Decision Support Systems in Neurosurgery: Current Applications and Future Directions
Abstract
1. Introduction
2. Decision Support in Neuro-Oncology
2.1. Diagnostic Application
2.1.1. Tumor Detection and Classification
2.1.2. Radiomics
2.1.3. Radiogenomics
2.1.4. Summary
2.2. Decision Support in Preoperative Planning
2.3. Decision Support During Neurosurgical Procedures
2.3.1. Intraoperative Neuronavigation
2.3.2. Stereotactic Biopsy Guidance
2.3.3. Intraoperative Brain Mapping
2.3.4. Summary
2.4. Decision Support in Post-Operative/Traumatic Intensive Care
2.5. Decision Support in Histopathology and Cancer Genetic Profiling
2.5.1. Applications in Histopathology
2.5.2. Applications in Genetic Testing
2.5.3. Summary
3. Decision Support in Vascular Neurosurgery
3.1. Intracranial Aneurysm
3.2. Other Vascular Conditions
4. Decision Support in Functional Neurosurgery
Decision Support for Deep Brain Stimulation for Parkinson’s Disease
5. Future Directions for DSSs in Neurosurgery
6. Summary and Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Acc | Accuracy |
| AI | Artificial Intelligence |
| AVM | Arteriovenous Malformation |
| CCEp | Cortico-cortical Evoked Potential |
| CCM | Cerebral Cavernous Malformations |
| CNN | Convolutional Neural Network |
| CT | Computed Tomography |
| CTA | Computed Tomography Angiography |
| DBS | Deep Brain Stimulation |
| DSA | Digital Substraction Angiography |
| DSS | Decision Support System |
| fMRI | Functional Magnetic Resonance Imaging |
| IA | Intracranial Aneurysm |
| ICP | Intracranial Pressure |
| LLM | Large Language Model |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| ROC AUC | Receiver Operating Characteristic Area Under the Curve |
| rs-MRI | resting-state MRI |
| SCS | Spinal Cord Stimulation |
| STN | Subthalamic Nucleus |
| SVM | Support Vector Machine |
| TBI | Traumatic Brain Injury |
| TCGA | The Cancer Genome Atlas |
References
- Feigin, V.L.; Vos, T.; Nichols, E.; Owolabi, M.O.; Carroll, W.M.; Dichgans, M.; Deuschl, G.; Parmar, P.; Brainin, M.; Murray, C. The global burden of neurological disorders: Translating evidence into policy. Lancet Neurol. 2020, 19, 255–265. [Google Scholar] [CrossRef]
- Salari, N.; Ghasemi, H.; Fatahian, R.; Mansouri, K.; Dokaneheifard, S.; Shiri, M.H.; Hemmati, M.; Mohammadi, M. The global prevalence of primary central nervous system tumors: A systematic review and meta-analysis. Eur. J. Med. Res. 2023, 28, 39. [Google Scholar] [CrossRef] [PubMed]
- Achrol, A.S.; Rennert, R.C.; Anders, C.; Soffietti, R.; Ahluwalia, M.S.; Nayak, L.; Peters, S.; Arvold, N.D.; Harsh, G.R.; Steeg, P.S.; et al. Brain metastases. Nat. Rev. Dis. Prim. 2019, 5, 5. [Google Scholar] [CrossRef]
- Delaidelli, A.; Moiraghi, A. Recent Advances in the Diagnosis and Treatment of Brain Tumors. Brain Sci. 2024, 14, 224. [Google Scholar] [CrossRef]
- Winther, R.R.; Hjermstad, M.J.; Skovlund, E.; Aass, N.; Helseth, E.; Kaasa, S.; Yri, O.E.; Vik-Mo, E.O. Surgery for brain metastases—Impact of the extent of resection. Acta Neurochir. 2022, 164, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- Asfaw, Z.K.; Young, T.; Brown, C.; Germano, I.M. Charting the success of neuronavigation in brain tumor surgery: From inception to adoption and evolution. J. Neuro-Oncol. 2024, 170, 1–10. [Google Scholar] [CrossRef]
- Schipmann-Miletić, S.; Stummer, W. Image-Guided Brain Surgery. Recent Results Cancer Res. 2020, 216, 813–841. [Google Scholar] [CrossRef]
- Chen, C.J.; Ding, D.; Derdeyn, C.P.; Lanzino, G.; Friedlander, R.M.; Southerland, A.M.; Lawton, M.T.; Sheehan, J.P. Brain arteriovenous malformations. Neurology 2020, 95, 917–927. [Google Scholar] [CrossRef]
- Grigore, F.N.; Amin-Hanjani, S. A3-A3 Bypass Surgery for Aneurysm: Technical Nuances. Oper. Neurosurg. 2019, 17, 277–285. [Google Scholar] [CrossRef]
- Fredrickson, V.L.; Russin, J.J.; Strickland, B.A.; Bakhsheshian, J.; Amar, A.P. Intraoperative Imaging for Vascular Lesions. Neurosurg. Clin. N. Am. 2017, 28, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Ciecierski, K.A.; Mandat, T. Classification of DBS microelectrode recordings using a residual neural network with attention in the temporal domain. Neural Netw. 2024, 170, 18–31. [Google Scholar] [CrossRef]
- Hariz, M. Pros and Cons of Ablation for Functional Neurosurgery in the Neurostimulation Age. Neurosurg. Clin. N. Am. 2023, 34, 291–299. [Google Scholar] [CrossRef]
- Lev-Tov, L.; Barbosa, D.A.; Ghanouni, P.; Halpern, C.H.; Buch, V.P. Focused ultrasound for functional neurosurgery. J. Neuro-Oncol. 2022, 156, 17–22. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, W.; Wang, L.; Zhao, Z.; Jia, X. TriQuery: A Query-Based Model for Surgical Triplet Recognition. Sensors 2025, 25, 5306. [Google Scholar] [CrossRef] [PubMed]
- Milana, C.; Ashta, A. Artificial intelligence techniques in finance and financial markets: A survey of the literature. Strateg. Change 2021, 30, 189–209. [Google Scholar] [CrossRef]
- Che, C.; Huang, Z.; Li, C.; Zheng, H.; Tian, X. Integrating Generative AI into Financial Market Prediction for Improved Decision Making. Appl. Comput. Eng. 2024, 64, 154–160. [Google Scholar] [CrossRef]
- Elbasi, E.; Mostafa, N.; Alarnaout, Z.; Zreikat, A.I.; Cina, E.; Varghese, G.; Shdefat, A.; Topcu, A.E.; Abdelbaki, W.; Mathew, S.; et al. Artificial Intelligence Technology in the Agricultural Sector: A Systematic Literature Review. IEEE Access 2023, 11, 171–202. [Google Scholar] [CrossRef]
- Mana, A.A.; Allouhi, A.; Hamrani, A.; Rahman, S.; el Jamaoui, I.; Jayachandran, K. Sustainable AI-based production agriculture: Exploring AI applications and implications in agricultural practices. Smart Agric. Technol. 2024, 7, 100416. [Google Scholar] [CrossRef]
- Santos, M.R.C.; Cagica Carvalho, L. AI-driven participatory environmental management: Innovations, applications, and future prospects. J. Environ. Manag. 2025, 373, 123864. [Google Scholar] [CrossRef]
- Uriarte-Gallastegi, N.; Arana-Landín, G.; Landeta-Manzano, B.; Laskurain-Iturbe, I. The Role of AI in Improving Environmental Sustainability: A Focus on Energy Management. Energies 2024, 17, 649. [Google Scholar] [CrossRef]
- Munser, L.; Sathyanarayanan, K.K.; Raecke, J.; Mansour, M.M.; Uland, M.E.; Streif, S. Precise and Continuous Biomass Measurement for Plant Growth Using a Low-Cost Sensor Setup. Sensors 2025, 25, 4770. [Google Scholar] [CrossRef] [PubMed]
- Grosan, C.; Abraham, A. Rule-Based Expert Systems. Intell. Syst. Ref. Libr. 2011, 17, 149–185. [Google Scholar] [CrossRef]
- Windermere, S.A.; Shah, S.; Hey, G.; McGrath, K.; Rahman, M. Applications of Artificial Intelligence in Neurosurgery for Improving Outcomes Through Diagnostics, Predictive Tools, and Resident Education. World Neurosurg. 2025, 197, 123809. [Google Scholar] [CrossRef]
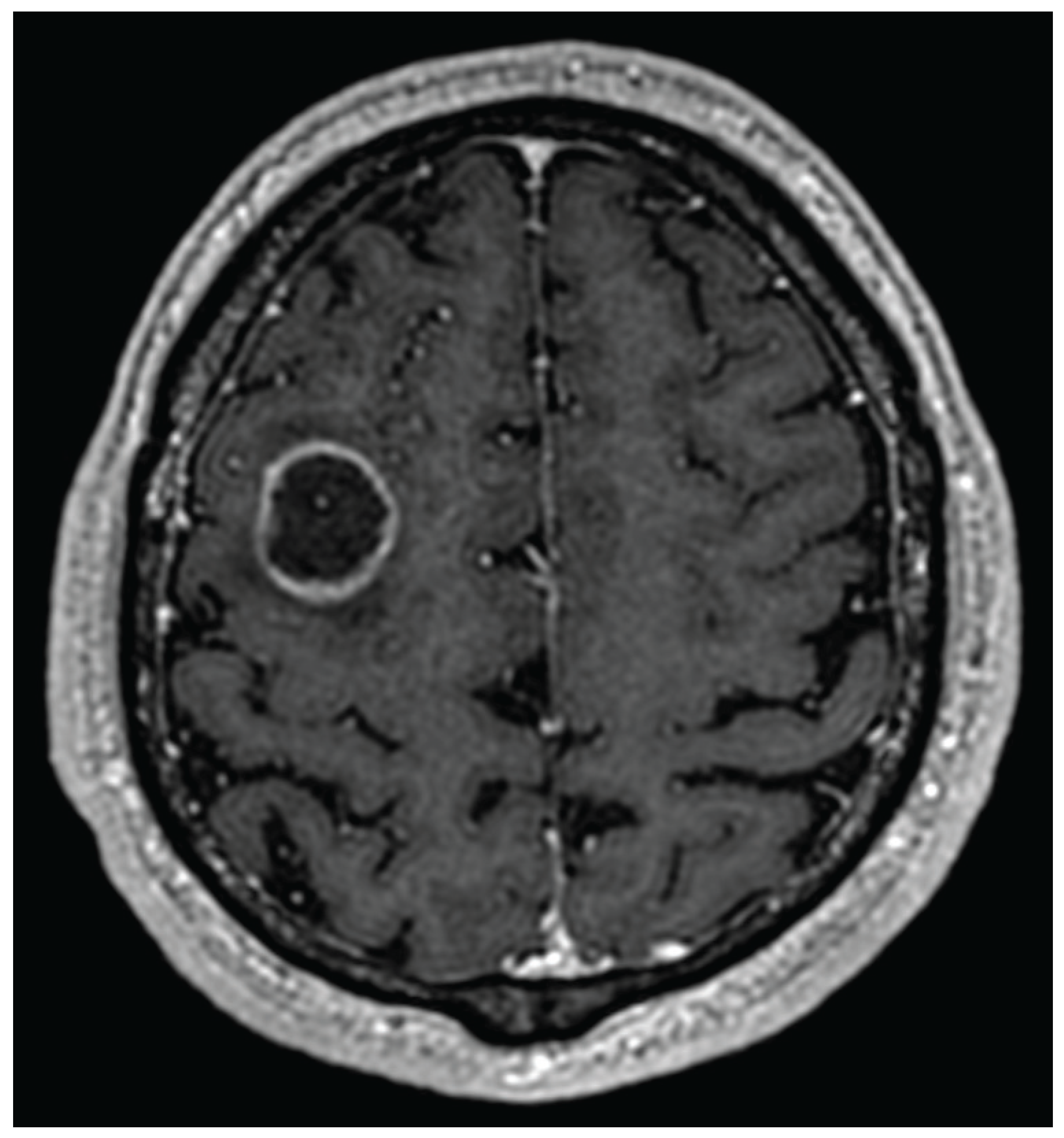
- Gaillard, F.; Sharma, R.; Le, L. Cerebral Ring Enhancing Lesions. Radiopaedia.org. 2009. Available online: https://radiopaedia.org/articles/cerebral-ring-enhancing-lesions-differential?lang=us (accessed on 24 June 2025).
- Jia, Z.; Chen, D. Brain Tumor Identification and Classification of MRI images using deep learning techniques. IEEE Access 2024, 13, 123783–123792. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V.; Saitta, L. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Anantharajan, S.; Gunasekaran, S.; Subramanian, T.; R, V. MRI brain tumor detection using deep learning and machine learning approaches. Meas. Sens. 2024, 31, 101026. [Google Scholar] [CrossRef]
- Mittal, M.; Goyal, L.M.; Kaur, S.; Kaur, I.; Verma, A.; Jude Hemanth, D. Deep learning based enhanced tumor segmentation approach for MR brain images. Appl. Soft Comput. J. 2019, 78, 346–354. [Google Scholar] [CrossRef]
- Abiwinanda, N.; Hanif, M.; Hesaputra, S.T.; Handayani, A.; Mengko, T.R. Brain tumor classification using convolutional neural network. In Proceedings of the IFMBE Proceedings, Banja Luka, Bosnia and Herzegovina, 16–18 May 2019; Springer: Berlin/Heidelberg, Germany, 2019; Volume 68, pp. 183–189. [Google Scholar] [CrossRef]
- Badža, M.M.; Barjaktarović, M.C. Classification of brain tumors from mri images using a convolutional neural network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A Deep Learning-Based Framework for Automatic Brain Tumors Classification Using Transfer Learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Cheng, J. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427/8 (accessed on 24 June 2025).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Li, D.; Hu, W.; Gan, T.; Liu, G.; Ma, L.; Ai, K.; Zhang, J. The MRI-Based 3D-ResNet-101 Deep Learning Model for Predicting Preoperative Grading of Gliomas: A Multicenter Study. Available online: https://archive.ismrm.org/2023/0522.html (accessed on 24 June 2025).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Hara, K.; Kataoka, H.; Satoh, Y. Learning Spatio-Temporal Features With 3D Residual Networks for Action Recognition. arXiv 2017, arXiv:1708.07632. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Rao, C.S.; Karunakara, K. A comprehensive review on brain tumor segmentation and classification of MRI images. Multimed. Tools Appl. 2021, 80, 17611–17643. [Google Scholar] [CrossRef]
- Kataria, P.; Dogra, A.; Sharma, T.; Goyal, B. Trends in DNN Model Based Classification and Segmentation of Brain Tumor Detection. Open Neuroimaging J. 2023, 16, e187444002303060. [Google Scholar] [CrossRef]
- Ghorbian, M.; Ghorbian, S.; Ghobaei-arani, M. A comprehensive review on machine learning in brain tumor classification: Taxonomy, challenges, and future trends. Biomed. Signal Process. Control 2024, 98, 106774. [Google Scholar] [CrossRef]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Kazerooni, A.F.; Khalili, N.; Liu, X.; Haldar, D.; Jiang, Z.; Anwar, S.M.; Albrecht, J.; Adewole, M.; Anazodo, U.; Anderson, H.; et al. The Brain Tumor Segmentation (BraTS) Challenge 2023: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs). arXiv 2024, arXiv:2305.17033v7. [Google Scholar]
- Kumar, A.; Jha, A.K.; Agarwal, J.P.; Yadav, M.; Badhe, S.; Sahay, A.; Epari, S.; Sahu, A.; Bhattacharya, K.; Chatterjee, A.; et al. Machine-Learning-Based Radiomics for Classifying Glioma Grade from Magnetic Resonance Images of the Brain. J. Pers. Med. 2023, 13, 920. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, Y.; Sun, Z.; Xu, K.; Fan, X.; Li, S.; Zhang, Z.; Jiang, T.; Liu, X.; Wang, Y. Radiogenomic analysis of PTEN mutation in glioblastoma using preoperative multi-parametric magnetic resonance imaging. Neuroradiology 2019, 61, 1229–1237. [Google Scholar] [CrossRef]
- Bonneau, D.; Longy, M. Mutations of the human PTEN gene. Hum. Mutat. 2000, 16, 109–122. [Google Scholar] [CrossRef]
- Winn, H.R. Youmans and Winn Neurological Surgery E-Book: 4-Volume Set; Elsevier Health Sciences: Philadelphia, PA, USA, 2022. [Google Scholar]
- Logothetis, N.K.; Pauls, J.; Augath, M.; Trinath, T.; Oeltermann, A. Neurophysiological investigation of the basis of the fMRI signal. Nature 2001, 412, 150–157. [Google Scholar] [CrossRef]
- Merboldt, K.D.; Hanicke, W.; Frahm, J. Self-diffusion NMR imaging using stimulated echoes. J. Magn. Reson. (1969) 1985, 64, 479–486. [Google Scholar] [CrossRef]
- Duffau, H.; Capelle, L.; Denvil, D.; Sichez, N.; Gatignol, P.; Lopes, M.; Mitchell, M.C.; Sichez, J.P.; Van Effenterre, R. Functional recovery after surgical resection of low grade gliomas in eloquent brain: Hypothesis of brain compensation. J. Neurol. Neurosurg. Psychiatry 2003, 74, 901–907. [Google Scholar] [CrossRef]
- Kuan, E.; Vegh, V.; Phamnguyen, J.; O’Brien, K.; Hammond, A.; Reutens, D. Language task-based fMRI analysis using machine learning and deep learning. Front. Radiol. 2024, 4, 1495181. [Google Scholar] [CrossRef]
- Rodríguez, J.J.; Kuncheva, L.I.; Alonso, C.J. Rotation forest: A New classifier ensemble method. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Cabello, N.; Naghizade, E.; Qi, J.; Kulik, L. Fast and accurate time series classification through supervised interval search. In Proceedings of the IEEE International Conference on Data Mining, ICDM, Sorrento, Italy, 17–20 November 2020; pp. 948–953. [Google Scholar] [CrossRef]
- Ismail Fawaz, H.; Lucas, B.; Forestier, G.; Pelletier, C.; Schmidt, D.F.; Weber, J.; Webb, G.I.; Idoumghar, L.; Muller, P.A.; Petitjean, F. InceptionTime: Finding AlexNet for time series classification. Data Min. Knowl. Discov. 2020, 34, 1936–1962. [Google Scholar] [CrossRef]
- Parker Jones, O.; Voets, N.L.; Adcock, J.E.; Stacey, R.; Jbabdi, S. Resting connectivity predicts task activation in pre-surgical populations. NeuroImage Clin. 2017, 13, 378–385. [Google Scholar] [CrossRef]
- Firouzi, M.H.; Kazemi, K.; Ahmadi, M.; Helfroush, M.S.; Aarabi, A. Enhanced ADHD classification through deep learning and dynamic resting state fMRI analysis. Sci. Rep. 2024, 14, 24473. [Google Scholar] [CrossRef] [PubMed]
- Huda, S.; Khan, D.M.; Masroor, K.; Warda; Rashid, A.; Shabbir, M. Advancements in automated diagnosis of autism spectrum disorder through deep learning and resting-state functional mri biomarkers: A systematic review. Cogn. Neurodynamics 2024, 18, 3585–3601. [Google Scholar] [CrossRef]
- Biswal, B.; Zerrin Yetkin, F.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef]
- Luckett, P.H.; Park, K.Y.; Lee, J.J.; Lenze, E.J.; Wetherell, J.L.; Eyler, L.T.; Snyder, A.Z.; Ances, B.M.; Shimony, J.S.; Leuthardt, E.C. Data Efficient Resting State Functional MRI Brain Mapping with Deep Learning. J. Neurosurg. 2023, 139, 1258. [Google Scholar] [CrossRef]
- Panesar, S.S.; Abhinav, K.; Yeh, F.C.; Jacquesson, T.; Collins, M.; Fernandez-Miranda, J. Tractography for Surgical Neuro-Oncology Planning: Towards a Gold Standard. Neurotherapeutics 2019, 16, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Neher, P.F.; Côté, M.A.; Houde, J.C.; Descoteaux, M.; Maier-Hein, K.H. Fiber tractography using machine learning. NeuroImage 2017, 158, 417–429. [Google Scholar] [CrossRef]
- Heker, M.; Amer, R.; Alexandroni, G.; Greenspan, H. Automated supervised segmentation of anatomical fiber tracts using an AdaBoost framework. In Proceedings of the 2016 IEEE International Conference on the Science of Electrical Engineering, ICSEE, Eilat, Israel, 16–18 November 2016. [Google Scholar] [CrossRef]
- Zhang, F.; Savadjiev, P.; Cai, W.; Song, Y.; Rathi, Y.; Tunç, B.; Parker, D.; Kapur, T.; Schultz, R.T.; Makris, N.; et al. Whole brain white matter connectivity analysis using machine learning: An application to autism. NeuroImage 2018, 172, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Jörgens, D.; Smedby, O.; Moreno, R. Learning a single step of streamline tractography based on neural networks. In Mathematics and Visualization; Springer: Berlin/Heidelberg, Germany, 2018; pp. 103–116. [Google Scholar] [CrossRef]
- Wegmayr, V.; Giuliari, G.; Buhmann, J.M. Entrack: A Data-Driven Maximum-Entropy Approach to Fiber Tractography. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); 11824 LNCS; Springer International Publishing: Cham, Switzerland, 2019; pp. 232–244. [Google Scholar] [CrossRef]
- Wegmayr, V.; Buhmann, J.M. Entrack: Probabilistic Spherical Regression with Entropy Regularization for Fiber Tractography. Int. J. Comput. Vis. 2021, 129, 656–680. [Google Scholar] [CrossRef]
- Wasserthal, J.; Neher, P.F.; Isensee, F.; Maier-Hein, K.H. Direct White Matter Bundle Segmentation using Stacked U-Nets. arXiv 2017, arXiv:1703.02036. [Google Scholar] [CrossRef]
- Gupta, V.; Thomopoulos, S.I.; Rashid, F.M.; Thompson, P.M. FiberNET: An ensemble deep learning framework for clustering white matter fibers. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); 10433 LNCS; Springer International Publishing: Cham, Switzerland, 2017; pp. 548–555. [Google Scholar] [CrossRef]
- Gupta, V.; Thomopoulos, S.I.; Corbin, C.K.; Rashid, F.; Thompson, P.M. Fibernet 2.0: An Automatic Neural Network Based Tool for Clustering White Matter Fibers in the Brain. bioRxiv 2017, 210781. [Google Scholar] [CrossRef]
- Wasserthal, J.; Neher, P.F.; Maier-Hein, K.H. Tract orientation mapping for bundle-specific tractography. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); 11072 LNCS; Springer International Publishing: Cham, Switzerland, 2018; pp. 36–44. [Google Scholar] [CrossRef]
- Kumar, P.R.; Shilpa, B.; Jha, R.K. BrainTract: Segmentation of white matter fiber tractography and analysis of structural connectivity using hybrid convolutional neural network. Neuroscience 2025, 580, 218–230. [Google Scholar] [CrossRef]
- Korycinski, M.; Ciecierski, K.A.; Niewiadomska-Szynkiewicz, E. HyTract: Advancing tractography for neurosurgical planning with a hybrid method integrating neural networks and a path search algorithm. Neural Netw. 2025, 190, 107624. [Google Scholar] [CrossRef]
- Buckner, R.L.; Roffman, J.L.; Smoller, J.W. Brain Genomics Superstruct Project (GSP). 2014. Available online: https://cir.nii.ac.jp/crid/1881991017814738176 (accessed on 24 June 2025).
- LaMontagne, P.J.; Benzinger, T.L.; Morris, J.C.; Keefe, S.; Hornbeck, R.; Xiong, C.; Grant, E.; Hassenstab, J.; Moulder, K.; Vlassenko, A.G.; et al. OASIS-3: Longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. medrxiv 2019. [Google Scholar] [CrossRef]
- Maier-Hein, K.H.; Neher, P.F.; Houde, J.C.; Côté, M.A.; Garyfallidis, E.; Zhong, J.; Chamberland, M.; Yeh, F.C.; Lin, Y.C.; Ji, Q.; et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 2017, 8, 1349. [Google Scholar] [CrossRef]
- Wasserthal, J.; Neher, P.; Maier-Hein, K.H. TractSeg-Fast and accurate white matter tract segmentation. Neuroimage 2018, 183, 239–253. [Google Scholar] [CrossRef]
- Mazzucchi, E.; La Rocca, G.; Hiepe, P.; Pignotti, F.; Galieri, G.; Policicchio, D.; Boccaletti, R.; Rinaldi, P.; Gaudino, S.; Ius, T.; et al. Intraoperative Integration of Multimodal Imaging to Improve Neuronavigation: A Technical Note. World Neurosurg. 2022, 164, 330–340. [Google Scholar] [CrossRef]
- Guo, X.; Xing, H.; Pan, H.; Wang, Y.; Chen, W.; Wang, H.; Zhang, X.; Liu, J.; Xu, N.; Wang, Y.; et al. Neuronavigation Combined With Intraoperative Ultrasound and Intraoperative Magnetic Resonance Imaging Versus Neuronavigation Alone in Diffuse Glioma Surgery. World Neurosurg. 2024, 192, e355–e365. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, C.; Ma, L.; Zhang, C. Artificial Intelligence Algorithm-Based Intraoperative Magnetic Resonance Navigation for Glioma Resection. Contrast Media Mol. Imaging 2022, 2022, 4147970. [Google Scholar] [CrossRef]
- Şahin, E.; TALU, M.F. AI-Guided Optimal Trajectory Selection in Stereotactic Brain Biopsy. Artif. Intell. Stud. 2025, 8, 1–19. [Google Scholar] [CrossRef]
- Ishankulov, T.; Danilov, G.; Pitskhelauri, D.; Titov, O.; Ogurtsova, A.; Buklina, S.; Gulaev, E.; Konakova, T.; Bykanov, A. Prediction of Postoperative Speech Dysfunction Based on Cortico-Cortical Evoked Potentials and Machine Learning. In Proceedings of the Studies in Health Technology and Informatics, Nice, France, 27–30 May 2022; IOS Press BV: Amsterdam, The Netherlands, 2022; Volume 289, pp. 33–36. [Google Scholar] [CrossRef]
- Schweingruber, N.; Mader, M.M.D.; Wiehe, A.; Röder, F.; Göttsche, J.; Kluge, S.; Westphal, M.; Czorlich, P.; Gerloff, C. A recurrent machine learning model predicts intracranial hypertension in neurointensive care patients. Brain 2022, 145, 2910–2919. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.W.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed]
- Petrov, D.; Miranda, S.P.; Balu, R.; Wathen, C.; Vaz, A.; Mohan, V.; Colon, C.; Diaz-Arrastia, R. Prediction of intracranial pressure crises after severe traumatic brain injury using machine learning algorithms. J. Neurosurg. 2023, 139, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, G.; Maas, A.; Lecky, F.; Manley, G.; Stocchetti, N.; Murray, G. The Glasgow Coma Scale at 40 years: Standing the test of time. Lancet Neurol. 2014, 13, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Ker, J.; Bai, Y.; Lee, H.Y.; Rao, J.; Wang, L. Automated brain histology classification using machine learning. J. Clin. Neurosci. 2019, 66, 239–245. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.; Maher, C.O.; et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef]
- Quan, H.; Li, X.; Hu, D.; Nan, T.; Cui, X. Dual-Channel Prototype Network for Few-Shot Pathology Image Classification. IEEE J. Biomed. Health Inform. 2024, 28, 4132–4144. [Google Scholar] [CrossRef]
- Nan, T.; Zheng, S.; Qiao, S.; Quan, H.; Gao, X.; Niu, J.; Zheng, B.; Guo, C.; Zhang, Y.; Wang, X.; et al. Deep learning quantifies pathologists’ visual patterns for whole slide image diagnosis. Nat. Commun. 2025, 16, 5493. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Nuechterlein, N.; Shapiro, L.G.; Holland, E.C.; Cimino, P.J. Machine learning modeling of genome-wide copy number alteration signatures reliably predicts IDH mutational status in adult diffuse glioma. Acta Neuropathol. Commun. 2021, 9, 191. [Google Scholar] [CrossRef]
- Hu, D.; Dong, Z.; Liang, K.; Yu, H.; Wang, S.; Liu, X. High-order Topology for Deep Single-Cell Multiview Fuzzy Clustering. IEEE Trans. Fuzzy Syst. 2024, 32, 4448–4459. [Google Scholar] [CrossRef]
- Hu, D.; Liang, K.; Dong, Z.; Wang, J.; Zhao, Y.; He, K. Effective multi-modal clustering method via skip aggregation network for parallel scRNA-seq and scATAC-seq data. Briefings Bioinform. 2024, 25, 102. [Google Scholar] [CrossRef]
- Javed, S.; Mahmood, A.; Fraz, M.M.; Koohbanani, N.A.; Benes, K.; Tsang, Y.W.; Hewitt, K.; Epstein, D.; Snead, D.; Rajpoot, N. Cellular community detection for tissue phenotyping in colorectal cancer histology images. Med. Image Anal. 2020, 63, 101696. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Halama, N.; Marx, A. 100,000 Histological Images of Human Colorectal Cancer and Healthy Tissue. 2018. Available online: https://cir.nii.ac.jp/crid/1883399490387124992 (accessed on 24 June 2025).
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and colon cancer histopathological image dataset (lc25000). arXiv 2019, arXiv:1912.12142. [Google Scholar] [CrossRef]
- Heo, J.; Park, S.J.; Kang, S.H.; Oh, C.W.; Bang, J.S.; Kim, T. Prediction of Intracranial Aneurysm Risk using Machine Learning. Sci. Rep. 2020, 10, 6921. [Google Scholar] [CrossRef]
- Zhu, G.; Luo, X.; Yang, T.; Cai, L.; Yeo, J.H.; Yan, G.; Yang, J. Deep learning-based recognition and segmentation of intracranial aneurysms under small sample size. Front. Physiol. 2022, 13, 1084202. [Google Scholar] [CrossRef]
- Yang, Y.; Chang, Z.; Nie, X.; Wu, J.; Chen, J.; Liu, W.; He, H.; Wang, S.; Zhu, C.; Liu, Q. Integrated Deep Learning Model for the Detection, Segmentation, and Morphologic Analysis of Intracranial Aneurysms Using CT Angiography. Radiol. Artif. Intell. 2025, 7. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Malik, K.M.; Ahmad, J.; Malik, G. StrokeNet: An automated approach for segmentation and rupture risk prediction of intracranial aneurysm. Comput. Med. Imaging Graph. 2023, 108, 102271. [Google Scholar] [CrossRef]
- Abdullah, I.; Javed, A.; Malik, K.M.; Malik, G. DeepInfusion: A dynamic infusion based-neuro-symbolic AI model for segmentation of intracranial aneurysms. Neurocomputing 2023, 551, 126510. [Google Scholar] [CrossRef]
- Silva, M.A.; Patel, J.; Kavouridis, V.; Gallerani, T.; Beers, A.; Chang, K.; Hoebel, K.V.; Brown, J.; See, A.P.; Gormley, W.B.; et al. Machine Learning Models can Detect Aneurysm Rupture and Identify Clinical Features Associated with Rupture. World Neurosurg. 2019, 131, e46–e51. [Google Scholar] [CrossRef]
- Kim, J.Y.; Choi, H.J.; Kim, S.H.; Ju, H. Improved differentiation of cavernous malformation and acute intraparenchymal hemorrhage on CT using an AI algorithm. Sci. Rep. 2024, 14, 11818. [Google Scholar] [CrossRef]
- Yun, T.J.; Choi, J.W.; Han, M.; Jung, W.S.; Choi, S.H.; Yoo, R.E.; Hwang, I.P. Deep learning based automatic detection algorithm for acute intracranial haemorrhage: A pivotal randomized clinical trial. NPJ Digit. Med. 2023, 6, 61. [Google Scholar] [CrossRef]
- Jabal, M.S.; Mohammed, M.A.; Kobeissi, H.; Lanzino, G.; Brinjikji, W.; Flemming, K.D. Quantitative image signature and machine learning-based prediction of outcomes in cerebral cavernous malformations. J. Stroke Cerebrovasc. Dis. 2024, 33, 107462. [Google Scholar] [CrossRef]
- Akiyama, Y.; Mikami, T.; Mikuni, N. Deep Learning-Based Approach for the Diagnosis of Moyamoya Disease. J. Stroke Cerebrovasc. Dis. 2020, 29, 105322. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Takaku, A. Cerebrovascular Moyamoya Disease: Disease Showing Abnormal Net-Like Vessels in Base of Brain. Arch. Neurol. 1969, 20, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Israel, Z.; Burchiel, K.J. Microelectrode Recording in Movement Disorder Surgery; Thieme: New York, NY, USA, 2011. [Google Scholar]
- Ciecierski, K.A. Mathematical methods of signal analysis applied in medical diagnostic. Int. J. Appl. Math. Comput. Sci. 2020, 30, 449–462. [Google Scholar] [CrossRef]
- Aerts, H.; Marinazzo, D. “BTC_preop”. 2022. Available online: https://openneuro.org/datasets/ds001226/versions/5.0.0 (accessed on 24 June 2025).
- Noh, S.H.; Cho, P.G.; Kim, K.N.; Kim, S.H.; Shin, D.A. Artificial Intelligence for Neurosurgery: Current State and Future Directions. J. Korean Neurosurg. Soc. 2022, 66, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Leveson, N.G.; Turner, C.S. An Investigation of the Therac-25 Accidents. Computer 1993, 26, 18–41. [Google Scholar] [CrossRef]

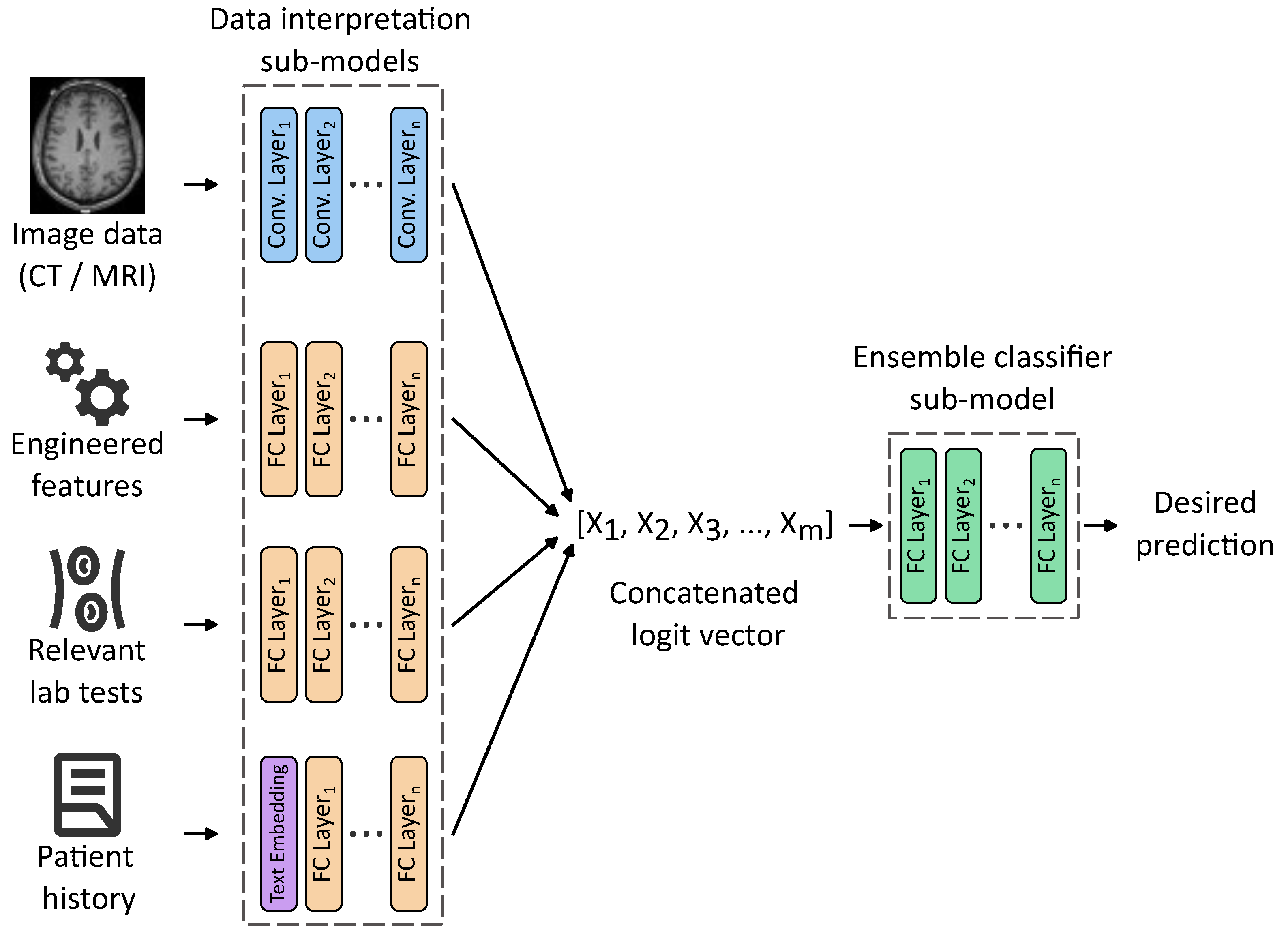

| Authors | Scope | Method | Input | Metrics | Dataset |
|---|---|---|---|---|---|
| Jia et al. [25] | Brain tumor detection | SVM | MRI slices | Acc = 98.51% | Multi-parametric MRI images |
| Anantharajan et al. [27] | Brain tumor detection | SVM | MRI slices | Acc = 97.93% sensitivity = 92% specificity = 98% | Kaggle open data 255 T1-mode MRI |
| Mittal et al. [28] | Brain tumor segmentation | Hybrid technique: SWT + RF + GCNN | MRI slices | MSE = 0.001 sensitivity = 98.23% precision = 98.81% | BRAINIX medical images |
| Abiwinanda et al. [29] | Brain tumor classification (3 classes): glioma, meningioma, pituitary | CNN | MRI slices (512 × 512) | Acc = 84.19% | Figshare [32] (3064 CE-MRI) |
| Badža et al. [30] | Brain tumor classification (3 classes): glioma, meningioma, pituitary | CNN | MRI slices (256 × 256) | Confusion matrices Acc = 88.48% | Figshare [32] augmented dataset (9192 CE-MRI) |
| Rehman et al. [31] | Brain tumor classification (3 classes): glioma, meningioma, pituitary | CNN: AlexNet, GoogLeNet, VGG16 | MRI slices (512 × 512) | Acc = 98.69% | ImageNet [33], Figshare [32] |
| Li et al. [35] | Glioma grading classification | 3D-ResNet101 | MRI image | Acc = 83% F1 score = 83% AUC = 0.89 | TCIA [38], 708 glioma patients from 2nd Hospital of Lanzhou University |
| Authors | Scope | Method | Input | Metrics | Dataset | Characteristics |
|---|---|---|---|---|---|---|
| Kuan et al. [51] | fMRI-based identification of brain regions subserving language | RF, WEASEL, TSFresh, RISE, sTSF, ARSENAL, Inception | fMRI EPI sequences, T1 MRI 256 × 256 | AUC = 0.97, ± = 0.03 | Patients of Royal Brisbane and Women’s Hospital | Evaluation of different ML/DL methods in predicting language related brain regions |
| Luckett et al. [59] | Mapping of resting-state networks in the brain | 3D CNN | T1w MRI 256 × 256; RS-fMRI scans, voxel 3 4 mm3; T1w MRI voxel 1 mm3; T2w MRI voxel 1 mm3 | LAN = 96.9%, MOT true-positive rate = 96.3% | 2252 patients from [73,74] | Demonstrates the utility of DL for accurate mapping of eloquent cortex using a reduced amount of RS-fMRI data |
| Neher et al. [61] | Tracking fiber pathways | RF + boosting by voting | 3D gradient data | Valid connections = 93%, bundle coverage rate = 94% | nitrc.org/projects/diffusion-data; https://tractometer.org/ismrm2015/home/; www.humanconnectome.org/data | RF-based fiber tractography using neighborhood information. |
| Heker et al. [62] | Segmentation of anatomical fiber tracts | AdaBoost, Viola Jones algorithm | DWI MRI 3D data | FP = 0.44%, detection rate = 98.77% | Human Connectome Project | Novel approach to the automatic segmentation of brain tracts |
| Zhang et al. [63] | Analysis of whole brain white matter for autism spectrum disorder detection | SVM with polynomial kernel | DWI MRI 3D data | Acc = 78.33% | 149 pediatric male children (70 wist ASD) from Center for Autism Research, Children’s Hospital of Philadelphia | Novel method of autism spectrum detection from DWI MRI data |
| Jörgens et al. [64] | Step-by-step prediction of streamline tractography from DWI MRI data | NN composed of fully connected layers | DWI MRI 3D data | Angular error below 5.5° | ISMRM 2015 tractography challenge data [75] | Streamline prediction using NN with linear interpolation of DWI data |
| Wegmayr et al. [65] | Entropy-based approach to fiber tractography | Maximum Entropy Principle, conditional distribution, NN composed of fully connected layers | DWI MRI 3D data | Valid bundles = 23 out of 25; valid connections = 51% | Human Connectome Project | Probabilistic framework for ML approaches to fiber tractography |
| Wegmayr et al. [66] | Entropy-based approach to fiber tractography | Maximum Entropy Principle, conditional distribution, entropy regularization, annealing, NN composed of fully connected layers | DWI MRI 3D data | Valid bundles = 24 out of 25; valid connections = 52% | Human Connectome Project | Probabilistic framework for ML approaches to fiber tractography |
| Wasserthal et al. [67] | White matter bundle segmentation | CNN (U-Net architecture) | DWI MRI 3D data | Dice score (CST, SLF) close to 0.85 | ISMRM 2015 tractography challenge data [75] | Human-comparable performance achieved on all bundles |
| Gupta et al. [68] | Detection and clustering of white matter fibers | 2D CNN | DWI MRI 3D data | Not given | Not specified | Sampling of fiber data with replacements for application of CNN |
| Gupta et al. [69] | Automatic clustering of white matter fibers | 2D CNN | DWI MRI 3D data | Acc above 97% | Parkinson’s Progression Markers Initiative (226 individuals) | False-positive fibers are removed from a fiber bundle segmented using ROI-based segmentation tools |
| Wasserthal et al. [70] | White matter fiber segmentation | CNN CNN (U-Net architecture) | DWI MRI 3D data | Outperforms the reference methods by 14 Dice points | Proprietary dataset [76] | Very good results for low-quality datasets |
| Kumar et al. [71] | White matter fiber segmentation | Fully connected CNN with skip connections and spatial attention, Gray Wolf Optimization to tune CNN classifier | DWI MRI 3D data | Acc = 97.10%, sensitivity = 95.74%, F1 = 94.79% | Human Connectome Project | Novel, efficient classifier |
| Korycinski et al. [72] | Fiber tracking | Hybrid technique: fully connected CNN + A* path search algorithm | DWI MRI 3D data | Mean Euclidean Distance (MED) to EuDX below 10 | Human Connectome Project | Combination of deep learning with classical ML |
| Authors | Scope | Method | Input | Metrics | Dataset |
|---|---|---|---|---|---|
| Intraoperative neuronavigation | |||||
| Mazzucchi et al. [77] | Multimodal imaging for improving intraoperative neuronavigation | Fusion of T1w, T2w, DTI, Flair and ultrasound | MRI: T1w, T2w, DTI and Flair modalities; intraoperative ultrasound | Descriptive | Proprietary |
| Guo et al. [78] | Intraoperative MRI and ultrasound in diffuse glioma surgery | Fusion of T1w, T2w, DTI, DWI and ultrasound | MRI: T1w, T2w, DTI and DWI modalities; intraoperative ultrasound | Increased extent of resection, more cases with a total resection; increased operative time | Proprietary |
| Wei et al. [79] | Intraoperative MRI navigation for glioma resection | Segmentation dictionary learning algorithm | MRI: T1, T2, Flair, DTI and BOLD modalities | Descriptive | Proprietary |
| Stereotactic biopsy guidance | |||||
| Şahin et al. [80] | Trajectory selection in stereotactic brain biopsy | 3D Residual U-Net | CT, MRI, MRA | Descriptive | Proprietary |
| Intraoperative brain mapping | |||||
| Ishankulov et al. [81] | Prediction of post-operative speech impairment based upon evoked potentials | RF, logistic regression, SVM | Cortico-Cortical Evoked Potentials | Descriptive | Proprietary |
| Post-operative/traumatic intensive care | |||||
| Schweingruber et al. [82] | ICP hypertension prediction in neuro ICU patients | LSTM | ICP monitoring | Descriptive | Proprietary |
| Petrov et al. [84] | Prediction of an ICP crisis in patients with TBI | RF, XGBoost, LGBM | ICP monitoring | Acc = 88%, AUC 0.87 | Proprietary |
| Histopathology | |||||
| Ker et al. [86] | Automatic brain histological classification | CNN (Inception V3 model) | Histology images 1600 × 1200 | Normal brain vs. high-grade glioma: F1 = 100%; normal brain vs. glioma: F1 = 99%; | Proprietary from Department of Pathology at Tan Tock Seng Hospital |
| Orringer et al. [88] | Fast intraoperative histology of unprocessed surgical specimens | Fully connected network (MLP) | SRS microscope images | Lesional vs. non-lesional: specificity = 94.1%, sensitivity = 94.5% | Proprietary |
| Quan et al. [89] | Few-shot pathology image classification | Transformers, ResNet | Histology images: 150 × 150, 224 × 224, 768 × 768 | Acc = 85.87%, AUC = 0.98 | CRCTP [95], NCTCRC [96], LC25000 [97] |
| Nan et al. [90] | DL patterns for whole-slide image diagnosis | Vision Transformer, ResNet50 | Histology images 224 × 224 | Acc = 93%, AUC = 0.984 | Proprietary from First Affiliated Hospital of China Medical University |
| Genetic testing | |||||
| Nuechterlein et al. [92] | Prediction of IDH mutational status in adult diffuse glioma | PCA, various scikit-learn classifiers | Genome-wide somatic copy number alteration, The Cancer Genome Atlas | Descriptive | https://xena.ucsc.edu, https://gdc.cancer.gov |
| Hu et al. [93] | Deep single-Cell multiview fuzzy clustering | KNN, graph random walk, node2vec, transformers | Transcriptome data | Descriptive | https://github.com/satijalab/seurat-data, https://www.10xgenomics.com/resources/datasets |
| Hu et al. [94] | Multi-modalclustering of scRNA-seq and scATAC-seq data | Transformers, embeddings, KL divergence, deep k-means clustering | scRNA-seq and scATAC-seq data | ARI, NMI metrics | https://www.ncbi.nlm.nih.gov/geo, https://www.10xgenomics.com/resources/datasets, https://github.com/YosefLab/totalVI_reproducibility |
| Authors | Scope | Method | Input | Metrics | Dataset |
|---|---|---|---|---|---|
| Intracranial aneurysm treatment | |||||
| Heo et al. [98] | Prediction of intracranial aneurysm risk | Logistic regression, RF, scalable tree boosting system, and ANN | Tabular data from NHIS-NSC | AUC = 0.76 | NHIS-NSC Korean database |
| Zhu et al. [99] | Detection and segmentation of intracranial aneurysms with a small sample size | 3D UNet, VNet, 3D Res-UNet | CTA images of 101 patients with 140 aneurysms | Sensitivity = 80.9% | Proprietary from First Affiliated Hospital of Xi’an Jiaotong University |
| Yang et al. [100] | Detection, segmentation and analysis of intracranial aneurysms using CT and angiography | CNN (U-Net architecture) | CT, angiography | Acc = 97% | Proprietary |
| Irfan et al. [101] | Segmentation and rupture risk prediction of intracranial aneurysm | CNN (U-Net architecture), decision tree | DSA images: 128 × 128, 256 × 256, 512 × 512 | Acc = 68& 87%, precision: 65% 84%, sensitivity = 67% 81% | 2D Digital Subtraction Angiography (DSA) images |
| Abdullah et al. [102] | Segmentation of intracranial aneurysms | Infusion-based AI model (U-Net) | DSA images: 128 × 128, 256 × 256, 512 × 512 | Acc = 99%, F1 = 63.6% | 2D Digital Subtraction Angiography (DSA) images |
| Vascular conditions treatment | |||||
| Kim at al. [104] | Differentiation of cavernous malformation and acute intraparenchymal hemorrhage | CNN | CT images | AI-assisted results (accuracy, sensitivity, specificity) significantly better than unassisted (p-value < 0.001) | Proprietary, not public |
| Yun et al. [105] | Automatic detection algorithm for acute intracranial haemorrhage | CNN, RNN, VAE, GAN | CT images | AI-assisted results (accuracy, sensitivity, specificity) significantly better than unassisted (p-value < 0.001) | Proprietary |
| Jabal at al. [106] | Prediction of outcomes in cerebral cavernous malformations | SHapley Additive exPlanations (SHAP) | MRI: T2w, FLAIR modalities | p-value results confirm quality of predictions | Proprietary |
| Akiyama et al. [107] | The Moyamoya Disease diagnosing. | Xception, VGG 16, VGG19, Inception V3, ResNet, DenseNet | MRI, MRA | Acc = 99% | Proprietary from Sapporo Medical University Hospital |
| Application Type | Method Type | References |
|---|---|---|
| Diagnosis | Machine Learning | [25,27,28,44,45,92,98,106] |
| Deep Learning | [23,27,28,29,30,31,35,42,43,89,90,93,94,98,99,100,101,102,103,104,105,106,107] | |
| Preoperative | Machine Learning | [51,61,62,63] |
| Deep Learning | [23,51,59,64,65,66,67,68,69,70,71,72] | |
| Intraoperative | Machine Learning | [79,81,110] |
| Deep Learning | [11,77,78,80,88] | |
| Postoperative | Machine Learning | [84] |
| Deep Learning | [82,86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koryciński, M.; Ciecierski, K.A.; Niewiadomska-Szynkiewicz, E. Decision Support Systems in Neurosurgery: Current Applications and Future Directions. Sensors 2025, 25, 7415. https://doi.org/10.3390/s25247415
Koryciński M, Ciecierski KA, Niewiadomska-Szynkiewicz E. Decision Support Systems in Neurosurgery: Current Applications and Future Directions. Sensors. 2025; 25(24):7415. https://doi.org/10.3390/s25247415
Chicago/Turabian StyleKoryciński, Mateusz, Konrad A. Ciecierski, and Ewa Niewiadomska-Szynkiewicz. 2025. "Decision Support Systems in Neurosurgery: Current Applications and Future Directions" Sensors 25, no. 24: 7415. https://doi.org/10.3390/s25247415
APA StyleKoryciński, M., Ciecierski, K. A., & Niewiadomska-Szynkiewicz, E. (2025). Decision Support Systems in Neurosurgery: Current Applications and Future Directions. Sensors, 25(24), 7415. https://doi.org/10.3390/s25247415






