DPM-UNet: A Mamba-Based Network with Dynamic Perception Feature Enhancement for Medical Image Segmentation
Abstract
1. Introduction
- (1)
- We propose a novel segmentation network named DPM-UNet, which integrates the local feature extraction capability of CNNs with the global information aggregation ability of Mamba, aiming to achieve precise medical image segmentation.
- (2)
- We design three key components: the Dual-path Residual Fusion Module (DRFM), the DPMamba Module, and the Multi-scale Aggregation Attention Network (MAAN). The DRFM enhances local feature extraction by fusing features from standard and dilated convolutions. The DPMamba Module leverages Mamba to generate global features and further enhances feature representation in critical channels through a Dynamic Perception Feature Enhancement Block (DPFE). Additionally, the MAAN is embedded in skip connection paths to optimize the transmission and fusion of multi-scale information.
- (3)
- Experimental results on three public medical image segmentation datasets demonstrate that DPM-UNet achieves state-of-the-art segmentation performance compared to existing methods, fully validating the effectiveness and strong generalization capability of our approach for medical image segmentation tasks.
2. Methods
2.1. DPMamba Module
2.1.1. VSS Block
2.1.2. Dynamic Perception Feature Enhancement Block (DPFE)
2.2. Dual-Path Residual Fusion Module (DRFM)
2.3. Multi-Scale Aggregation Attention Network (MAAN)
3. Experiments
3.1. Datasets
3.2. Evaluation Metrics and Baselines
3.3. Implementation Details
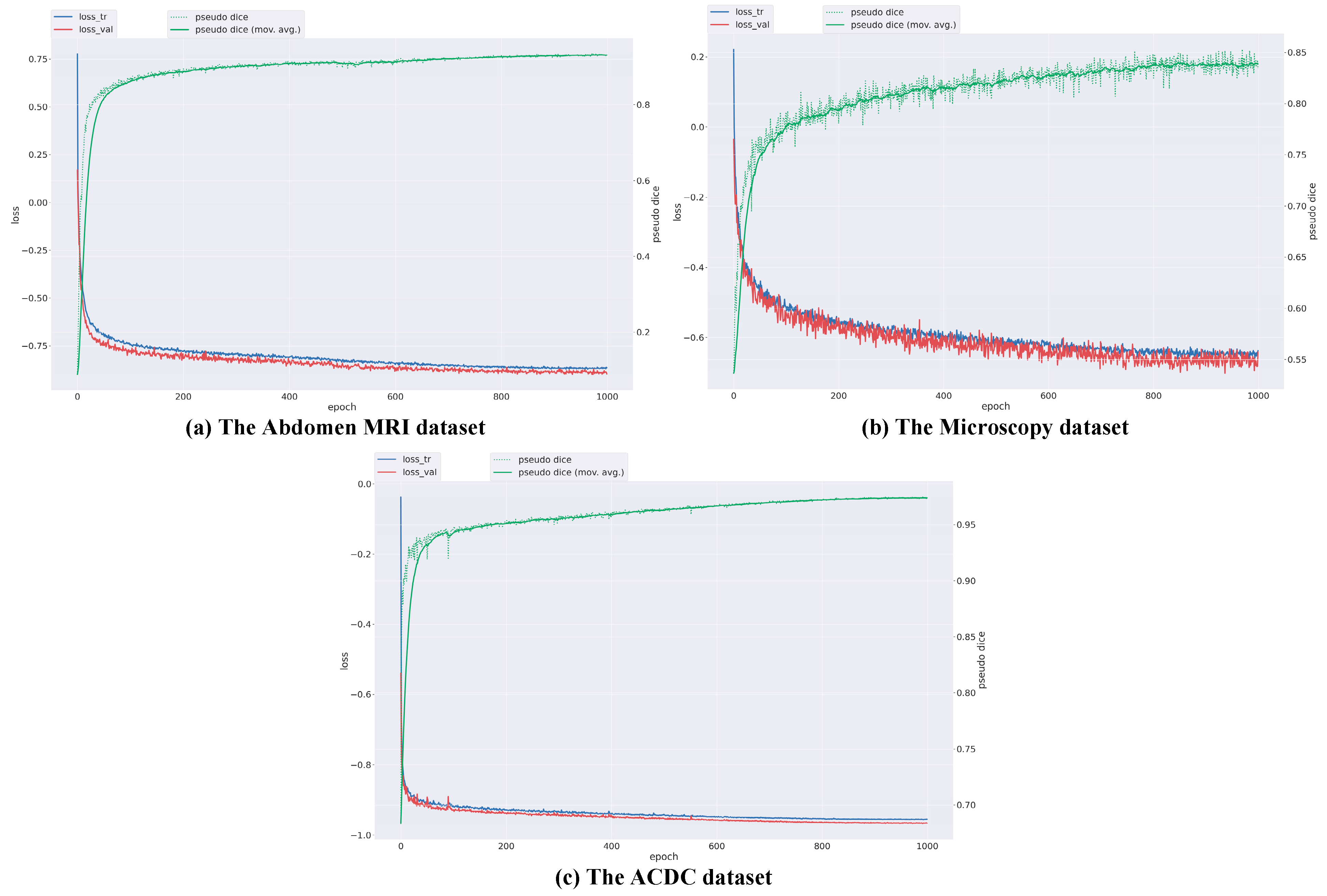
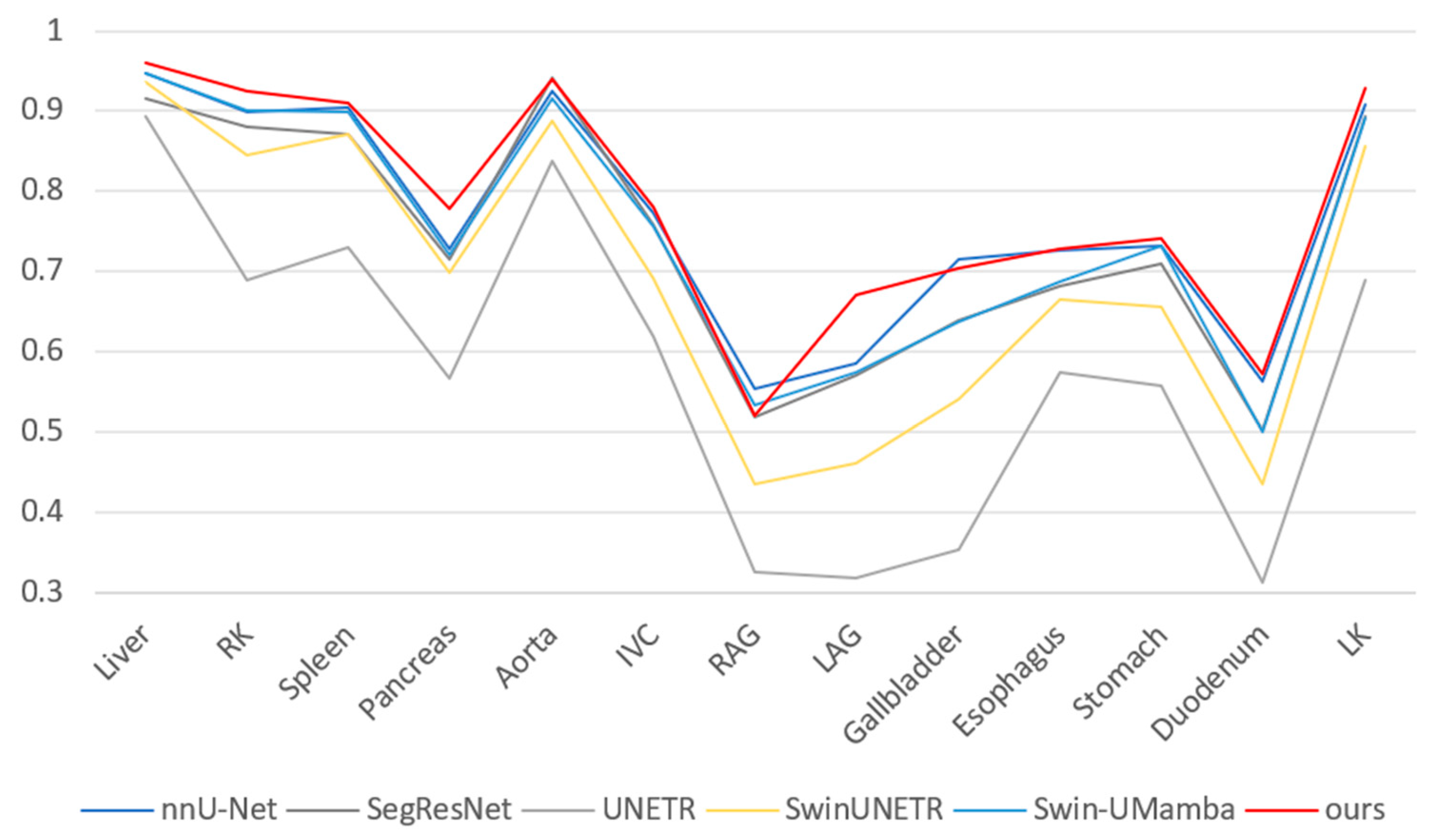
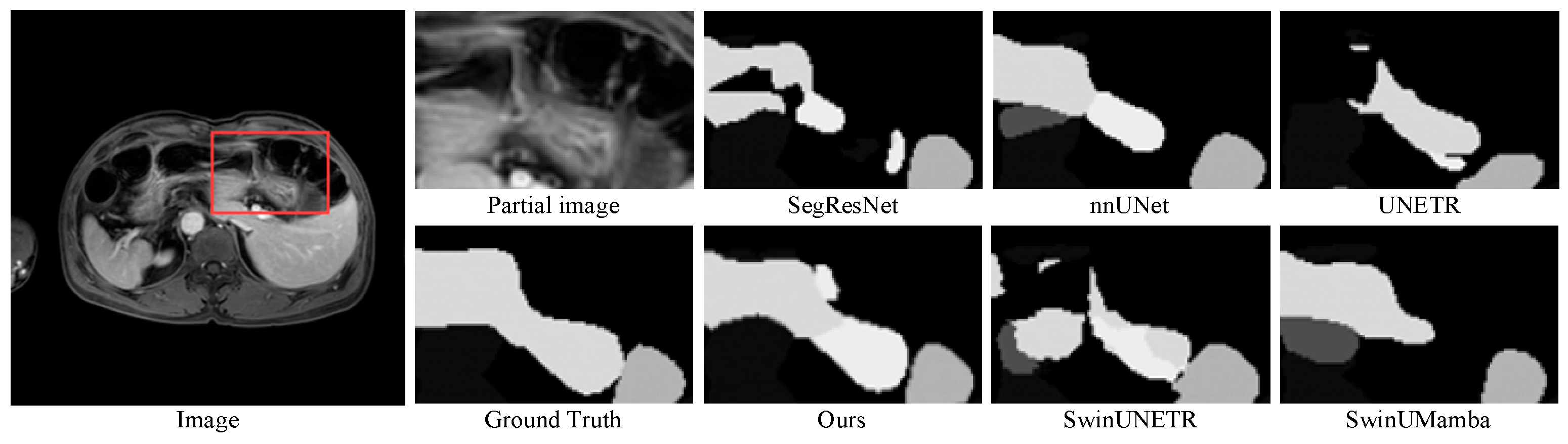
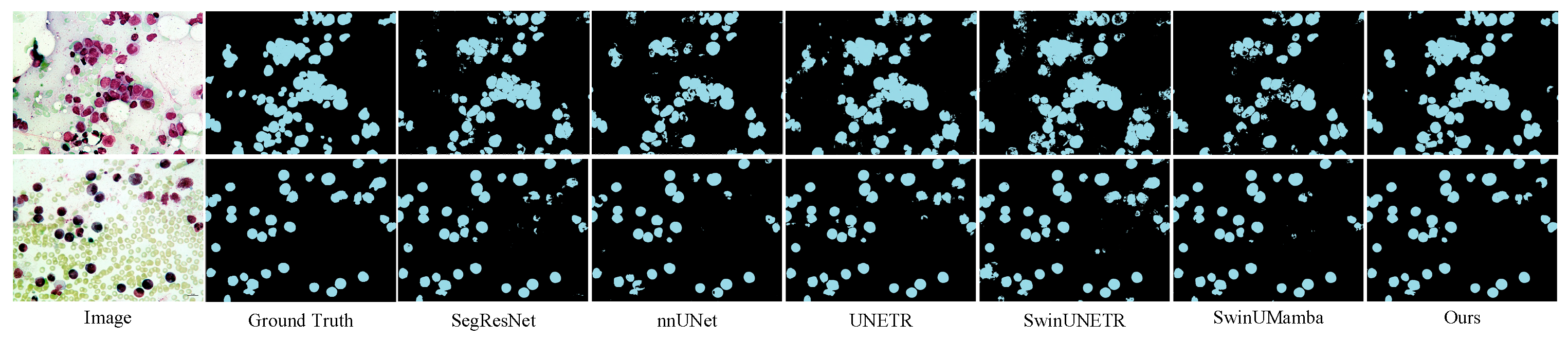
3.4. Experimental Results
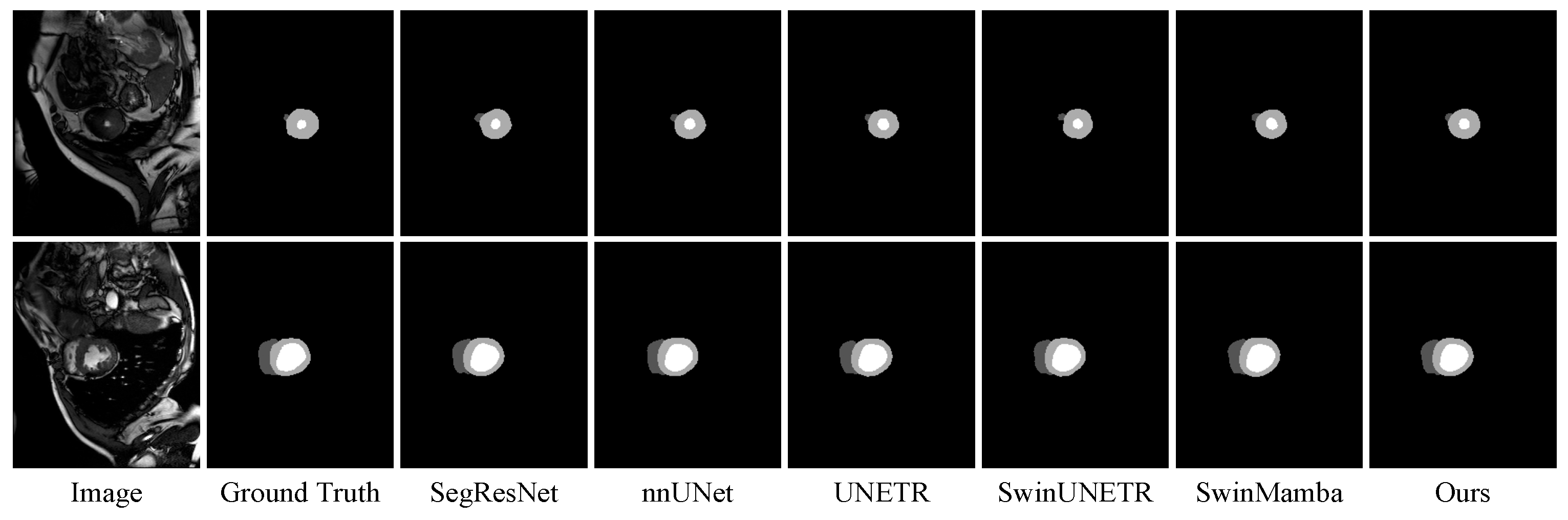
3.5. Further Analysis
3.5.1. Ablation Study
3.5.2. Model Complexity
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, L.; Sheng, B.; Chen, T.; Wu, Q.; Liu, R.; Cai, C.; Wu, L.; Yang, D.; Hamzah, H.; Liu, Y.; et al. A deep learning system for predicting time to progression of diabetic retinopathy. Nat. Med. 2024, 30, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef] [PubMed]
- Bai, W.; Suzuki, H.; Huang, J.; Francis, C.; Wang, S.; Tarroni, G.; Guitton, F.; Aung, N.; Fung, K.; Petersen, S.E.; et al. A population-based phenome-wide association study of cardiac and aortic structure and function. Nat. Med. 2020, 26, 1654–1662. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.-C.; Diao, K.-y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Jungo, A.; Meier, R.; Ermis, E.; Blatti-Moreno, M.; Herrmann, E.; Wiest, R.; Reyes, M. On the Effect of Inter-Observer Variability for a Reliable Estimation of Uncertainty of Medical Image Segmentation; Springer: Cham, Switzerland, 2018; pp. 682–690. [Google Scholar]
- Graham, S.; Vu, Q.D.; Raza, S.E.A.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 2019, 58, 101563. [Google Scholar] [CrossRef]
- Sun, M.; Zou, W.; Wang, Z.; Wang, S.; Sun, Z. An Automated Framework for Histopathological Nucleus Segmentation with Deep Attention Integrated Networks. IEEE ACM Trans. Comput. Biol. Bioinform. 2024, 21, 995–1006. [Google Scholar] [CrossRef]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic Multi-Organ Segmentation on Abdominal CT with Dense V-Networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef]
- Qi, X.; Wu, Z.; Zou, W.; Ren, M.; Gao, Y.; Sun, M.; Zhang, S.; Shan, C.; Sun, Z. Exploring Generalizable Distillation for Efficient Medical Image Segmentation. IEEE J. Biomed. Health Inform. 2024, 28, 4170–4183. [Google Scholar] [CrossRef]
- Khened, M.; Kollerathu, V.A.; Krishnamurthi, G. Fully convolutional multi-scale residual DenseNets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Med. Image Anal. 2019, 51, 21–45. [Google Scholar] [CrossRef]
- Jiang, X.; Hoffmeister, M.; Brenner, H.; Muti, H.S.; Yuan, T.; Foersch, S.; West, N.P.; Brobeil, A.; Jonnagaddala, J.; Hawkins, N.; et al. End-to-end prognostication in colorectal cancer by deep learning: A retrospective, multicentre study. Lancet Digit. Health 2024, 6, e33–e43. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Wang, W.; Dai, J.; Chen, Z.; Huang, Z.; Li, Z.; Zhu, X.; Hu, X.; Lu, T.; Lu, L.; Li, H.; et al. InternImage: Exploring Large-Scale Vision Foundation Models with Deformable Convolutions. arXiv 2022, arXiv:2211.05778. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization; Springer: Cham, Switzerland, 2019; pp. 311–320. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, L.; Tang, J. Augmented FCN: Rethinking context modeling for semantic segmentation. Sci. China Inf. Sci. 2023, 66, 142105. [Google Scholar] [CrossRef]
- Raghu, M.; Unterthiner, T.; Kornblith, S.; Zhang, C.; Dosovitskiy, A. Do Vision Transformers See Like Convolutional Neural Networks? In Proceedings of the Neural Information Processing Systems, Virtual, 6–10 December 2021. [Google Scholar]
- Hatamizadeh, A.; Yin, H.; Kautz, J.; Molchanov, P. Global Context Vision Transformers. In Proceedings of the International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2022. [Google Scholar]
- Gu, A.; Dao, T. Mamba: Linear-Time Sequence Modeling with Selective State Spaces. arXiv 2023, arXiv:2312.00752. [Google Scholar] [CrossRef]
- Gu, A.; Goel, K.; Ré, C. Efficiently Modeling Long Sequences with Structured State Spaces. arXiv 2021, arXiv:2111.00396. [Google Scholar]
- Gu, A.; Johnson, I.; Goel, K.; Saab, K.K.; Dao, T.; Rudra, A.; Ré, C. Combining Recurrent, Convolutional, and Continuous-time Models with Linear State-Space Layers. In Proceedings of the Neural Information Processing Systems, Virtual, 6–10 December 2021. [Google Scholar]
- Liu, Y.; Tian, Y.; Zhao, Y.; Yu, H.; Xie, L.; Wang, Y.; Ye, Q.; Liu, Y. VMamba: Visual State Space Model. arXiv 2024, arXiv:2401.10166. [Google Scholar]
- Huang, T.; Pei, X.; You, S.; Wang, F.; Qian, C.; Xu, C. LocalMamba: Visual State Space Model with Windowed Selective Scan. In Proceedings of the ECCV Workshops, Milan, Italy, 29 September–4 October 2024. [Google Scholar]
- Ma, J.; Li, F.; Wang, B.J.A. U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation. arXiv 2024, arXiv:2401.04722. [Google Scholar]
- Xing, Z.; Ye, T.; Yang, Y.; Liu, G.; Zhu, L. SegMamba: Long-range Sequential Modeling Mamba For 3D Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024. [Google Scholar]
- Liu, J.; Yang, H.; Zhou, H.-Y.; Xi, Y.; Yu, L.; Yu, Y.; Liang, Y.; Shi, G.; Zhang, S.; Zheng, H.; et al. Swin-UMamba: Mamba-based UNet with ImageNet-based pretraining. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024. [Google Scholar]
- Rajagopal, A.; Nirmala, V. Convolutional Gated MLP: Combining Convolutions & gMLP. arXiv 2021, arXiv:2111.03940. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Ma, J.; Xie, R.; Ayyadhury, S.; Ge, C.; Gupta, A.; Gupta, R.; Gu, S.; Zhang, Y.; Lee, G.; Kim, J.; et al. The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions. arXiv 2023, arXiv:2308.05864. [Google Scholar]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. arXiv 2021, arXiv:2103.10504. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. arXiv 2022, arXiv:2201.01266. [Google Scholar] [CrossRef]





| Methods | DSC | NSD |
|---|---|---|
| nnU-Net | 76.63 | 83.51 |
| SegResNet | 73.84 | 80.13 |
| UNETR | 57.46 | 62.82 |
| SwinUNETR | 69.08 | 74.73 |
| Swin-UMamba | 74.56 | 81.15 |
| DPM-UNet | 78.15 | 84.67 |
| Methods | DSC | F1 |
|---|---|---|
| nnU-Net | 69.55 | 54.65 |
| SegResNet | 68.51 | 54.02 |
| UNETR | 71.69 | 40.57 |
| SwinUNETR | 66.69 | 37.08 |
| Swin-UMamba | 67.60 | 49.00 |
| DPM-UNet | 73.25 | 60.23 |
| Methods | DSC | NSD | RV | Myo | LV |
|---|---|---|---|---|---|
| nnU-Net | 91.81 | 97.88 | 89.44 | 90.60 | 95.38 |
| SegResNet | 91.71 | 97.99 | 89.64 | 90.27 | 95.23 |
| UNETR | 89.34 | 95.49 | 86.76 | 87.46 | 93.80 |
| SwinUNETR | 91.50 | 97.61 | 89.41 | 90.03 | 95.06 |
| Swin-UMamba | 91.69 | 98.04 | 90.00 | 89.78 | 95.30 |
| DPM-UNet | 91.95 | 98.09 | 89.46 | 90.76 | 95.64 |
| DRFM | DPMamba | CBAM | MAAN | DSC ↑ | NSD ↑ |
|---|---|---|---|---|---|
| ✘ | ✘ | ✘ | ✘ | 75.17 | 81.52 |
| ✔ | ✘ | ✘ | ✘ | 75.62 | 82.40 |
| ✔ | ✔ | ✘ | ✘ | 76.02 | 82.81 |
| ✔ | ✔ | ✔ | ✘ | 76.84 | 83.54 |
| ✔ | ✔ | ✘ | ✔ | 78.15 | 84.67 |
| Dilated Convolution | Fusion | Residual | DSC ↑ | NSD ↑ |
|---|---|---|---|---|
| ✘ | ✘ | ✘ | 76.04 | 82.80 |
| ✔ | ✘ | ✘ | 76.35 | 83.06 |
| ✔ | ✔ | ✘ | 77.53 | 84.32 |
| ✔ | ✔ | ✔ | 78.15 | 84.67 |
| Methods | FLOPs (G) ↓ | Param. (M) ↓ | Training Time (H) | DSC ↑ | NSD ↑ |
|---|---|---|---|---|---|
| nnU-Net | 23 | 33 | 6 | 76.63 | 83.51 |
| SegResNet | 24 | 6 | 8 | 73.84 | 80.13 |
| UNETR | 41 | 87 | 17 | 57.46 | 62.82 |
| SwinUNETR | 29 | 25 | 20 | 69.08 | 74.73 |
| Swin-UMamba | 63 | 59 | 30 | 74.56 | 81.15 |
| DPM-UNet | 31 | 38 | 24 | 78.15 | 84.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Liu, X.; Lei, H.; Hui, B. DPM-UNet: A Mamba-Based Network with Dynamic Perception Feature Enhancement for Medical Image Segmentation. Sensors 2025, 25, 7053. https://doi.org/10.3390/s25227053
Xu S, Liu X, Lei H, Hui B. DPM-UNet: A Mamba-Based Network with Dynamic Perception Feature Enhancement for Medical Image Segmentation. Sensors. 2025; 25(22):7053. https://doi.org/10.3390/s25227053
Chicago/Turabian StyleXu, Shangyu, Xiaohang Liu, Hongsheng Lei, and Bin Hui. 2025. "DPM-UNet: A Mamba-Based Network with Dynamic Perception Feature Enhancement for Medical Image Segmentation" Sensors 25, no. 22: 7053. https://doi.org/10.3390/s25227053
APA StyleXu, S., Liu, X., Lei, H., & Hui, B. (2025). DPM-UNet: A Mamba-Based Network with Dynamic Perception Feature Enhancement for Medical Image Segmentation. Sensors, 25(22), 7053. https://doi.org/10.3390/s25227053






