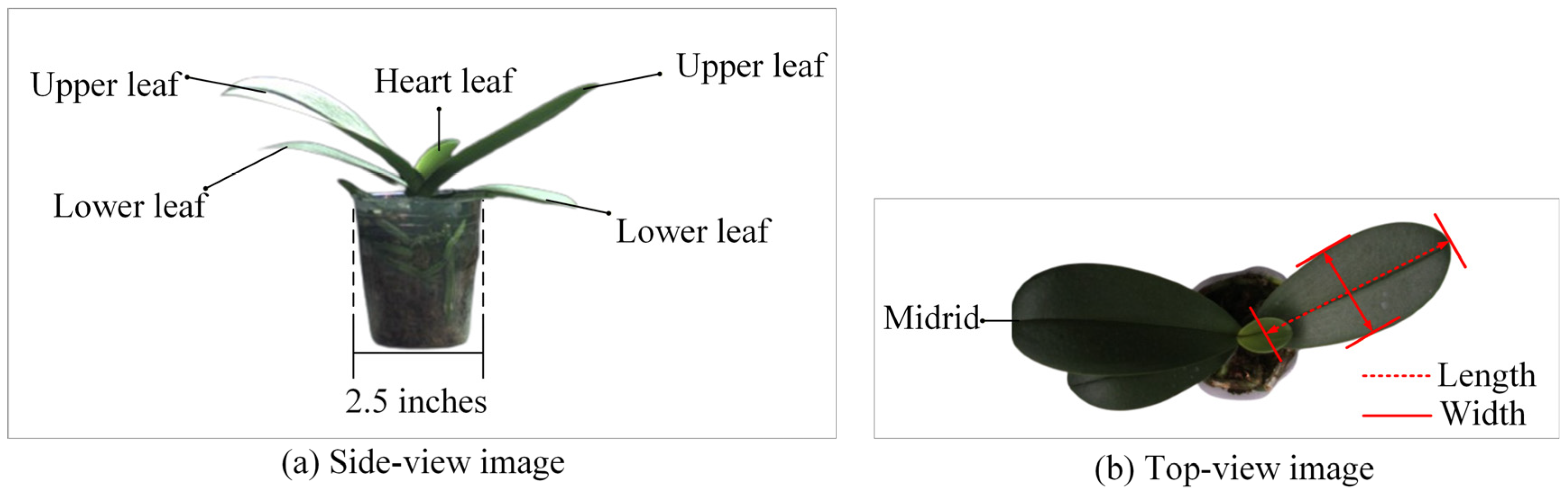
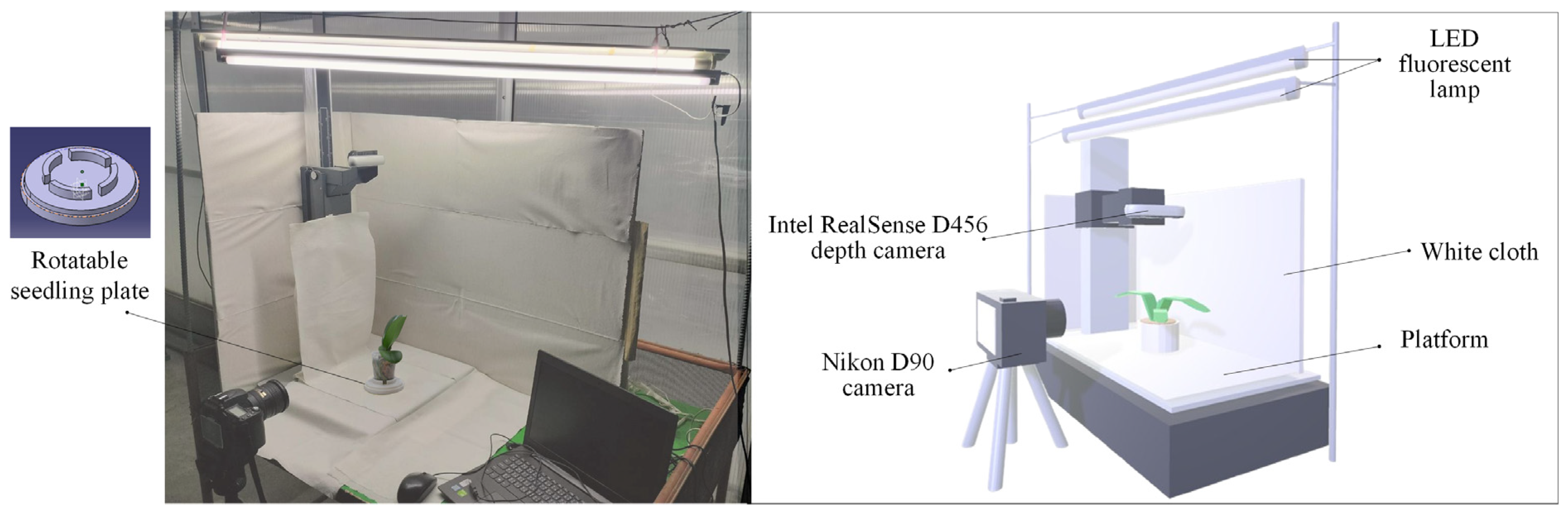
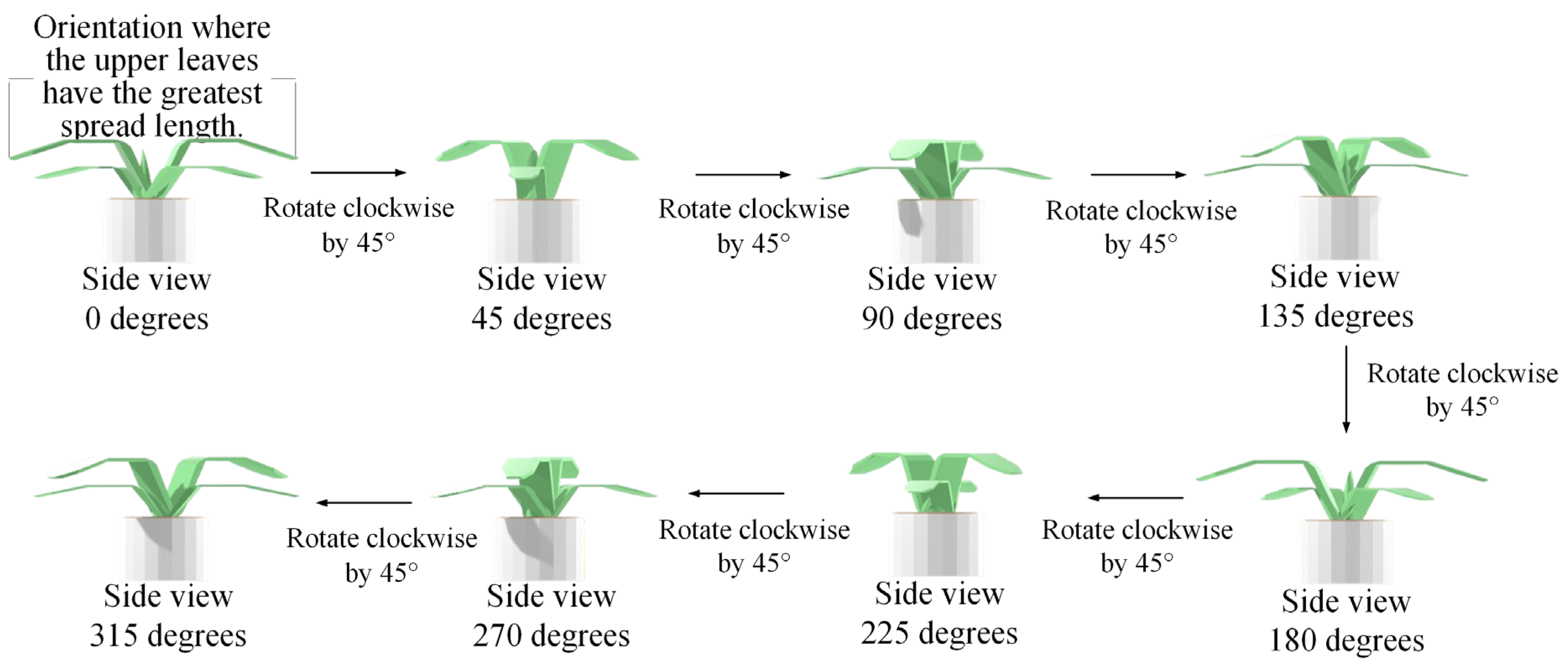
3.1. Image Acquisition of Potted Seedlings
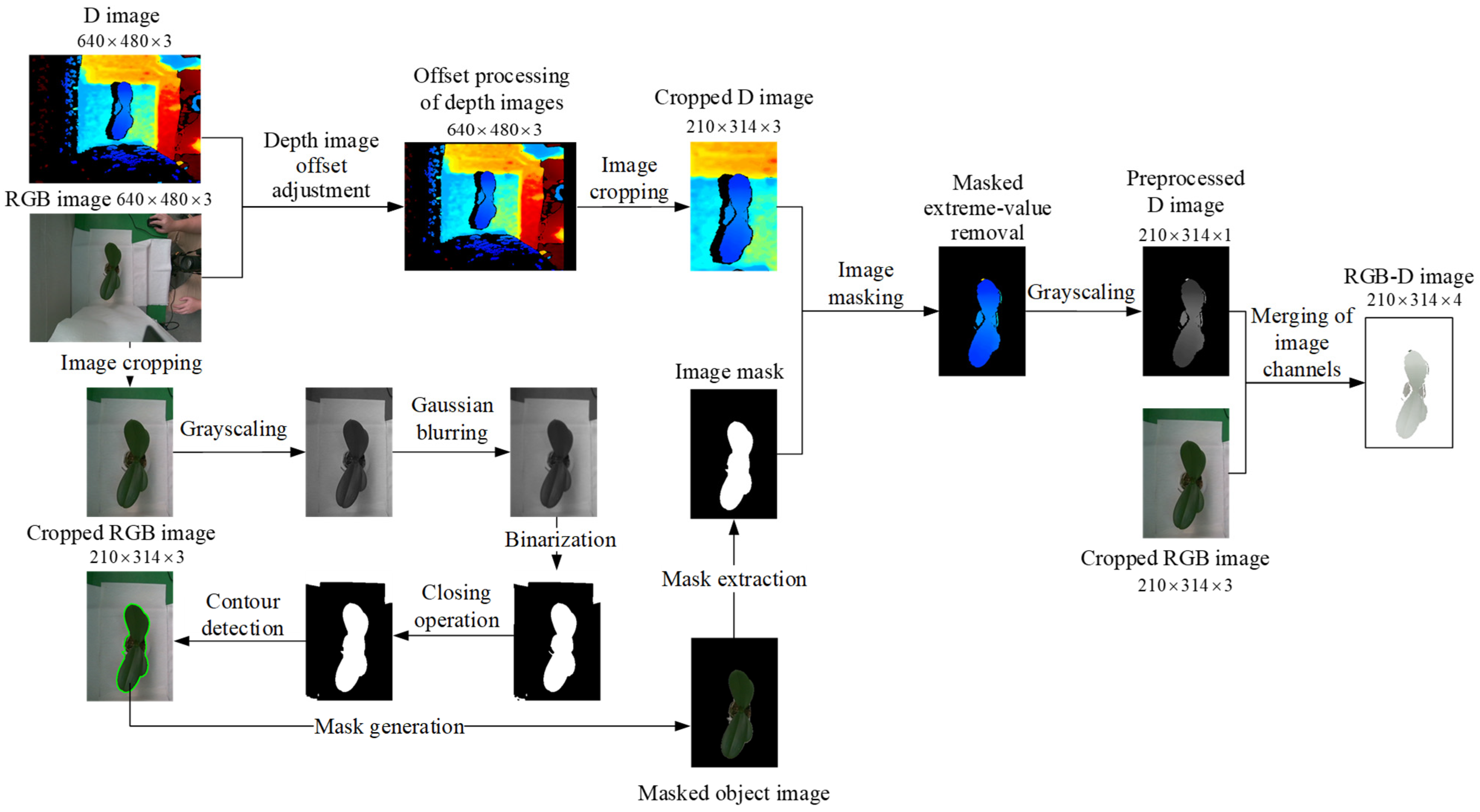
3.2. Image Pre-Processing of Potted Seedlings
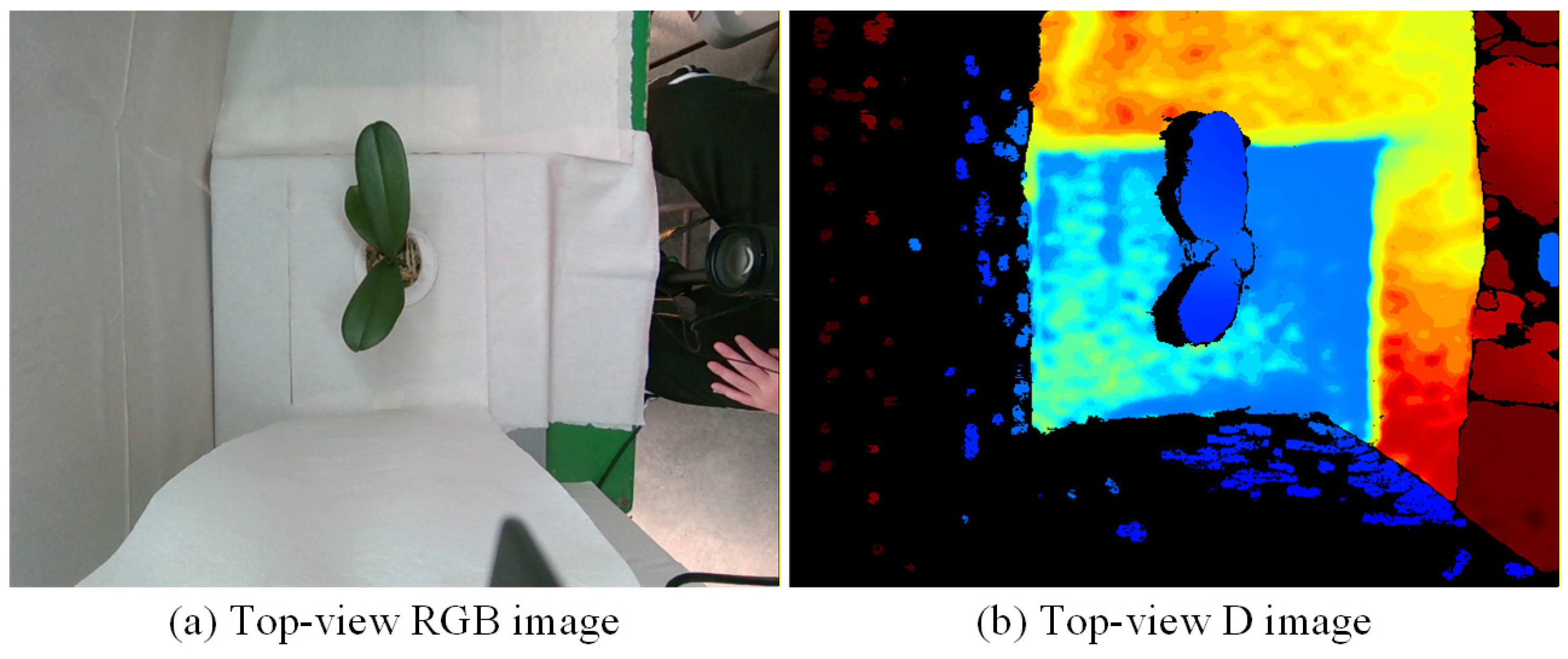
3.2.1. Image Pre-Processing for Top-View Images
3.2.2. Image Pre-Processing for Side-View Images
3.3. Feature Vector Transformation and Labeling
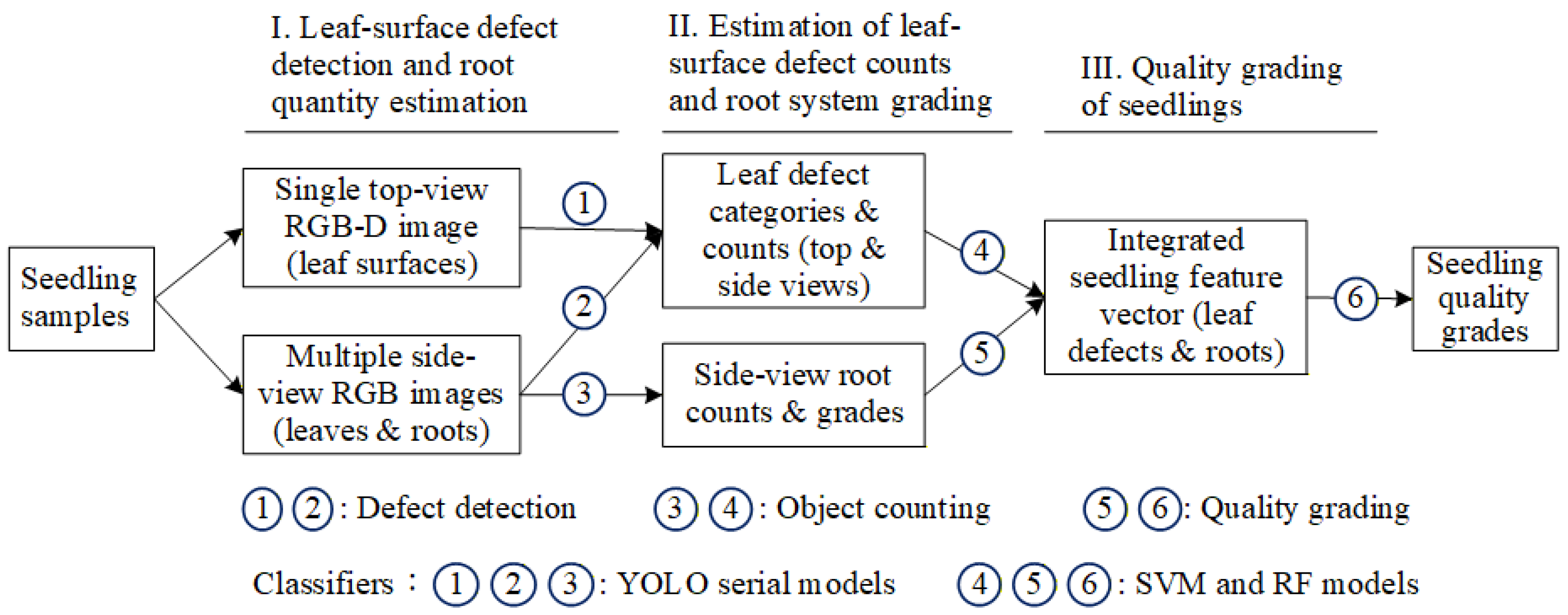
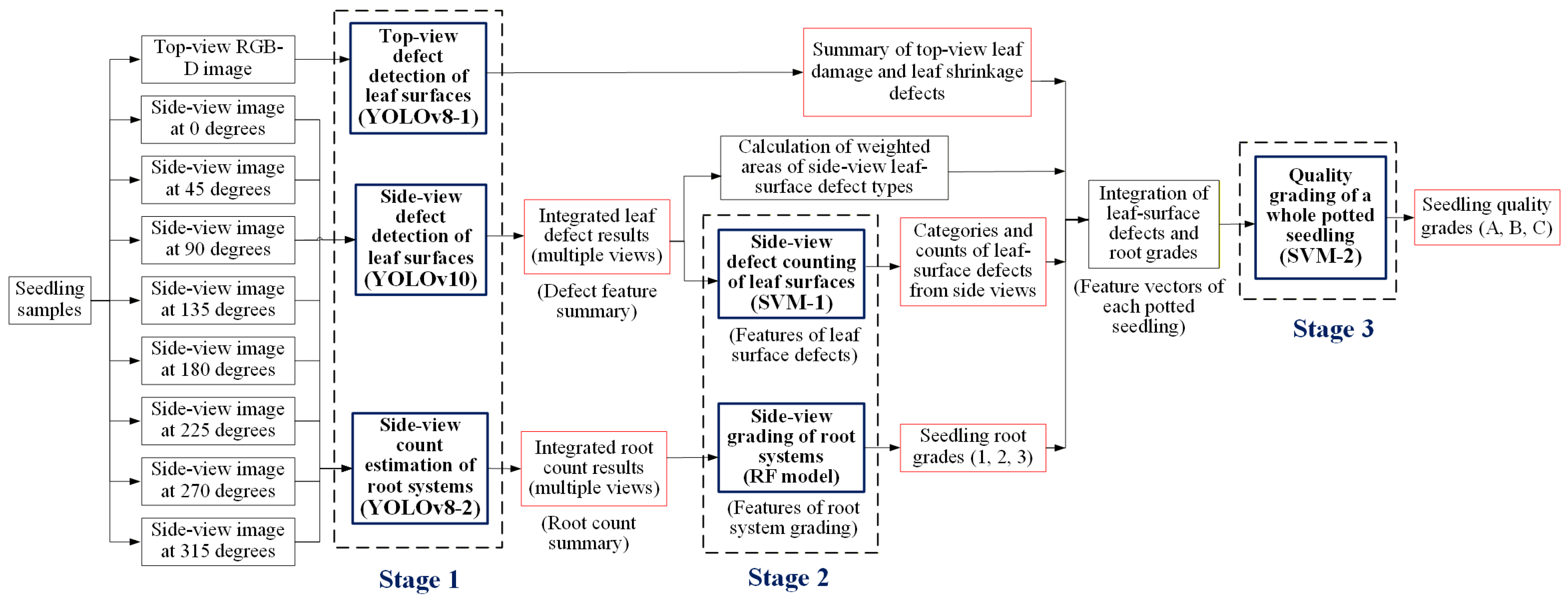
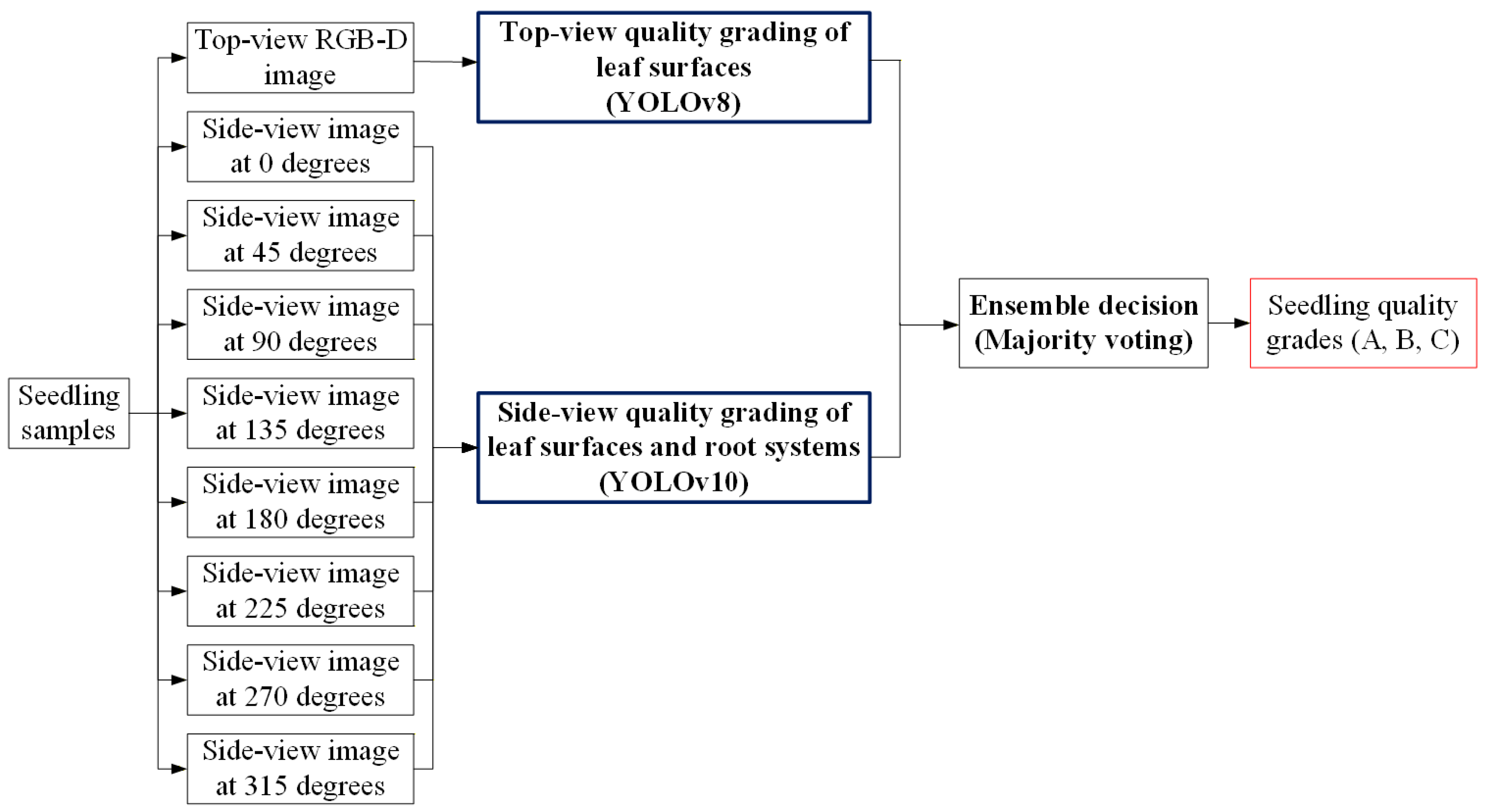
3.4. Proposed Three-Stage Grading Method in This Study
3.4.1. YOLOv8 Configuration with Four-Channel RGB-D Image Input
3.4.2. YOLOv10 Configuration for Multi-View Images Using Transfer Learning
3.4.3. YOLOv8 Model for Estimating Root Quantity
3.4.4. SVM Model for Estimating the Number of Leaf-Surface Defects in Potted Seedlings
3.4.5. RF Model for Determining the Root System Grade of Potted Seedlings
3.4.6. SVM Model for Grading the Quality of Whole Potted Seedlings
3.4.7. Mathematical Definition of Defect Weighting and Grade Boundaries
- Grade A: S < TA or good quality grade (G = 3),
- Grade B: TA ≤ S < TC or moderate quality grade (G = 2),
- Grade C: S ≥ TC or poor quality grade (G = 1),
3.5. Direct Grading Method in This Study
4. Results and Discussion
4.1. Experimental Hardware, Captured Images, and User Interface
4.2. Dataset Description and Experimental Workflow
4.3. Performance Evaluation Indices
4.4. Parameter Settings of Deep Learning and Machine Learning Models
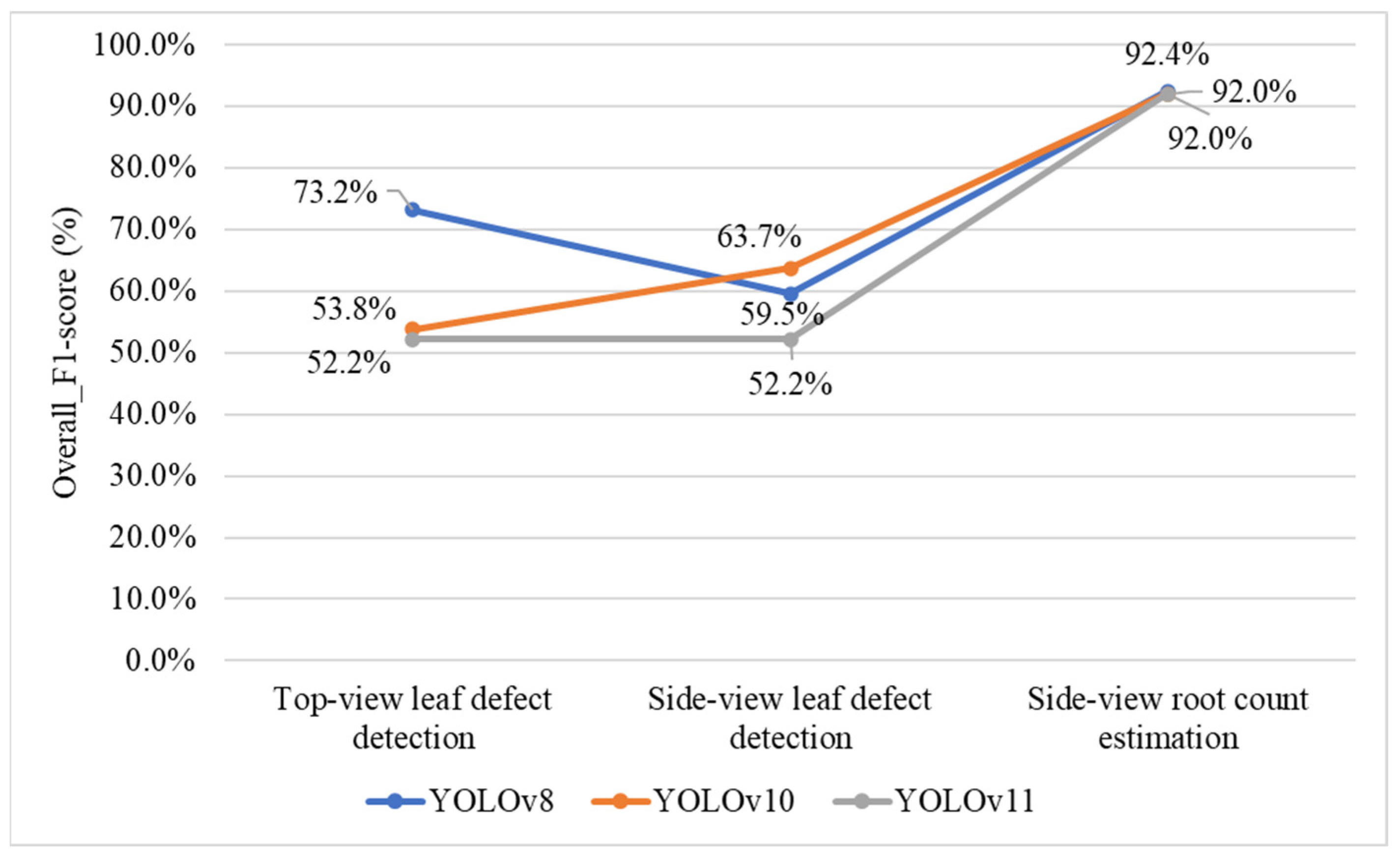
4.5. Selecting YOLO Models for Leaf Defect Detection and Root Count Estimation
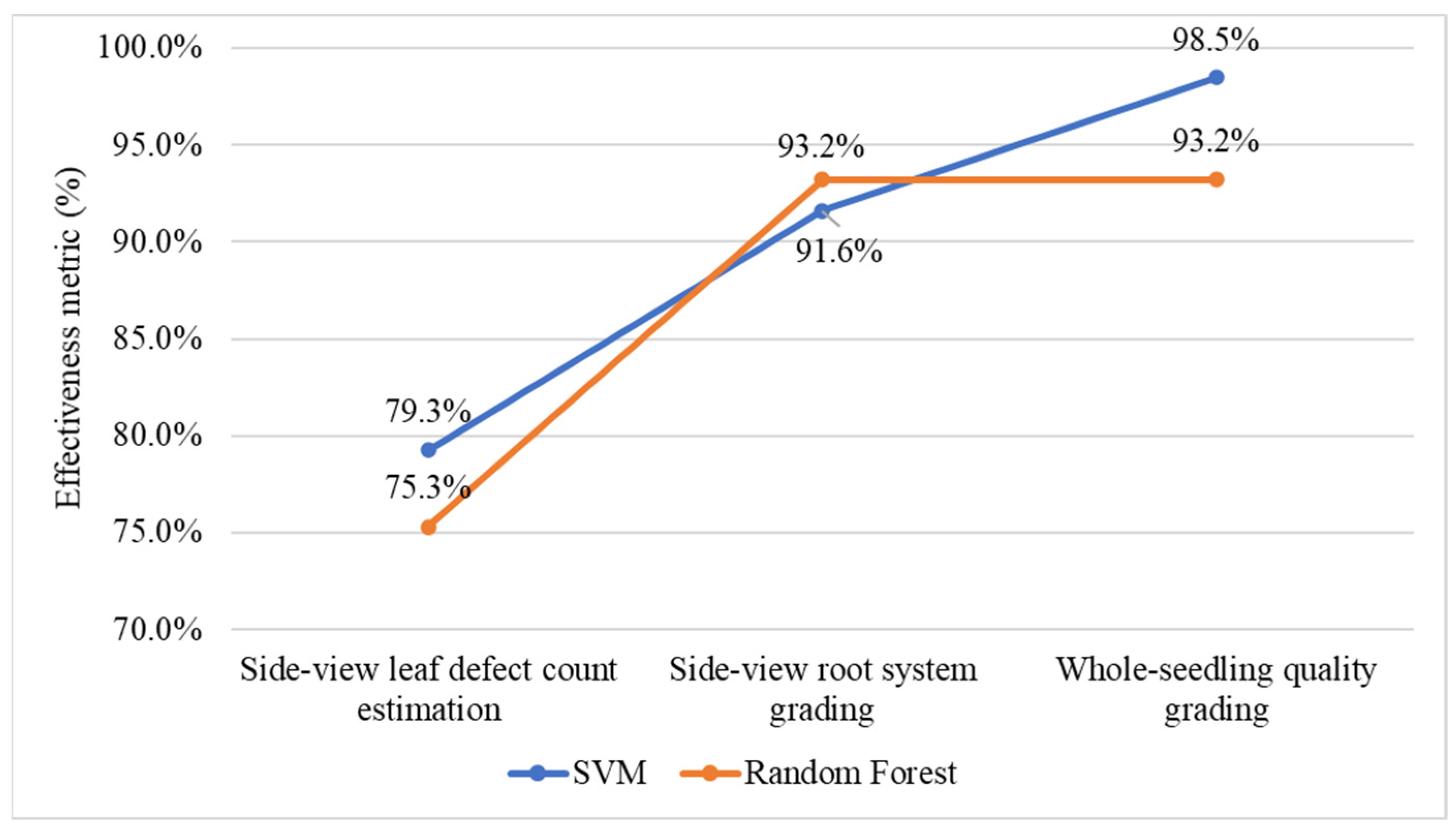
4.6. Selecting Machine-Learning Models for Leaf Count Estimation and Quality Grading
4.7. Performance Evaluation of Three-Stage and Direct Grading Methods in This Study
4.8. Misclassification Analyses and Failure Cases
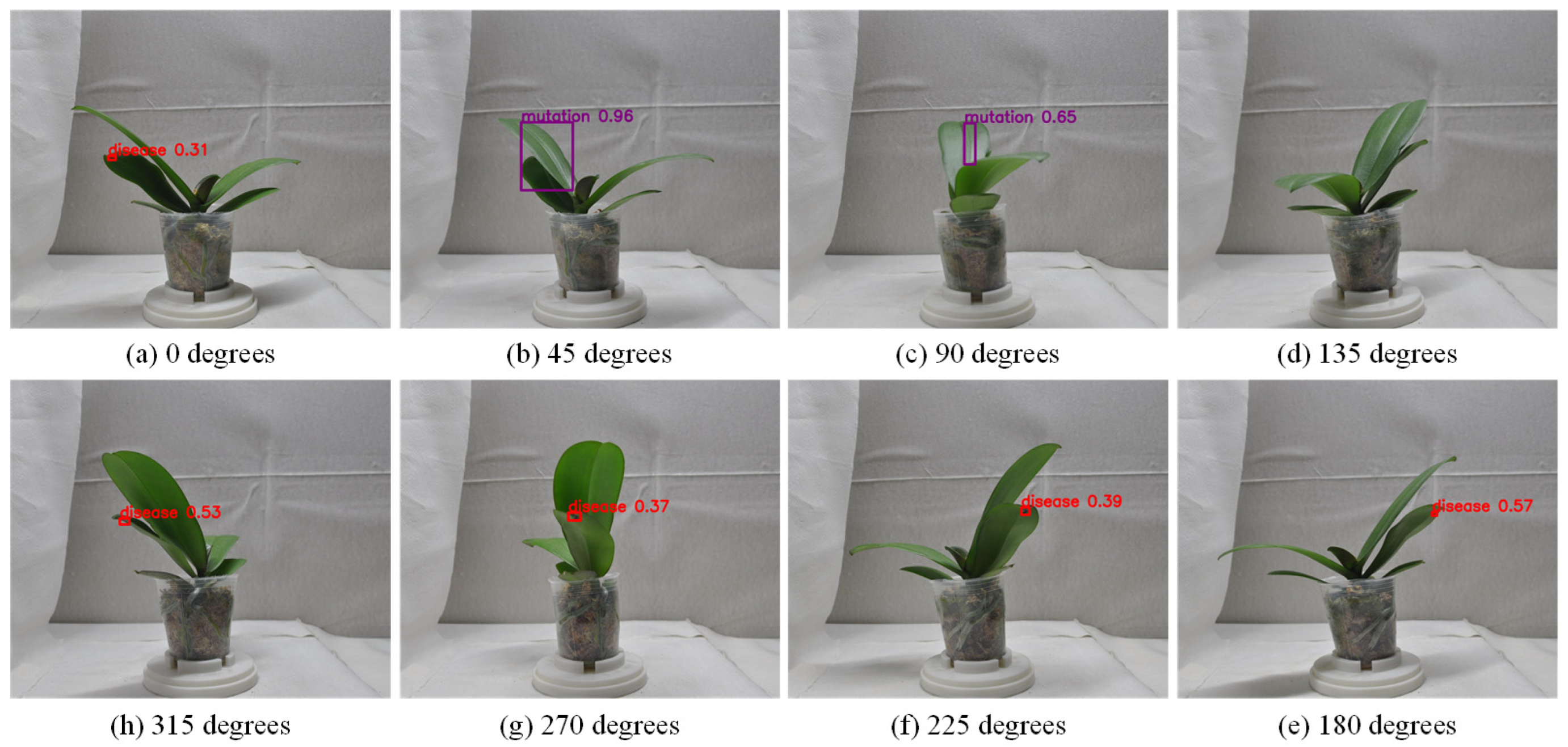
4.8.1. Misclassification Cases of Single-Sided Leaf Defect Detection in Stage 1
4.8.2. Failure Case Analyses in Final Quality Grading
4.9. Robustness Analysis of the Proposed Method
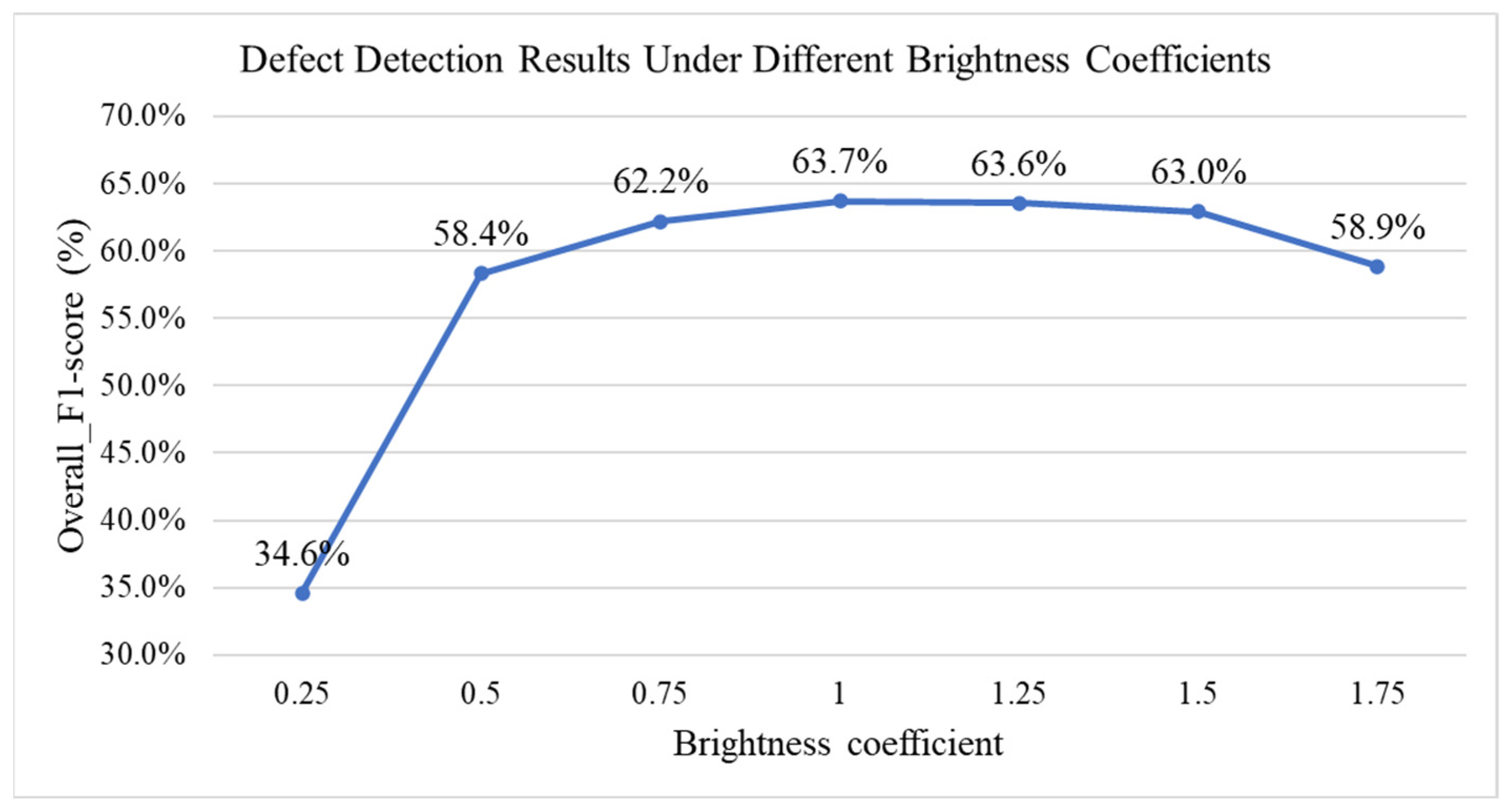
4.9.1. Impact of Environmental Lighting Changes on Detection Performance
4.9.2. Impact of Repeatedly Counting the Same Defect from Different Viewpoints on Detection Performance
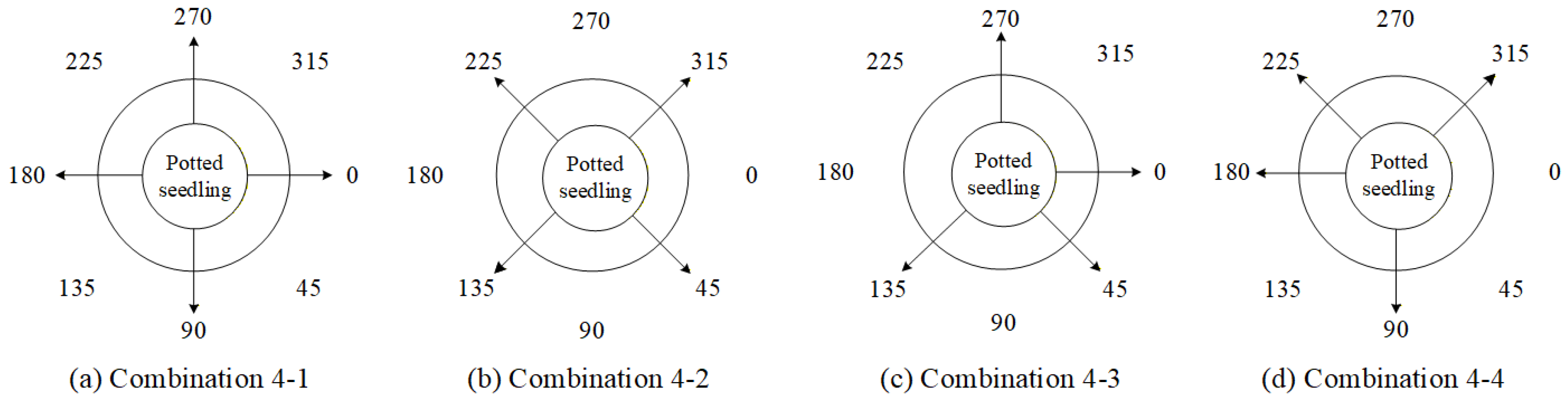
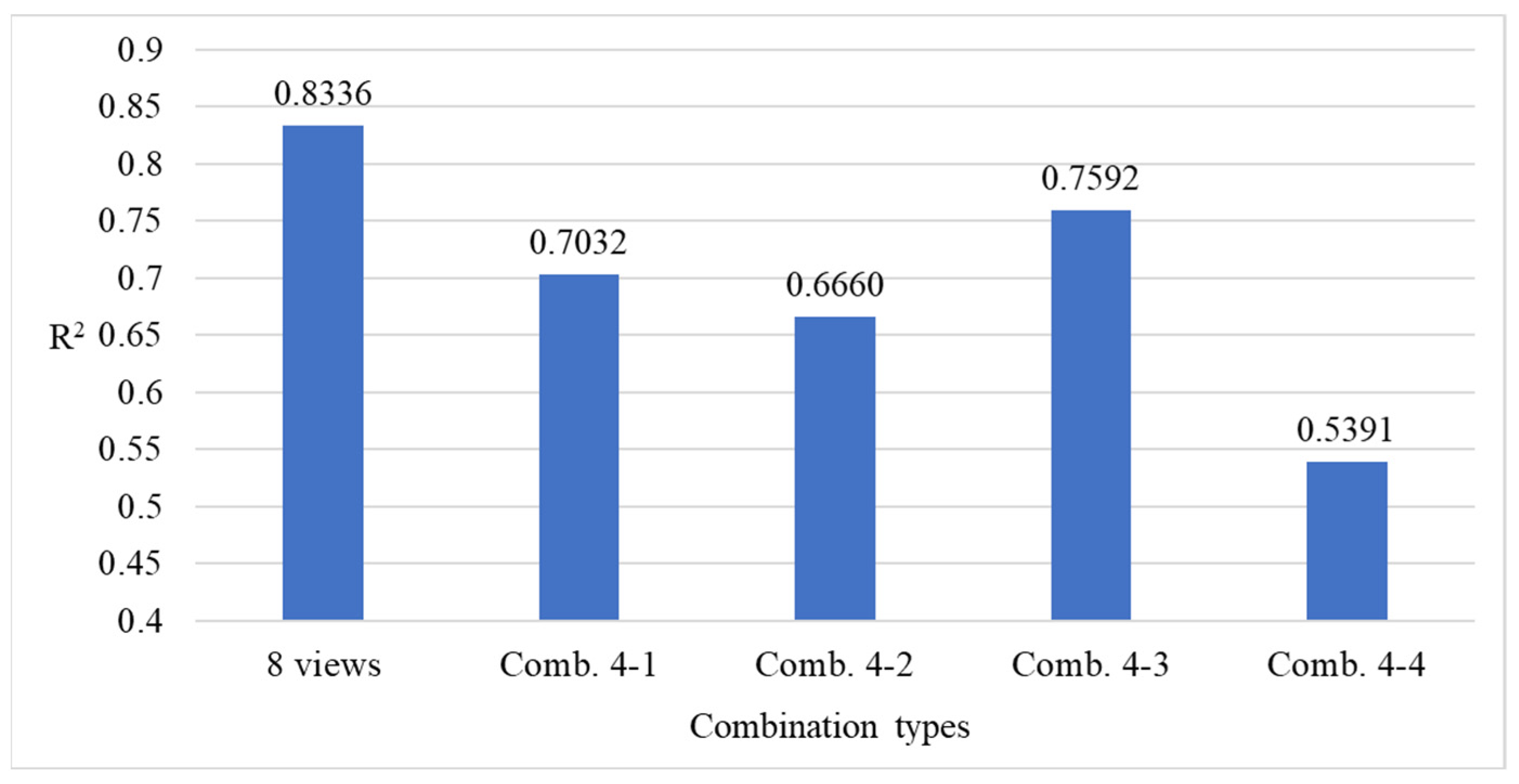
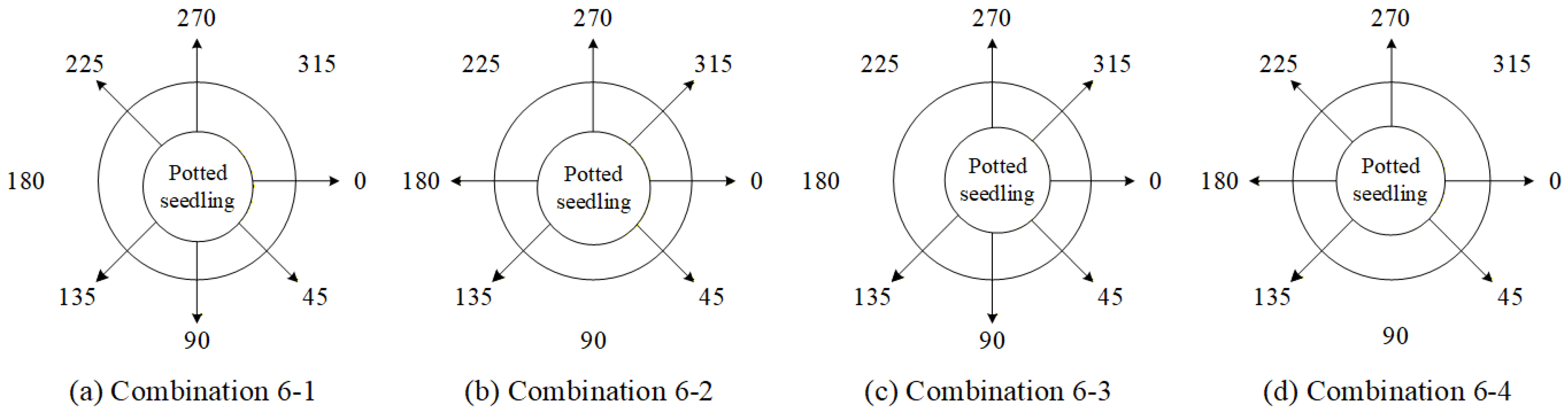
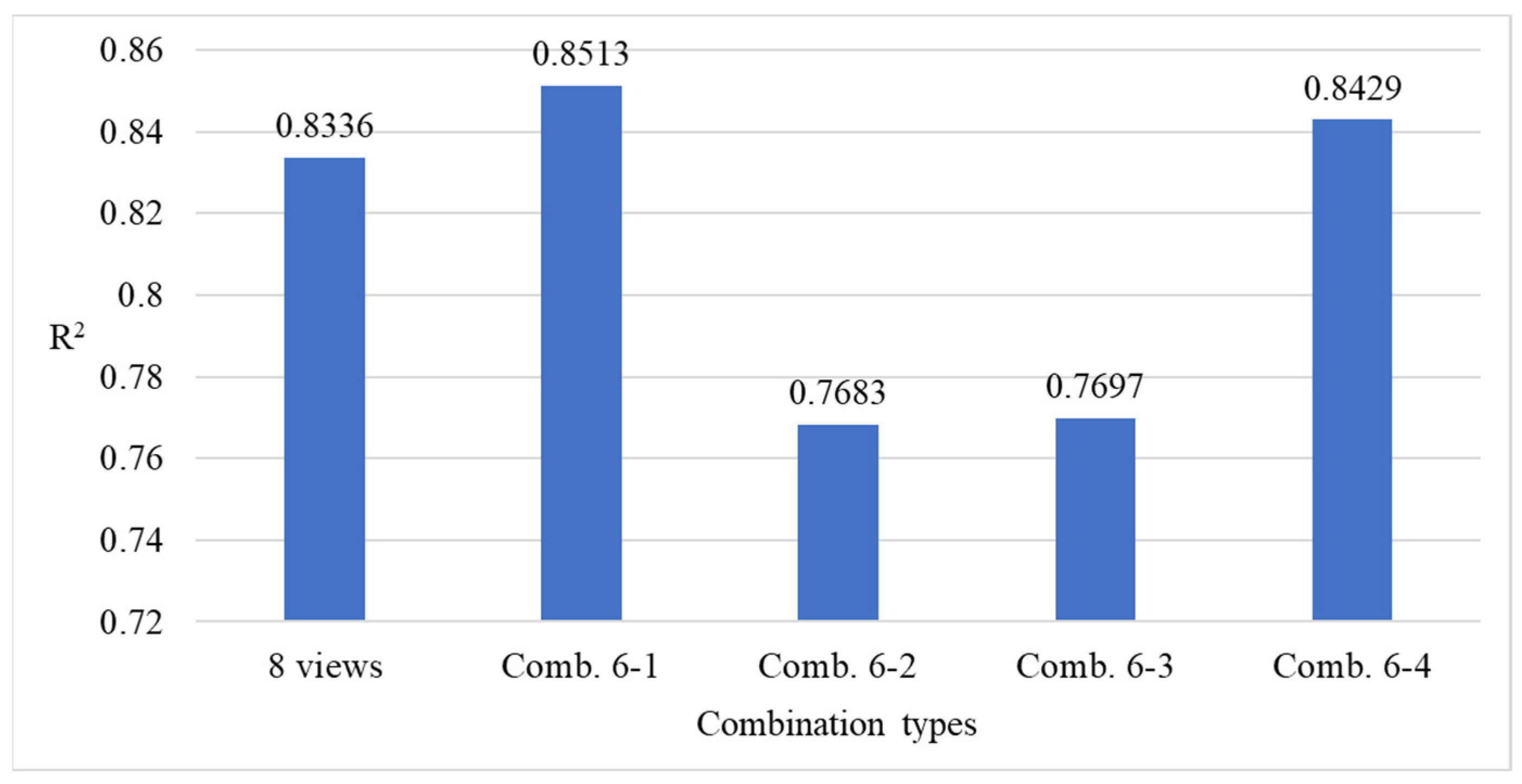
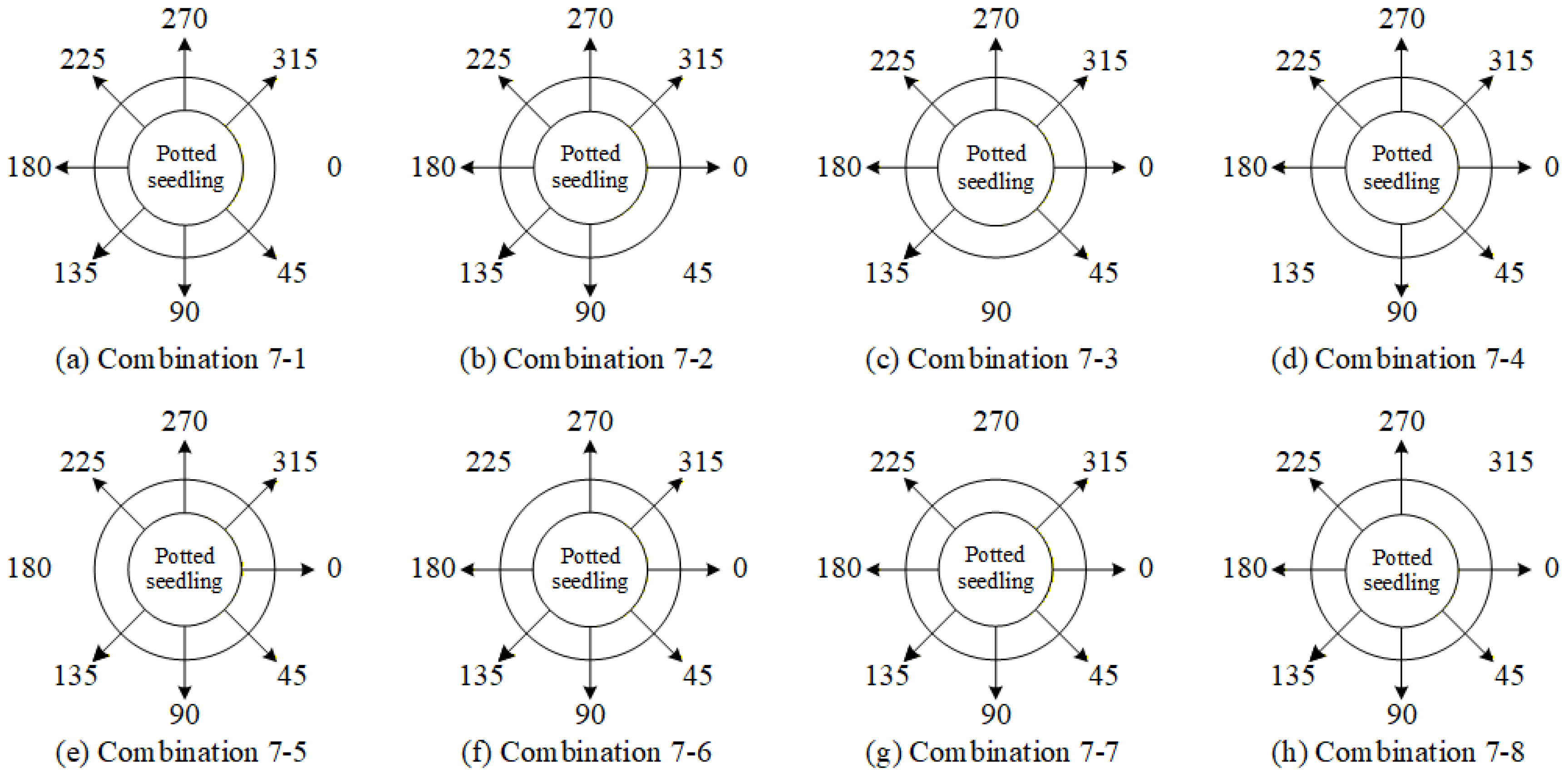
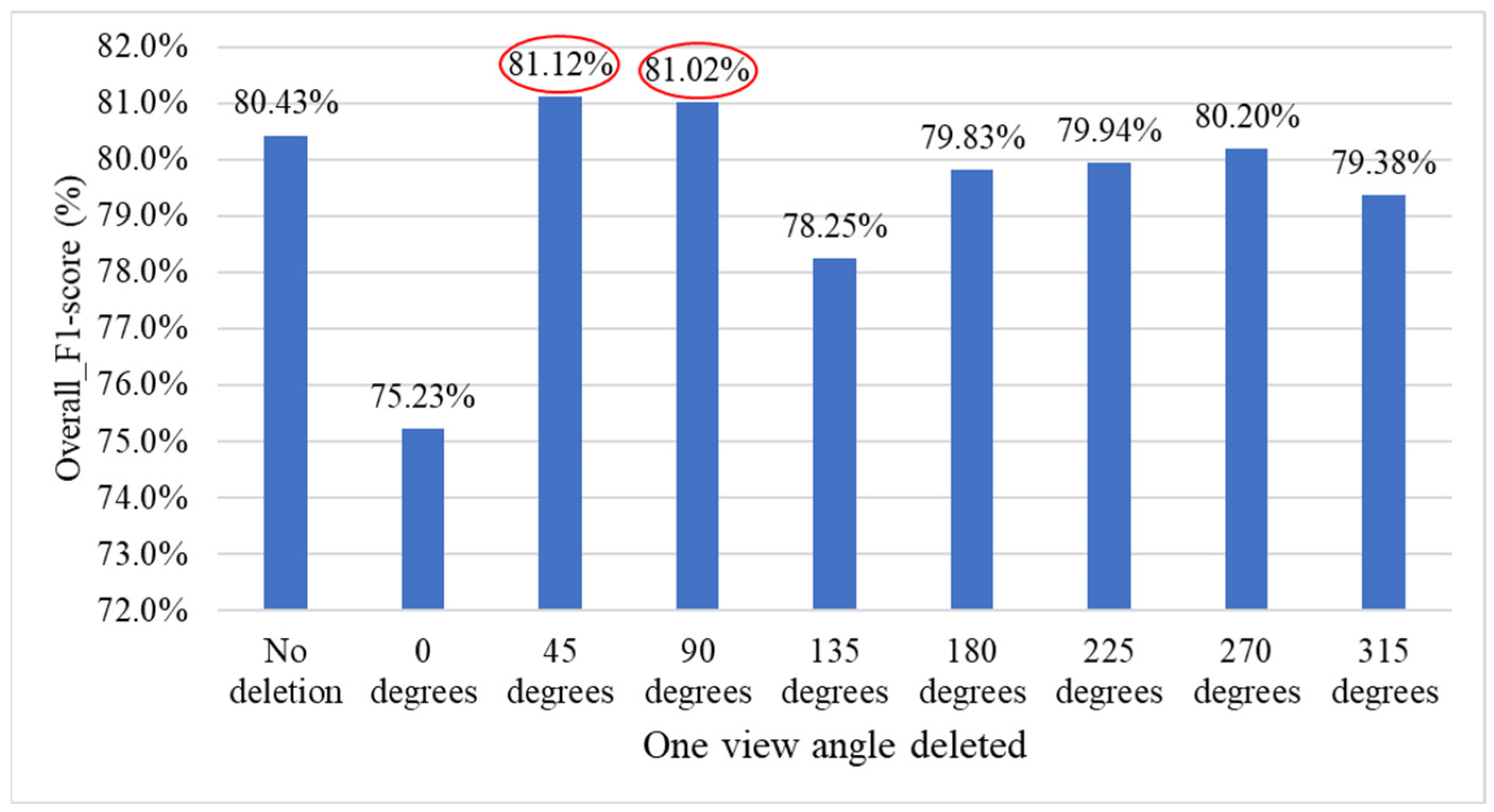
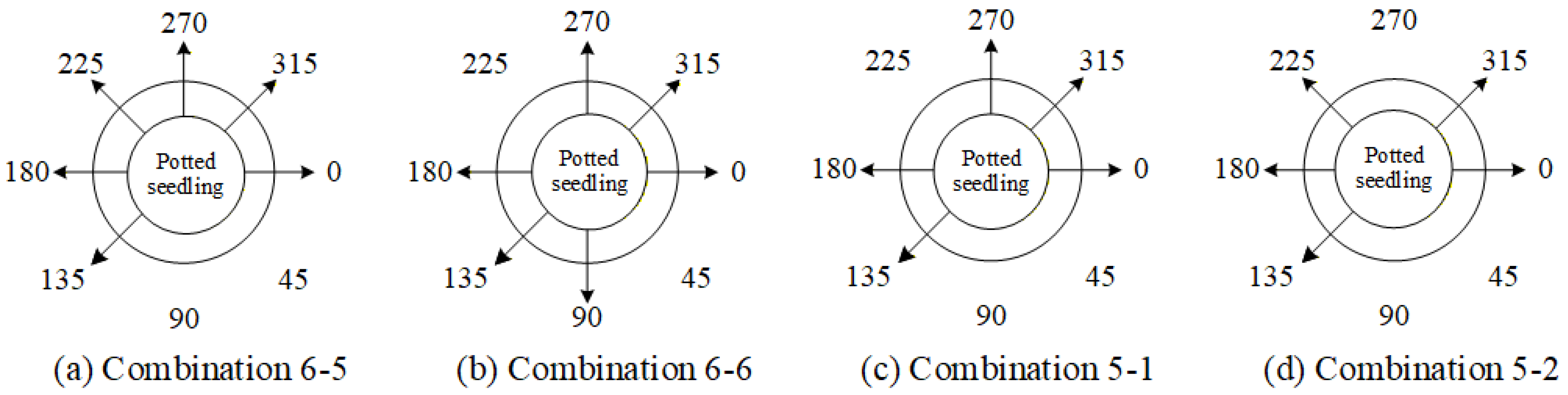
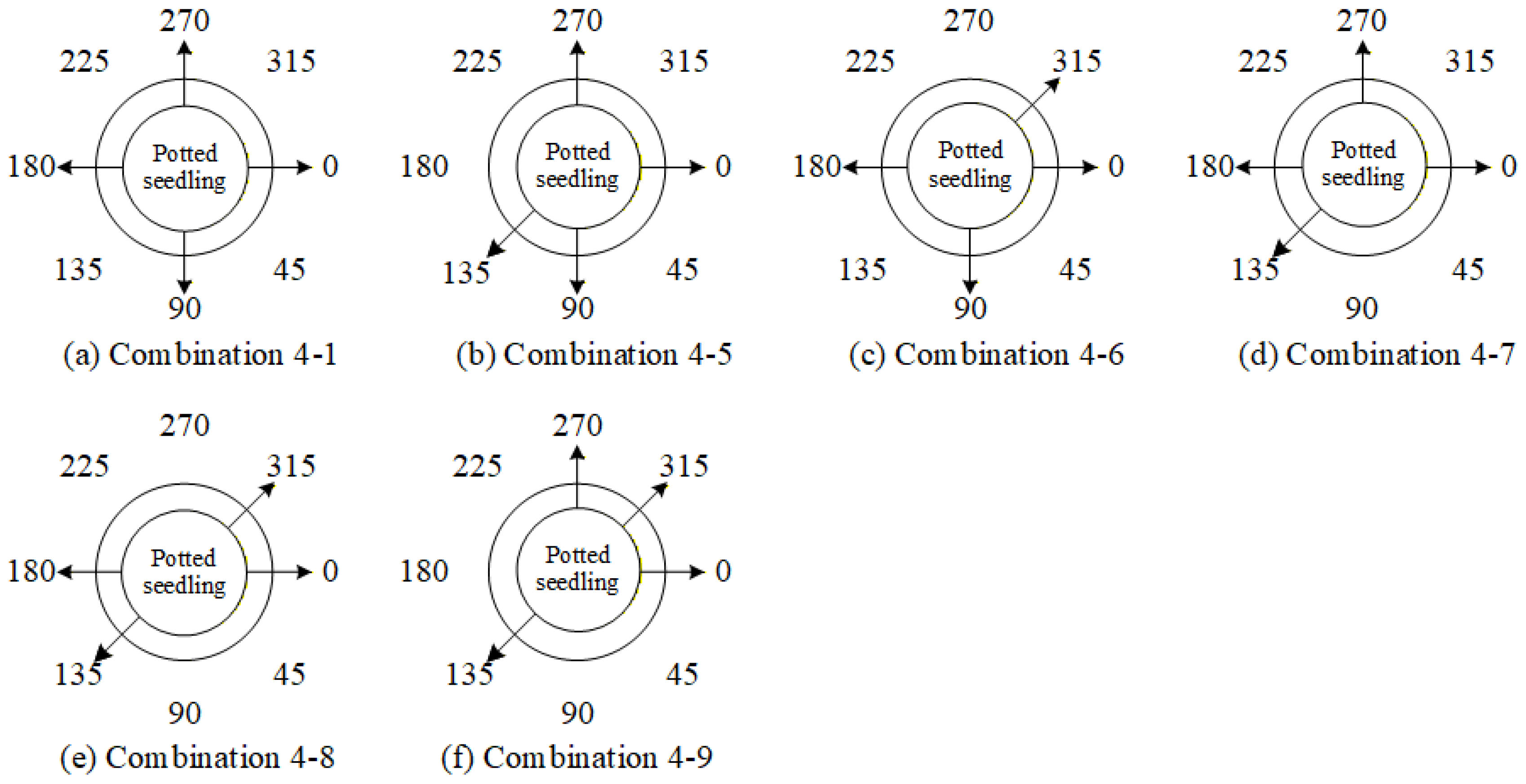
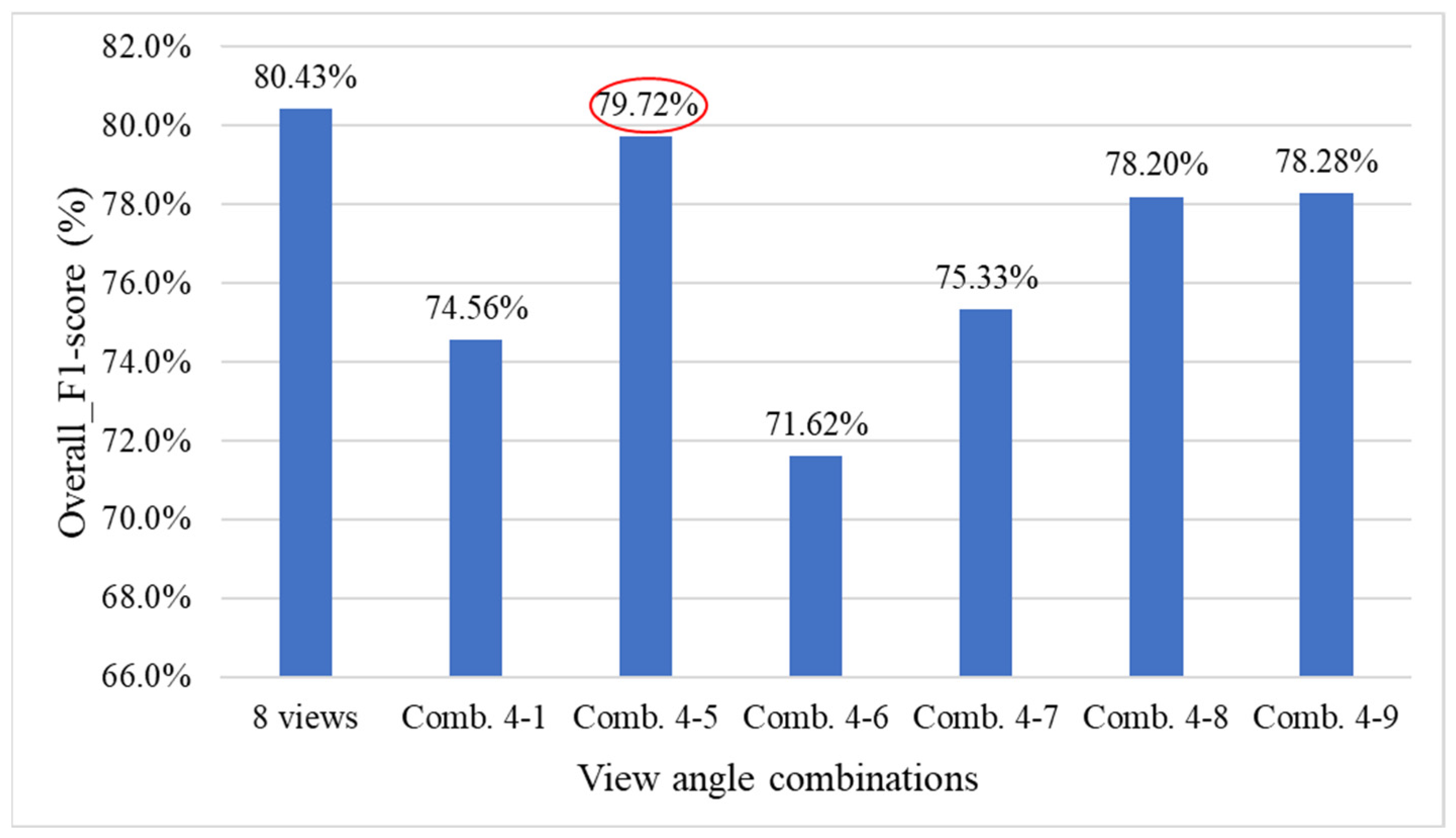
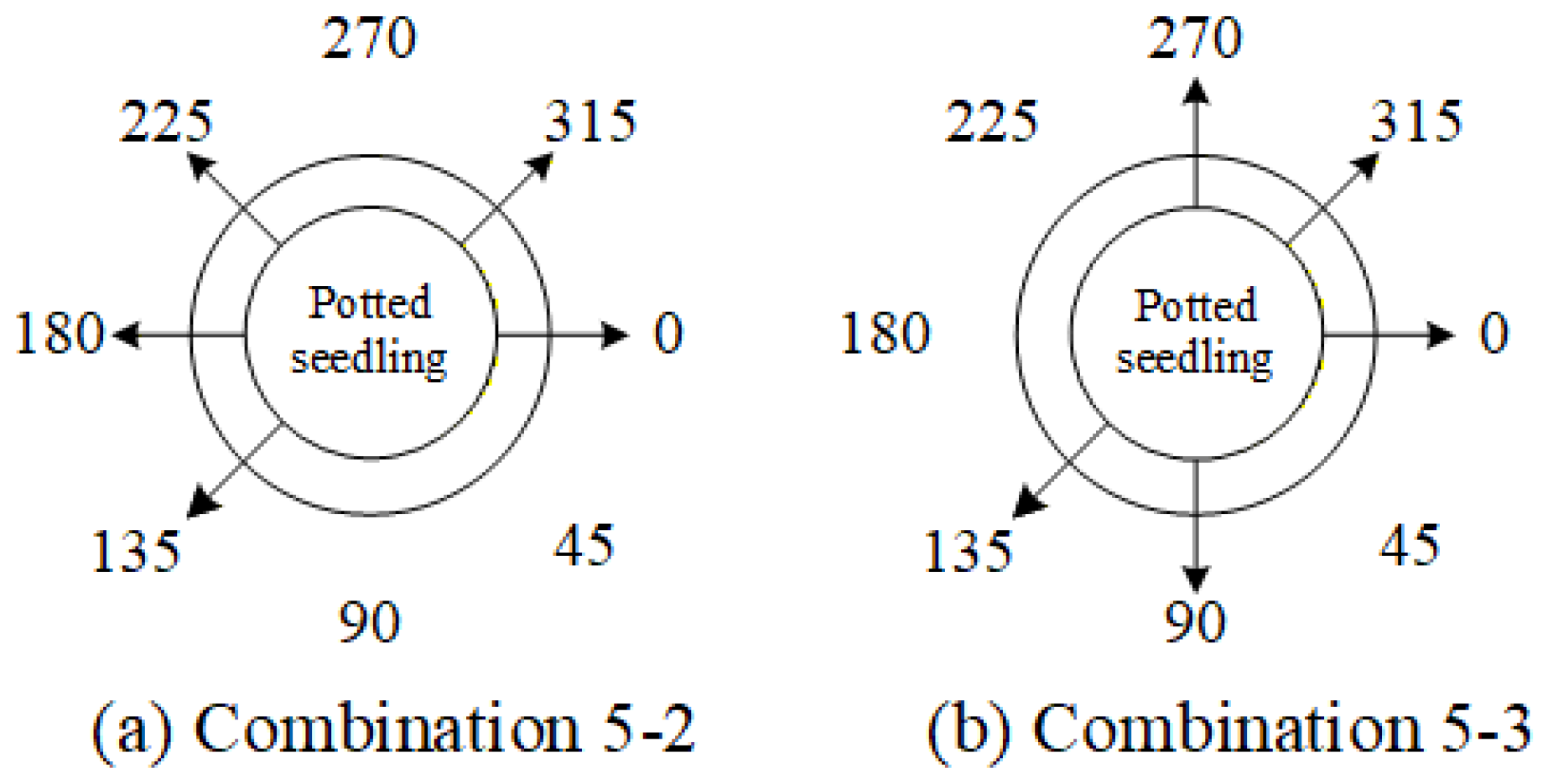
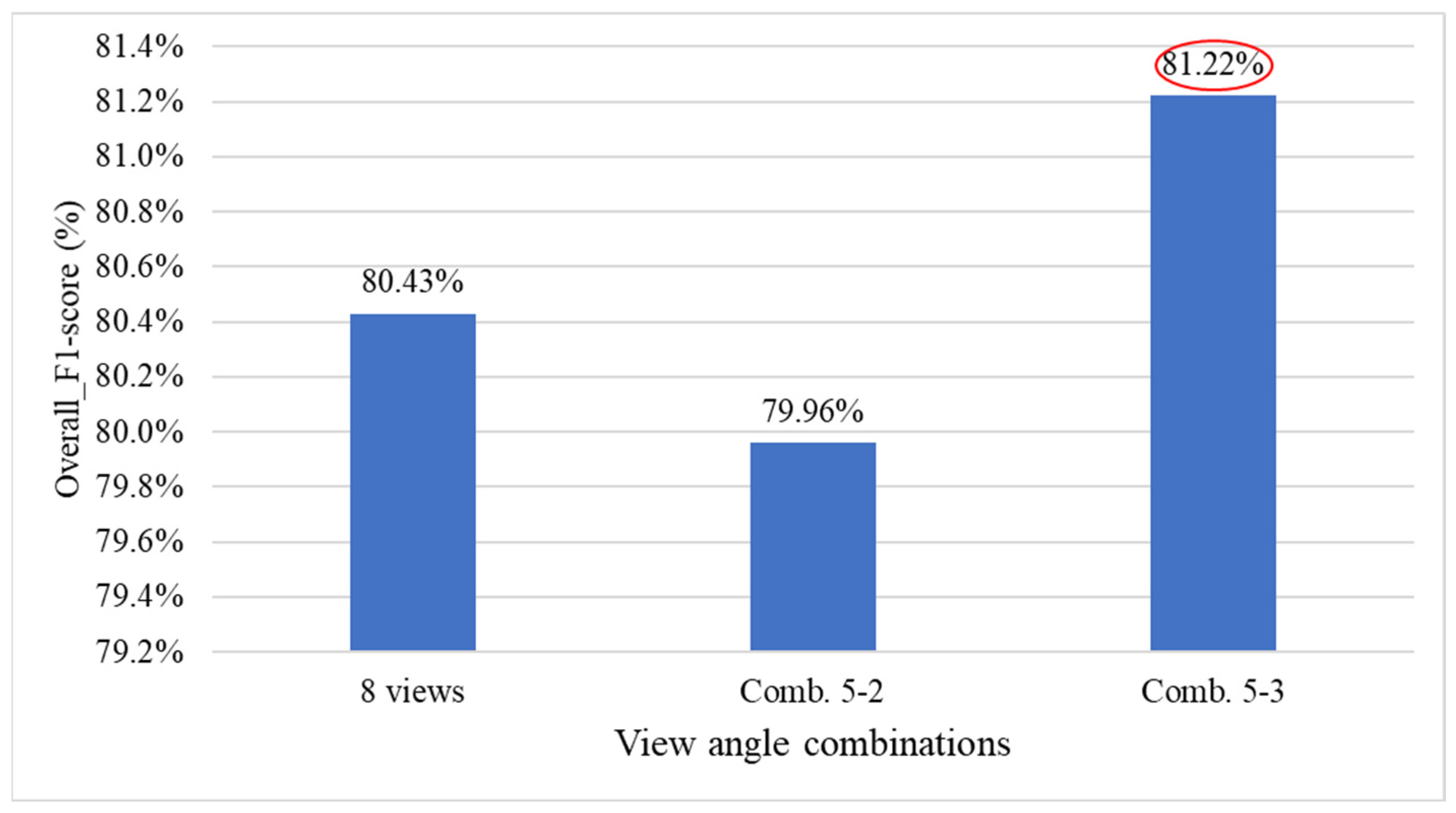
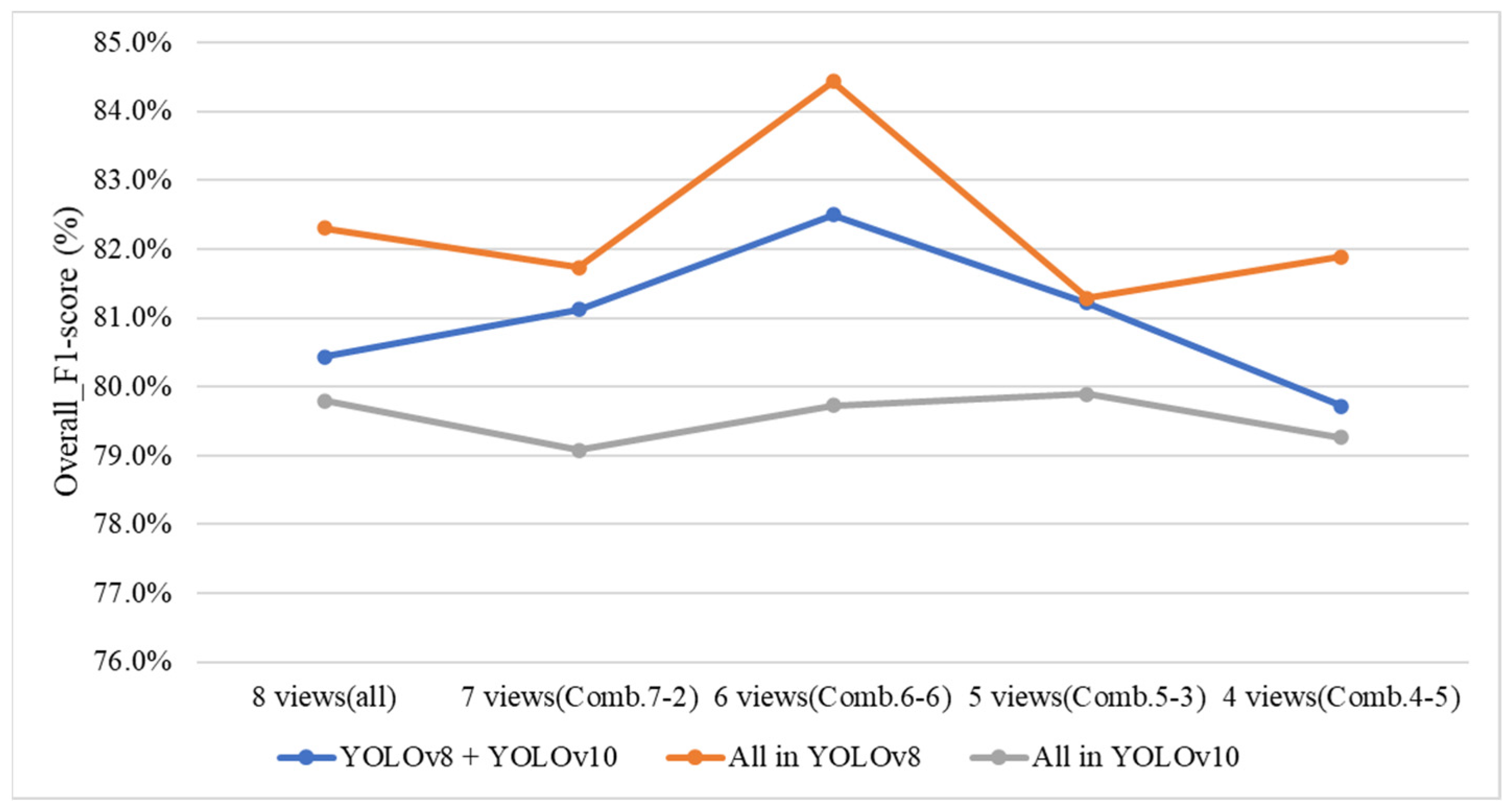
4.9.3. Ablation Experiments to Evaluate How Various Viewpoint Combinations Influence Overall System Performance
4.9.4. Effect of Changing the YOLO Model in the Defect-Detection Stage on Overall System Performance
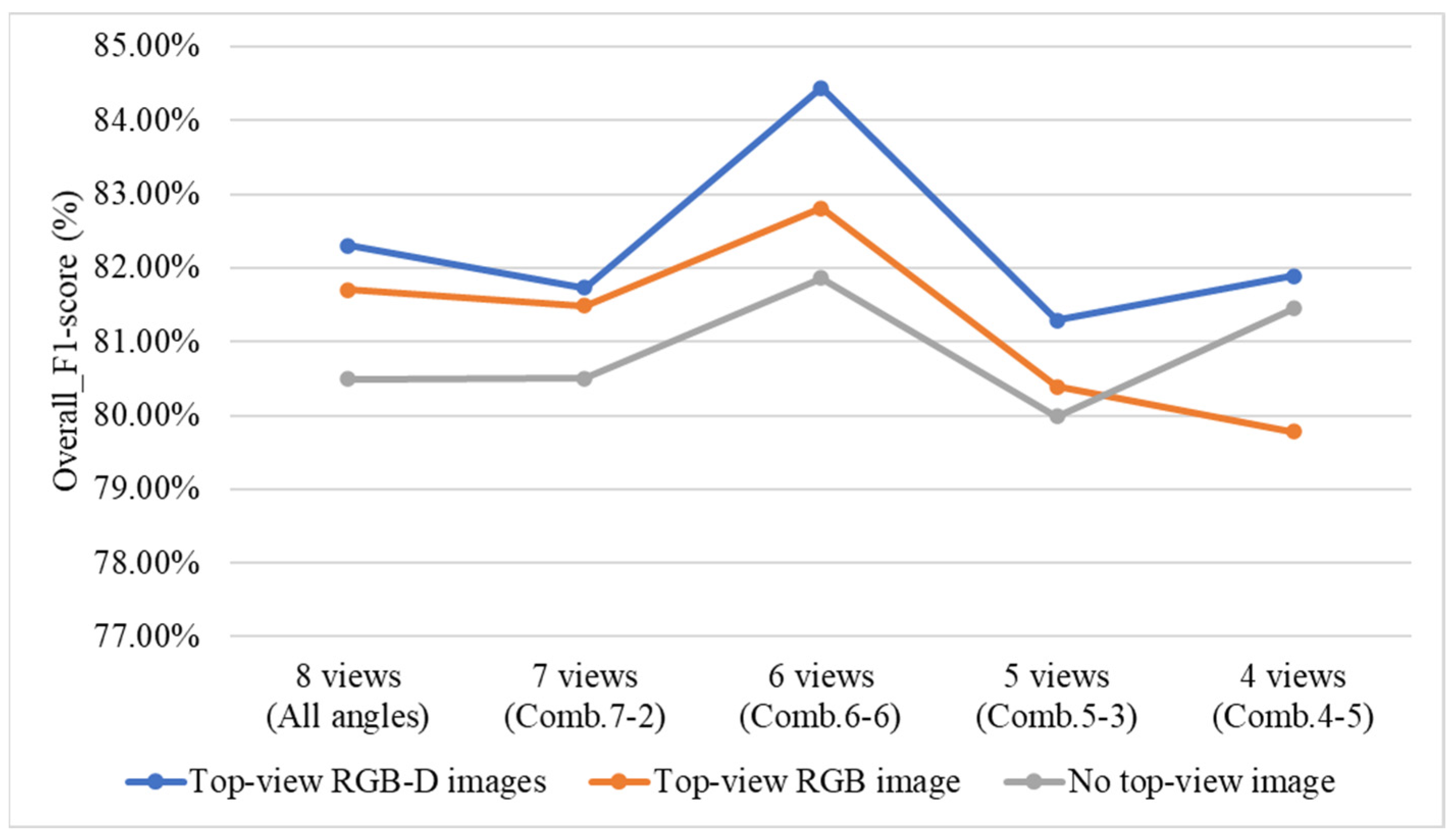
4.9.5. Effect of Including or Excluding Top-View Images and Top-View Depth (D) Images on Overall System Performance
4.10. Discussion and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan, S.C.; Lekawatana, S.; Amore, T.D.; Chen, F.C.; Chin, S.W.; Vega, D.M.; Wang, Y.T. The global orchid market. In The Orchid Genome; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–28. [Google Scholar] [CrossRef]
- Iiyama, C.M.; Vilcherrez-Atoche, J.A.; Germanà, M.A.; Vendrame, W.A.; Cardoso, J.C. Breeding of ornamental orchids with focus on Phalaenopsis: Current approaches, tools, and challenges for this century. Heredity 2024, 132, 163–178. [Google Scholar] [CrossRef]
- Taida Horticultural Co., Ltd. About Taida Orchids. Available online: https://www.taidaorchids.com.tw/en/about (accessed on 6 October 2025).
- Xiao, K.; Zhou, L.; Yang, H.; Yang, L. Phalaenopsis growth phase classification using convolutional neural network. Smart Agric. Technol. 2022, 2, 100060. [Google Scholar] [CrossRef]
- Sanjaya, K.W.V.; Vijesekara, H.M.S.S.; Wickramasinghe, I.M.A.C.; Amalraj, C.R. Orchid classification, disease identification and healthiness prediction system. Int. J. Sci. Technol. Res. 2015, 4, 215–220. [Google Scholar]
- Tsai, C.F.; Huang, C.H.; Wu, F.H.; Lin, C.H.; Lee, C.H.; Yu, S.S.; Chan, Y.K.; Jan, F.J. Intelligent image analysis recognizes important orchid viral diseases. Front. Plant Sci. 2022, 13, 1051348. [Google Scholar] [CrossRef]
- Meena, N.K.; Mani, M. Pests and their management in orchids. In Trends in Horticultural Entomology; Springer: Singapore, 2022; pp. 1239–1254. [Google Scholar] [CrossRef]
- Meena, N.K.; Medhi, R.P.; Mani, M. Orchids. In Mealybugs and Their Management in Agricultural and Horticultural Crops; Springer: New Delhi, India, 2016; pp. 525–534. [Google Scholar] [CrossRef]
- Klein, J.; Battistus, A.G.; Hoffman, V.; Vilanova da Costa, N.; Kestring, D.; Bulegon, L.G.; Guimarães, V.F. Chemical management of Pilea microphylla in orchid seedlings. Rev. Bras. Herbicidas 2015, 14, 15–20. [Google Scholar] [CrossRef]
- Khasim, S.M.; Hegde, S.N.; González-Arnao, M.T.; Thammasiri, K. (Eds.) Orchid Biology: Recent Trends & Challenges; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Shekhar, V.; Stöckle, D.; Thellmann, M.; Vermeer, J.E. The role of plant root systems in evolutionary adaptation. Curr. Top. Dev. Biol. 2019, 131, 55–80. [Google Scholar] [CrossRef]
- Naik, S.; Patel, B. Machine vision based fruit classification and grading—A review. Int. J. Comput. Appl. 2017, 170, 22–34. [Google Scholar] [CrossRef]
- Upadhyay, A.; Chandel, N.S.; Singh, K.P.; Chakraborty, S.K.; Nandede, B.M.; Kumar, M.; Subeesh, A.; Upendar, K.; Salem, A.; Elbeltagi, A. Deep learning and computer vision in plant disease detection: A comprehensive review. Artif. Intell. Rev. 2025, 58, 92. [Google Scholar] [CrossRef]
- Duan, Z.; Liu, W.; Zeng, S.; Zhu, C.; Chen, L.; Cui, W. Research on a real-time, high-precision end-to-end sorting system for fresh-cut flowers. Agriculture 2024, 14, 1532. [Google Scholar] [CrossRef]
- Fei, Y.; Li, Z.; Zhu, T.; Ni, C. A lightweight attention-based convolutional neural network for fresh-cut flower classification. IEEE Access 2023, 11, 17283–17293. [Google Scholar] [CrossRef]
- Xia, T.; Sheng, W.; Song, R.; Li, H.; Zhang, M. A review of three-dimensional multispectral imaging in plant phenotyping. In Sensing Technologies for Field and In-House Crop Production; Springer: Singapore, 2023; pp. 1–18. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Hussain, M. YOLOv5, YOLOv8 and YOLOv10: The go-to detectors for real-time vision. arXiv 2024, arXiv:2407.02988. [Google Scholar]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep learning for plant stress phenotyping: Trends and future perspectives. Trends Plant Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Yang, M.; Cho, S.I. High-resolution 3D crop reconstruction and automatic analysis of phenotyping index using machine learning. Agriculture 2021, 11, 1010. [Google Scholar] [CrossRef]
- Qiu, R.; Wei, S.; Zhang, M.; Li, H.; Sun, H.; Liu, G.; Li, M. Sensors for measuring plant phenotyping: A review. Int. J. Agric. Biol. Eng. 2018, 11, 1–17. [Google Scholar] [CrossRef]
- Vit, A.; Shani, G. Comparing RGB-D sensors for close range outdoor agricultural phenotyping. Sensors 2018, 18, 4413. [Google Scholar] [CrossRef]
- Garbouge, H.; Rasti, P.; Rousseau, D. Enhancing the tracking of seedling growth using RGB–depth fusion and deep learning. Sensors 2021, 21, 8425. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, G.S.; Silva, L.A.; Marengoni, M. 3D reconstruction of non-rigid plants and sensor data fusion for agricultural phenotyping. Sensors 2021, 21, 4115. [Google Scholar] [CrossRef]
- Koirala, A.; Walsh, K.B.; Wang, Z.; McCarthy, C. Deep learning for real-time fruit detection and orchard fruit load estimation: Benchmarking of ‘MangoYOLO’. Precis. Agric. 2019, 20, 1107–1135. [Google Scholar] [CrossRef]
- Gené-Mola, J.; Vilaplana, V.; Rosell-Polo, J.R.; Morros, J.R.; Ruiz-Hidalgo, J.; Gregorio, E. Multi-modal deep learning for Fuji apple detection using RGB-D cameras and their radiometric capabilities. Comput. Electron. Agric. 2019, 162, 689–698. [Google Scholar] [CrossRef]
- Patil, R.R.; Kumar, S. Rice-fusion: A multimodality data fusion framework for rice disease diagnosis. IEEE Access 2022, 10, 5207–5222. [Google Scholar] [CrossRef]
- Zhang, N.; Wu, H.; Zhu, H.; Deng, Y.; Han, X. Tomato disease classification and identification method based on multimodal fusion deep learning. Agriculture 2022, 12, 2014. [Google Scholar] [CrossRef]
- Ubbens, J.R.; Stavness, I. Deep plant phenomics: A deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 2017, 8, 1190. [Google Scholar] [CrossRef]
- Chen, M.; Tang, Y.; Zou, X.; Huang, K.; Huang, Z.; Zhou, H.; Wang, C.; Lian, G. Three-dimensional perception of orchard banana central stock enhanced by adaptive multi-vision technology. Comput. Electron. Agric. 2020, 174, 105508. [Google Scholar] [CrossRef]
- Yang, Z.X.; Li, Y.; Wang, R.F.; Hu, P.; Su, W.H. Deep learning in multimodal fusion for sustainable plant care: A comprehensive review. Sustainability 2025, 17, 5255. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Automation in Agriculture by Machine and Deep Learning Techniques: A Review of Recent Developments. Precis. Agric. 2021, 22, 2053–2091. [Google Scholar] [CrossRef]
- Rahnemoonfar, M.; Sheppard, C. Deep count: Fruit counting based on deep simulated learning. Sensors 2017, 17, 905. [Google Scholar] [CrossRef]
- Ryo, M. Explainable artificial intelligence and interpretable machine learning for agricultural data analysis. Artif. Intell. Agric. 2022, 6, 257–265. [Google Scholar] [CrossRef]
- Kaur, P.; Mishra, A.M.; Goyal, N.; Gupta, S.K.; Shankar, A.; Viriyasitavat, W. A novel hybrid CNN methodology for automated leaf disease detection and classification. Expert Syst. 2024, 41, e13543. [Google Scholar] [CrossRef]
- Taji, K.; Sohail, A.; Shahzad, T.; Khan, B.S.; Khan, M.A.; Ouahada, K. An ensemble hybrid framework: A comparative analysis of metaheuristic algorithms for CNN feature ensembles in plant disease classification. IEEE Access 2024, 12, 61886–61906. [Google Scholar] [CrossRef]
- Sivaranjani, A.; Senthilrani, S.; Ashok Kumar, B.; Senthil Murugan, A. An overview of computer vision–based grading systems for agricultural products. J. Hortic. Sci. Biotechnol. 2022, 97, 137–159. [Google Scholar] [CrossRef]
- Mehmood, A.; Ahmad, M.; Ilyas, Q.M. Enhanced automated fruit disease identification using a new ensemble classification method. Agriculture 2023, 13, 500. [Google Scholar] [CrossRef]
- Saberi Anari, M. A hybrid model for leaf disease classification using modified deep transfer learning and ensemble approaches for agricultural AIoT monitoring. Comput. Intell. Neurosci. 2022, 2022, 6504616. [Google Scholar] [CrossRef]
- Tahir, H.A.; Alayed, W.; Hassan, W.U. A federated explainable AI framework for smart agriculture: Enhancing transparency and sustainability. IEEE Access 2025, 13, 97567–97584. [Google Scholar] [CrossRef]
- Ngugi, H.N.; Akinyelu, A.A.; Ezugwu, A.E. Machine learning and deep learning for crop disease diagnosis: Performance analysis and review. Agronomy 2024, 14, 3001. [Google Scholar] [CrossRef]
- Ultralytics. YOLOv8: The Next-Generation YOLO Object Detection Model. Available online: https://docs.ultralytics.com (accessed on 15 January 2025).
- Wang, C.; Zhang, H.; Ren, H.; Zhang, X.; Li, S.; Xu, X.; Zhao, Y.; Zhang, J.; Luo, P. YOLOv10: Real-time end-to-end object detection. arXiv 2024, arXiv:2405.14458. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rana, S.; Gatti, A. High-fidelity synthetic data generation for agricultural RGB/IR imaging using modified WGAN-GP. MethodsX 2025, 14, 103309. [Google Scholar] [CrossRef] [PubMed]

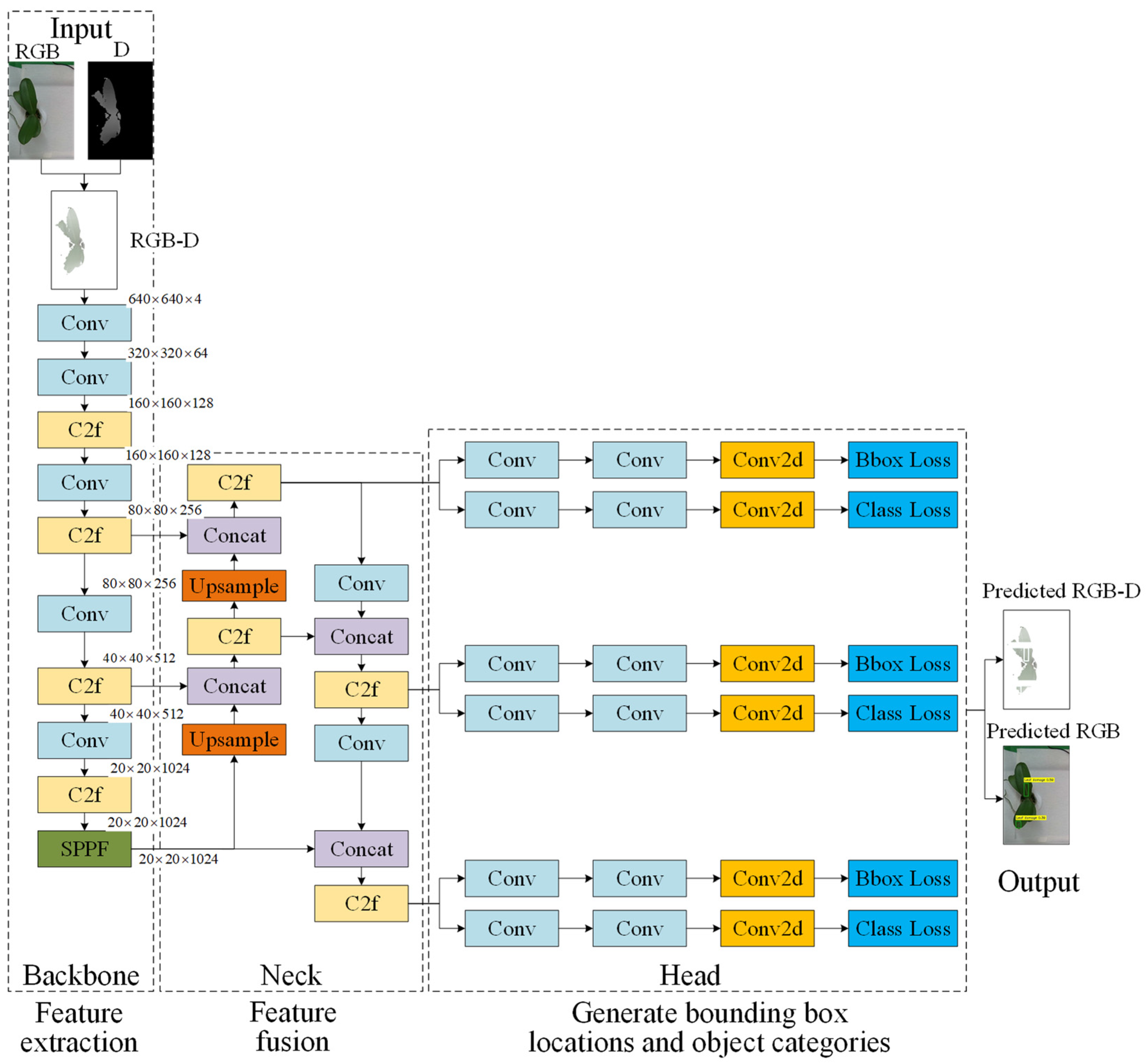
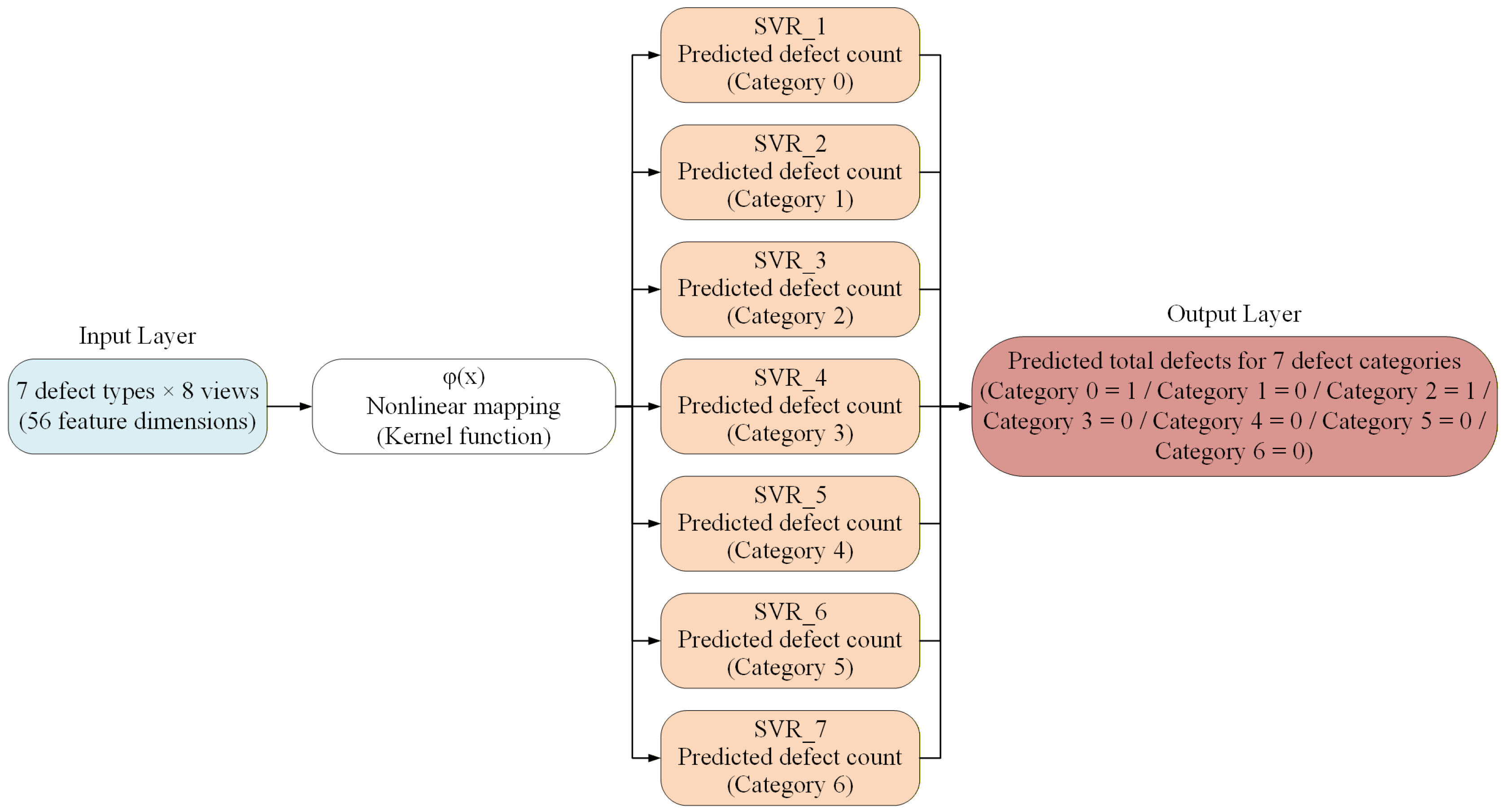
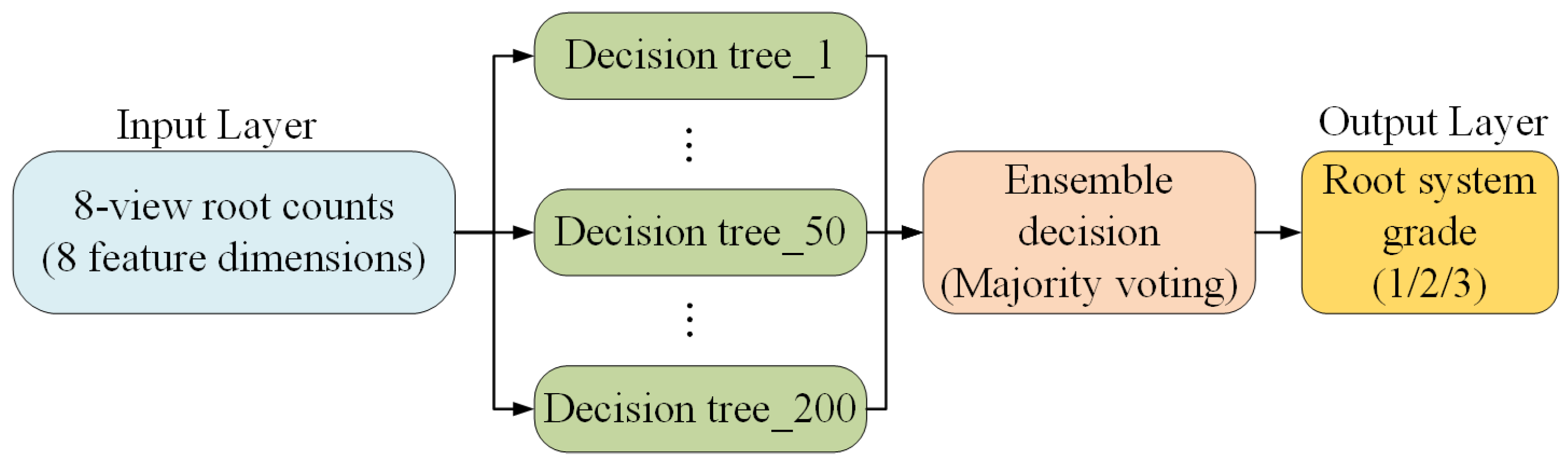
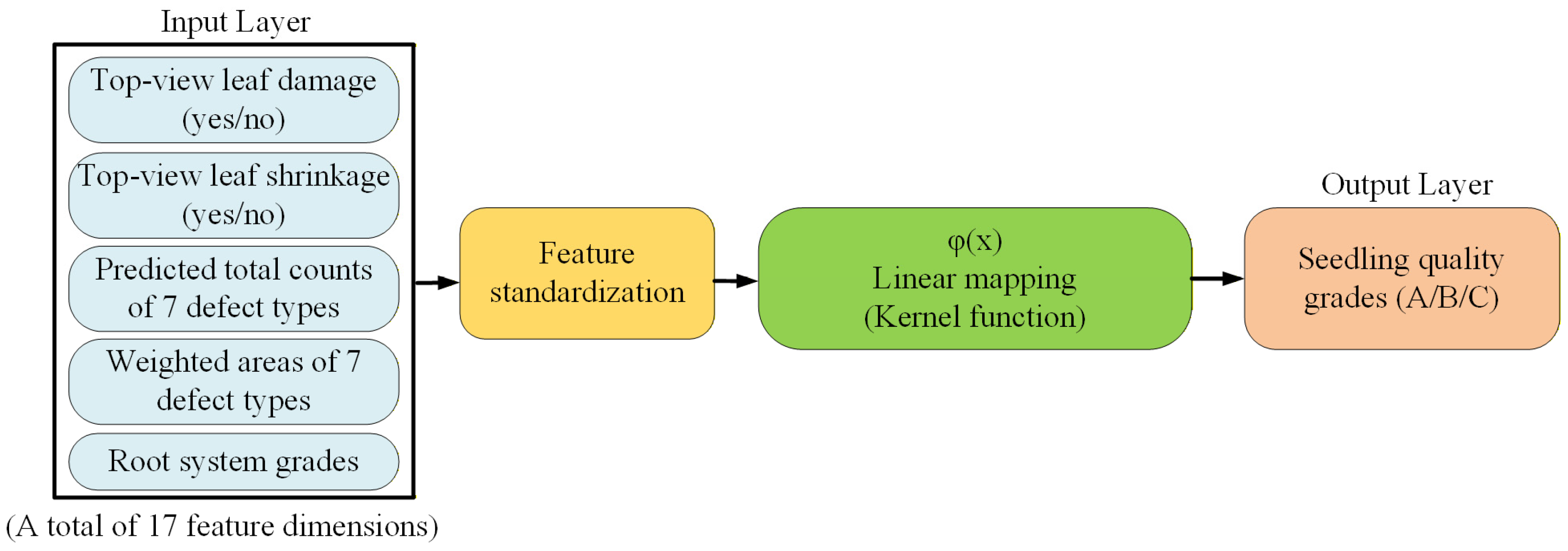


| Defect Types | A Grade | B Grade | C Grade |
|---|---|---|---|
| (1) Diseases (a. Anthracnose, b. Yellow leaf disease, c. Phytophthora disease, d. Southern blight, e. Bacterial soft rot, f. Unknown spots) | None | b. Fine black lines appear on the stem. | b.: Slight yellowing spreads outward starting from the black lines on the stem. d.: White mycelium appears on the stem. c., e.: The leaf surface rapidly decays with a water-soaked appearance; in severe cases, fungal slime may develop. a., e., f.: Black circular spots with slightly yellow margins. a., c., f.: Dark green circular spots, light brown speckles, or irregularly shaped lesions appear on the leaf surface. a., c., f.: Circular spots appear on the leaf surface. |
| (2) Pest damage (Insect bite) | None | Circular or irregular bite marks appear on the lower leaves, regardless of size. | Circular or irregular bite marks appear on the upper leaves, regardless of size. |
| (3) Phytotoxicity (Pesticide damage) | None | Area of abnormal coloration on the upper leaf surface is <10–15% of the entire leaf area; Symmetrical deformation of the leaf outline affects <10–15% of the entire leaf area. | Area of abnormal coloration on the upper leaf surface is ≥10–15% of the entire leaf area; Symmetrical deformation of the leaf outline affects ≥10–15% of the entire leaf contour; Twisting and deformation of the leaf outline. |
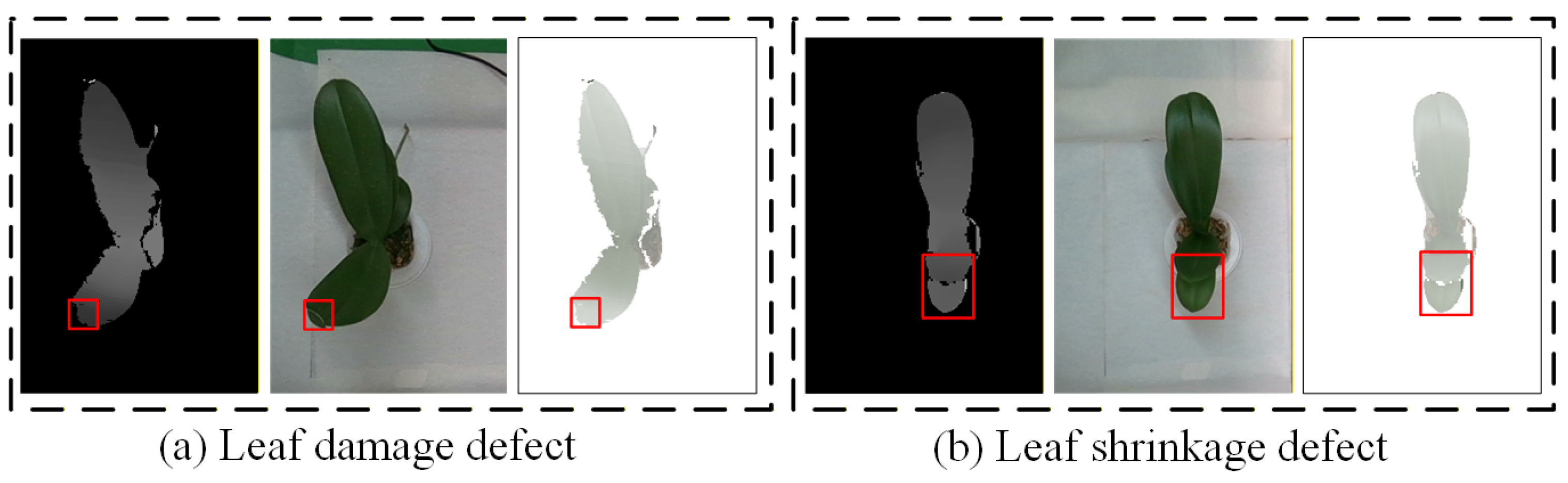
| (4) Leaf damage | Length of midrib damage on the surface of the lower leaves is <1.5 cm. | Length of damage along the contour of a single leaf is <1.5 cm; Length of midrib damage on the surface of the upper leaves is <1.5 cm. | Length of damage along the contour of a single leaf is ≥1.5 cm; Length of midrib damage on the surface of the upper leaves is ≥1.5 cm. |
| (5) Leaf shrinkage | None | Upper leaves are shorter than the lower leaves, regardless of size. | None |
| (6) Leaf variation | None | Embossed patterns (leaf surface protrusions or depressions) or linear markings appear on the leaf surface, regardless of leaf size. | Leaf surface becomes twisted and deformed. |
| (7) Lower-leaf yellowing | None | Yellowing appears at the tips of the lower leaf surface, regardless of leaf size. | None |
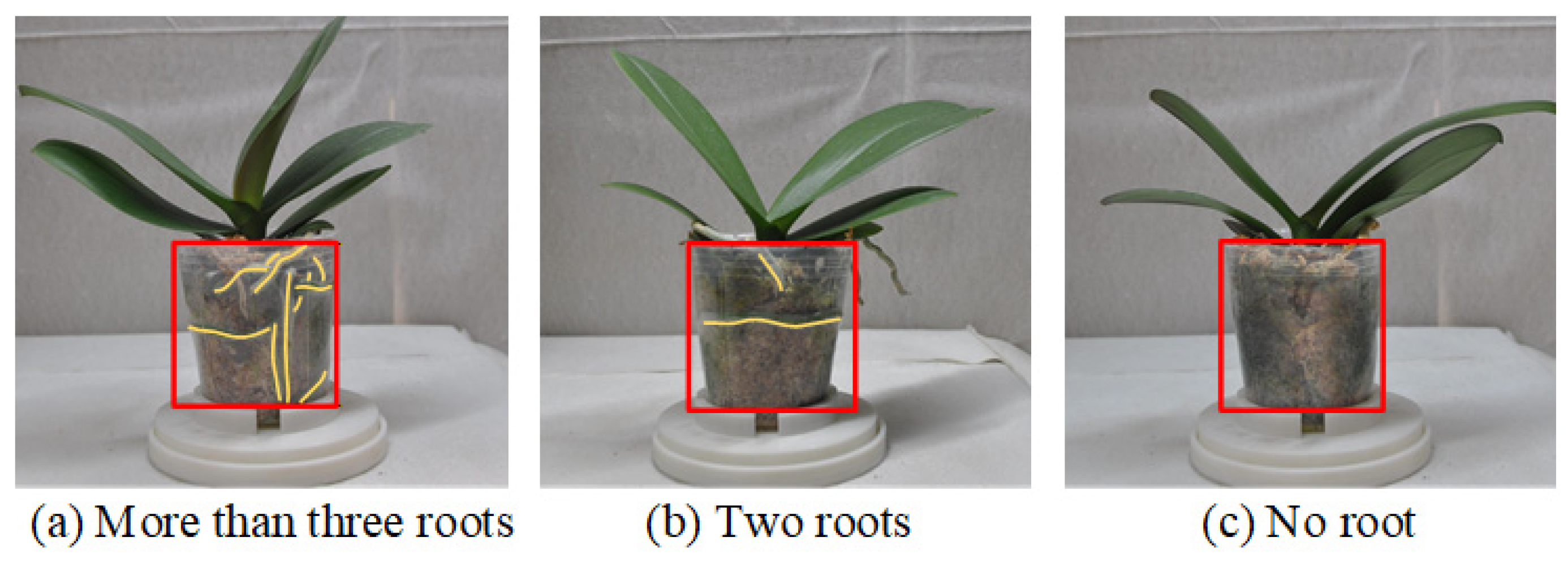
| (8) Root system conditions | Root system ≥ 70% | 50% < Root system < 70% | Root system ≤ 50% |
| Root Grade 3 | Root Grade 2 | Root Grade 1 |
|---|---|---|
| ≥3 roots and visual in =8 views | ≥3 roots and visual in =7 views 0–1 root and visual in =1 view | ≥3 roots and visual in ≤6 views 0–1 root and visual in ≥2 views |
| ≥3 roots and visual in =7 views 2 roots and visual in =1 view | ≥3 roots and visual in =6 views 2 roots and visual in =1 view 0–1 root and visual in =1 view | ≥3 roots and visual in =4 views 2 roots and visual in =2 views 0–1 root and visual in =2 views |
| ≥3 roots and visual in ≤6 views 2 roots and visual in ≥2 views 0–1 root and visual in =0 views | ≥3 roots and visual in ≤4 views 2 roots and visual in ≥2 views 0–1 root and visual in ≥2 views |
| The ith category (defect type, root grade, or quality grade) | |
| Number of samples in category i correctly classified as category i | |
| Number of samples from other categories incorrectly classified as category i | |
| Number of samples in category i incorrectly classified as other categories | |
| Total number of samples in category i |
| The jth category (regression type) | |
| Actual value of the jth category | |
| Predicted value of the jth category by the model | |
| Mean value of the jth category | |
| Number of samples |
| Stage 1 | Stage 2 | Stage 3 | |||
|---|---|---|---|---|---|
| Top-view leaf defect detection | Side-view leaf defect detection | Side-view root count estimation | Side-view leaf defect count estimation | Side-view root system grading | Whole-seedling quality grading |
| YOLOv8 | YOLOv10 | YOLOv8 | SVM-1 | RF | SVM-2 |
| 73.20% | 63.70% | 92.40% | 0.7026 | 89.43% | 80.43% |
| Category | Precision | Recall | F1-Score |
|---|---|---|---|
| Flawless | 64% | 86% | 73% |
| Disease | 87% | 44% | 58% |
| Pest damage | 63% | 20% | 30% |
| Pesticide damage | 87% | 50% | 64% |
| Leaf damage | 79% | 63% | 70% |
| Leaf shrinkage | 81% | 66% | 72% |
| Variation | 84% | 70% | 77% |
| Lower-leaf yellowing | 80% | 77% | 78% |
| Actual\Predicted | A | B | C |
|---|---|---|---|
| A | 15 | 4 | 2 |
| B | 7 | 84 | 5 |
| C | 3 | 9 | 24 |
| Actual\Predicted | A | B | C |
|---|---|---|---|
| A | 17 | 1 | 3 |
| B | 4 | 89 | 3 |
| C | 4 | 2 | 30 |
| Method | Class | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Three-Stage | A | 0.6000 | 0.7143 | 0.6512 |
| B | 0.8660 | 0.8750 | 0.8705 | |
| C | 0.7742 | 0.6667 | 0.7164 | |
| Overall_F1 score | — | — | 0.8043 | |
| Direct Method | A | 0.6800 | 0.8095 | 0.7380 |
| B | 0.9674 | 0.9263 | 0.9464 | |
| C | 0.8333 | 0.8333 | 0.8333 | |
| Overall_F1 score | — | — | 0.8916 |
| Viewing Angle Combination | 0 Degrees | 45 Degrees | 90 Degrees | 135 Degrees | 180 Degrees | 225 Degrees | 270 Degrees | 315 Degrees |
|---|---|---|---|---|---|---|---|---|
| 4-1 | ✓ | - | ✓ | - | ✓ | - | ✓ | - |
| 4-2 | - | ✓ | - | ✓ | - | ✓ | - | ✓ |
| 4-3 | ✓ | ✓ | - | ✓ | - | - | ✓ | - |
| 4-4 | - | - | ✓ | - | ✓ | ✓ | - | ✓ |
| Viewing Angle Combination | 0 Degrees | 45 Degrees | 90 Degrees | 135 Degrees | 180 Degrees | 225 Degrees | 270 Degrees | 315 Degrees |
|---|---|---|---|---|---|---|---|---|
| 6-1 | ✓ | ✓ | ✓ | ✓ | - | ✓ | ✓ | - |
| 6-2 | ✓ | ✓ | - | ✓ | ✓ | - | ✓ | ✓ |
| 6-3 | ✓ | ✓ | ✓ | ✓ | - | - | ✓ | ✓ |
| 6-4 | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | - |
| Viewing Angle Combination | 0 Degrees | 45 Degrees | 90 Degrees | 135 Degrees | 180 Degrees | 225 Degrees | 270 Degrees | 315 Degrees |
|---|---|---|---|---|---|---|---|---|
| 6-5 | ✓ | - | - | ✓ | ✓ | ✓ | ✓ | ✓ |
| 6-6 | ✓ | - | ✓ | ✓ | ✓ | - | ✓ | ✓ |
| 5-1 | ✓ | - | - | ✓ | ✓ | - | ✓ | ✓ |
| 5-2 | ✓ | - | - | ✓ | ✓ | ✓ | - | ✓ |
| Viewing Angle Combination | 0 Degrees | 45 Degrees | 90 Degrees | 135 Degrees | 180 Degrees | 225 Degrees | 270 Degrees | 315 Degrees |
|---|---|---|---|---|---|---|---|---|
| 4-1 | ✓ | - | ✓ | - | ✓ | - | ✓ | - |
| 4-5 | ✓ | - | ✓ | ✓ | - | - | ✓ | - |
| 4-6 | ✓ | - | ✓ | - | ✓ | - | - | ✓ |
| 4-7 | ✓ | - | - | ✓ | ✓ | - | ✓ | - |
| 4-8 | ✓ | - | - | ✓ | ✓ | - | - | ✓ |
| 4-9 | ✓ | - | - | ✓ | - | - | ✓ | ✓ |
| Viewing Angle Combination | 0 Degrees | 45 Degrees | 90 Degrees | 135 Degrees | 180 Degrees | 225 Degrees | 270 Degrees | 315 Degrees |
|---|---|---|---|---|---|---|---|---|
| 5-2 | ✓ | - | - | ✓ | ✓ | ✓ | - | ✓ |
| 5-3 | ✓ | - | ✓ | ✓ | - | - | ✓ | ✓ |
| Number of Deleted Side-View Angles | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Viewing angle combination | 7-2 | 6-6 | 5-3 | 4-5 |
| F1-score | 81.12% | 82.50% | 81.22% | 79.72% |
| Performance Indices | All in YOLOv8 | All in YOLOv10 | YOLOv8 + YOLOv10 |
|---|---|---|---|
| Overall_F1-score (%) | 82.30% | 79.80% | 80.43% |
| Total training time (Min.) | 394.02 | 407.92 | 436.18 |
| Testing time/seedling (s) | 1.9284 | 1.9752 | 1.9784 |
| Performance Indices | All in YOLOv8 | All in YOLOv10 | YOLOv8 + YOLOv10 | YOLOv10 + YOLOv8 |
|---|---|---|---|---|
| Overall_F1-score (%) | 73.86% | 89.79% | 89.79% | 73.29% |
| Total training time (Min.) | 239.48 | 295.07 | 276.26 | 299.44 |
| Testing time/seedling (s) | 1.2181 | 0.8272 | 0.8364 | 1.2442 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-D.; Zhang, Z.-Y.; Lin, C.-H. Smart Image-Based Deep Learning System for Automated Quality Grading of Phalaenopsis Seedlings in Outsourced Production. Sensors 2025, 25, 7502. https://doi.org/10.3390/s25247502
Lin H-D, Zhang Z-Y, Lin C-H. Smart Image-Based Deep Learning System for Automated Quality Grading of Phalaenopsis Seedlings in Outsourced Production. Sensors. 2025; 25(24):7502. https://doi.org/10.3390/s25247502
Chicago/Turabian StyleLin, Hong-Dar, Zheng-Yuan Zhang, and Chou-Hsien Lin. 2025. "Smart Image-Based Deep Learning System for Automated Quality Grading of Phalaenopsis Seedlings in Outsourced Production" Sensors 25, no. 24: 7502. https://doi.org/10.3390/s25247502
APA StyleLin, H.-D., Zhang, Z.-Y., & Lin, C.-H. (2025). Smart Image-Based Deep Learning System for Automated Quality Grading of Phalaenopsis Seedlings in Outsourced Production. Sensors, 25(24), 7502. https://doi.org/10.3390/s25247502







