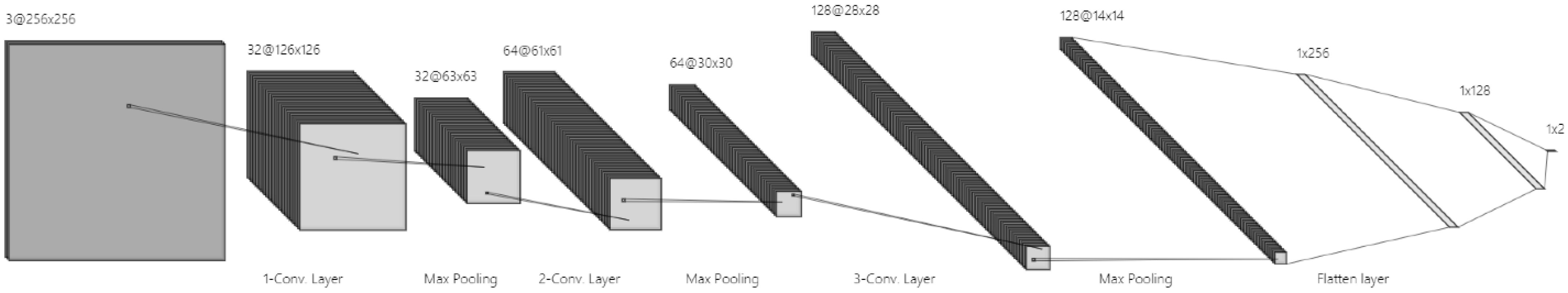
3.1. Fuzzy Enhancement Method
Our research relies heavily on medical picture databases, which allow neural networks to be trained and evaluated for lung disease detection. To guarantee the diversity and representativeness required for efficient model performance, the dataset must undergo a number of crucial preprocessing processes as shown In
Figure 1. Advanced sensors are essential for obtaining high-quality data in medical imaging, which is the foundation of deep-learning models. Primary imaging modalities used in this study include CT scans and chest X-rays, which rely on advanced sensor technology to provide high-resolution pictures. The precision and caliber of these sensors have a big impact on how well computer-aided diagnostic systems work.
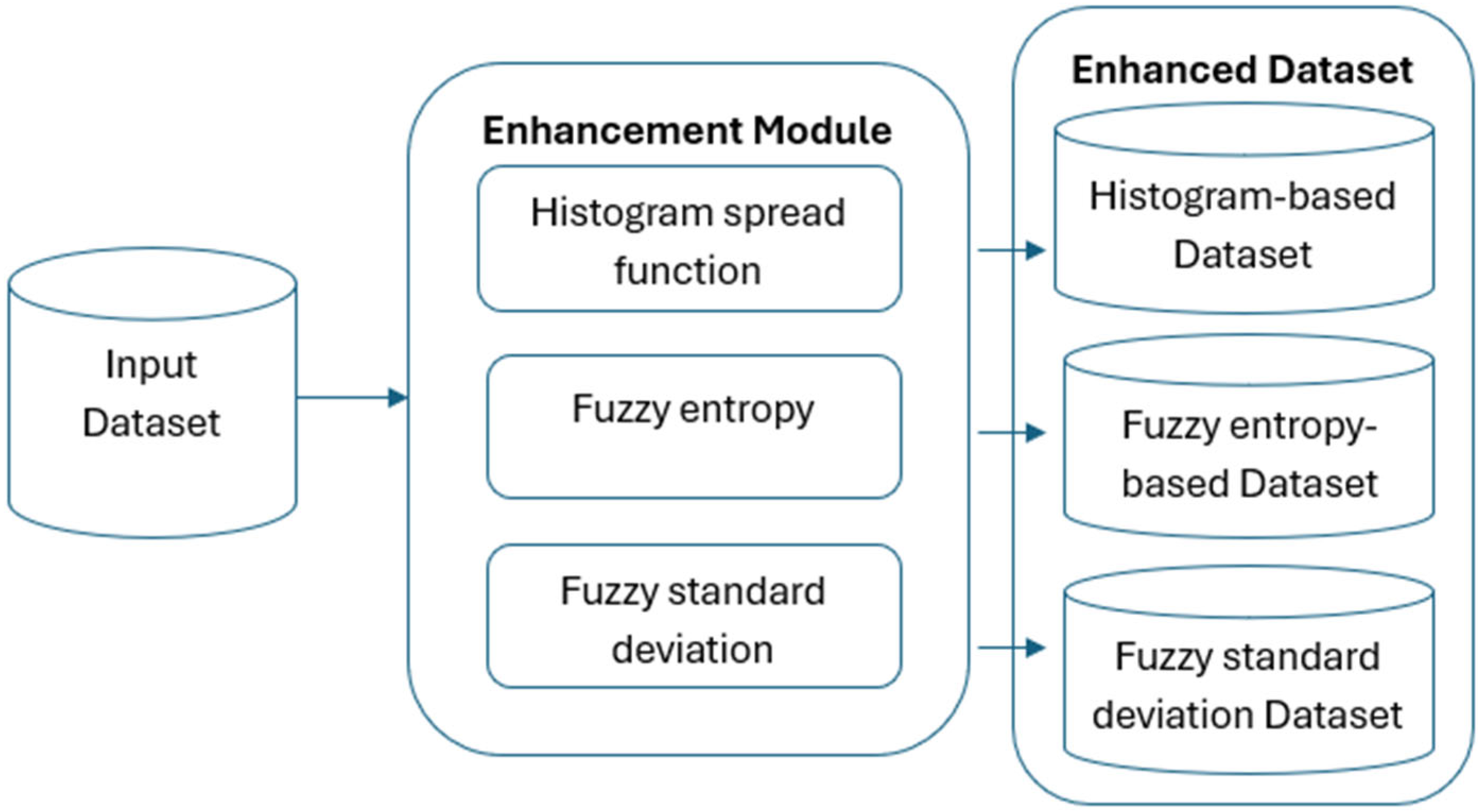
Input Dataset: This dataset acts as the initial input for the enhancement process.
Image Enhancement: Enhancing every input image using a fuzzy inference system (FIS) is the second crucial step. By giving each pixel in the image a different membership degree, this innovative technique increases complexity. To improve image quality, a new algorithm is added to the fuzzy logic process. Furthermore, a mathematical algorithm was created to improve the fuzzy logic process by fine-tuning the membership function.
Generating Transformed Datasets: The same FIS-enhanced images are used in three new datasets that are produced by using three different kinds of local contrast characteristics. The size of all the photos triples after the fuzzy enhancement process.
The high-fidelity data produced by contemporary sensors is directly employed by the picture improvement techniques used in this study, such as fuzzy entropy and standard deviation-based approaches. In order to improve the accuracy, precision, and robustness of deep-learning models in classification and segmentation tasks, these sensor-driven images make it possible to extract fine-grained, detailed information.
The improved datasets serve as a basis for convolutional neural network (CNN) training and evaluation, with the aim of determining how model performance is affected by images altered by the Fuzzy Inference System (FIS). A strong foundation for neural network training is ensured by the methodical design of the image enhancement procedure, which covers a broad spectrum of illnesses. Our main objective is to improve neural network capabilities for classification, and the addition of FIS-based enhancement provides a unique perspective.
In order to improve image quality inside the fuzzy logic framework, a fuzzy image enhancement approach is presented. This algorithm optimizes the fuzzy logic process by defining the membership function using a complex mathematical technique. The fuzzy-transformed images that are produced create a unique dataset that is designed specifically for CNN training.
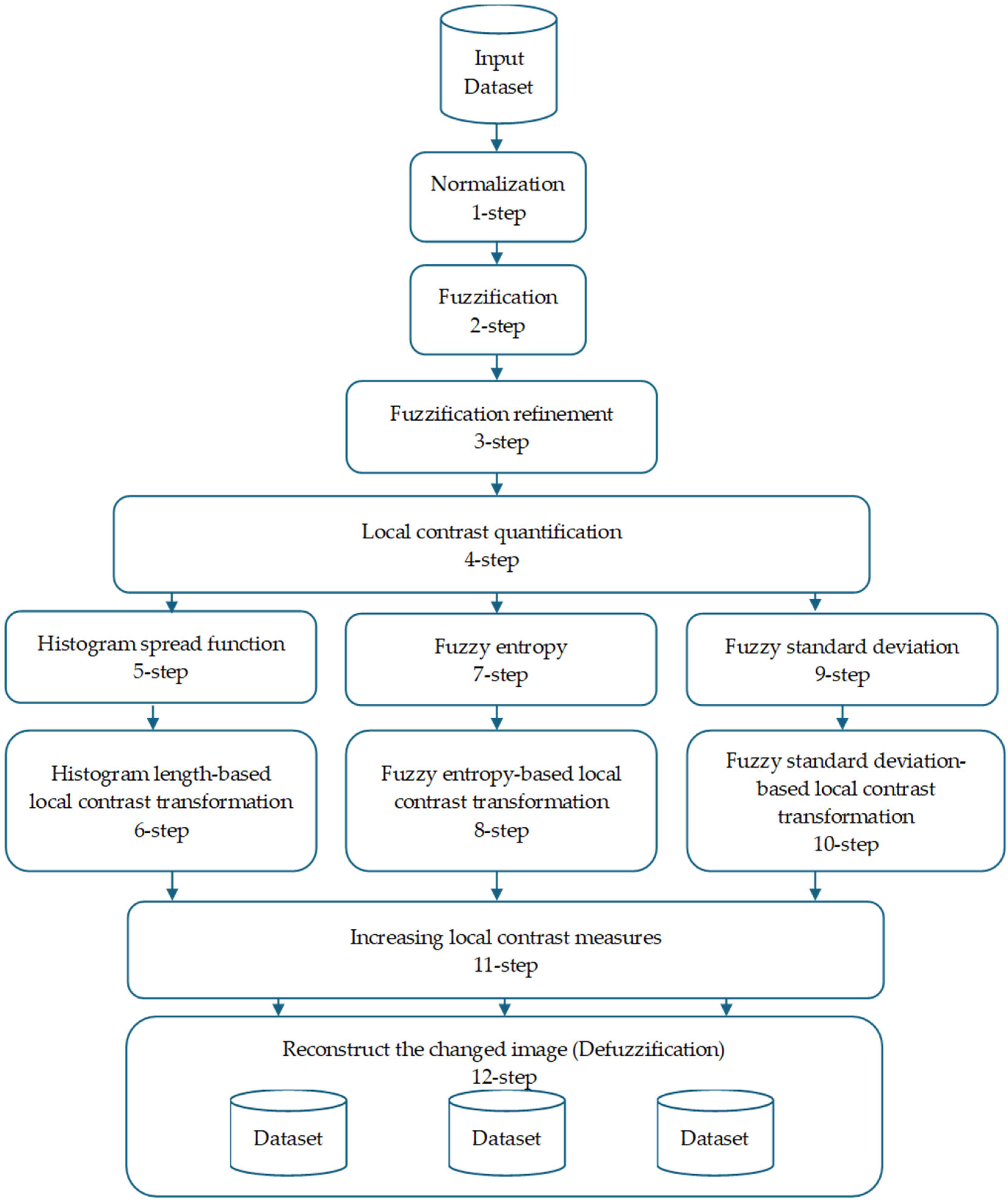
Figure 2 shows the image enhancement algorithm, which consists of 12 phases.
By giving each pixel in an image a different level of membership, the idea of fuzziness in image processing adds more complexity. This method takes into consideration ambiguity in the items’ depiction in the picture. Each pixel in the fuzzy framework model of image “F” is assigned a membership degree that indicates its association with particular groups or categories.
This approach provides a more adaptable and detailed representation for analyzing and interpreting images, recognizing and addressing the inherent ambiguity and imprecision often found in real-world images.
where
represents the membership degree of the pixel at coordinates (
x,
y) to a specific set, determined by the image’s properties.
Fuzzy images are particularly effective for image processing tasks involving ambiguity. Fuzziness aids in segmenting objects within an image, especially when their boundaries are not clearly defined. Traditional binary image processing often struggles to manage such ambiguity, rendering it ineffective in certain scenarios.
Step 1: Normalization
u(
x,
y)—fuzzy membership value of pixel (
x,
y) after applying the fuzzy membership function;
f(
x,
y)—grayscale intensity of pixel (
x,
y) in the input image (after enhancement, if applicable);
fmin,
fmax—minimum and maximum grayscale intensity values within the local window (for local operations) or across the whole image (for global operations, depending on context).
Step 3: Fuzzification Refinement
Step 4: Local Contrast Quantification
In image processing, measuring local contrast is crucial for assessing contrast variations across different regions of an image. Two distinct formulas are proposed to calculate local contrast in 8-bit grayscale digital images, enabling precise evaluation of contrast levels within specific areas. These methods allow for the quantification of contrast variations, supporting more informed and targeted image processing decisions:
Local Contrast Calculation:
Global Contrast Calculation:
where and are the maximum and minimum brightness values in the vicinity of pixels within a local neighborhood.
To fully grasp local contrast and its applications, it is necessary to analyze different categories of local neighborhoods based on their pixel luminance smoothness:
Homogeneous Neighborhood: A local area where pixel brightness values are similar or identical, indicating high uniformity. Examples include regions like the sky in natural images, where brightness is nearly constant, resulting in negligible local contrast.
Binary Neighborhood: A local area with pixels exhibiting extreme luminance values (e.g., black and white pixels), occupying opposite ends of the spectrum. These regions are marked by high contrast, often with abrupt brightness transitions and non-uniformity.
Varied Brightness Neighborhood: A local area containing pixels with diverse luminance values but smooth transitions, lacking sharp boundaries. Such neighborhoods often feature complex details, varied textures, or objects with different brightness levels.
Understanding the composition and characteristics of local neighborhoods is vital for calculating local contrast, as different neighborhood types may require distinct contrast estimation techniques or processing settings to achieve the desired results. Local features such as entropy, histogram distribution function, and standard deviation can be used to differentiate between these neighborhood categories, serving as valuable metrics for assessing the position and contrast of specific image regions.
Step 5: Histogram Spread Function
This step employs the Cumulative Distribution Function (CDF) to measure the proportion of pixels in an image with brightness values at or below a specified threshold:
where
is the local histogram length value computed for the fuzzy-enhanced image,
fmin and
fmax are the minimum and maximum brightness values in a sliding neighborhood W centered at coordinates (
x,
y), and
hmax is the maximum histogram value in W. In homogeneous regions, this feature is minimal, while in binary regions, it reaches its maximum. The CDF exhibits a near-linear pattern in homogeneous regions due to uniform brightness, shows distinct steps in binary neighborhoods with dominant brightness levels, and demonstrates a gradual progression in neighborhoods with varying brightness values and smooth transitions.
Step 6: Histogram Length-Based Local Contrast Transformation
This step uses histogram length functions to determine the degree of local contrast transformation:
—enhancement control parameter.
, —minimum and maximum bounds for α.
—local histogram length (FIS-based) at the current pixel.
—reference histogram length value (constant).
s > 0—scaling exponent controlling enhancement strength.
π—mathematical constant (~3.1416). See
Figure 3.
Step 7: Entropy
Entropy serves as a measure of variation or uncertainty in pixel values within a neighborhood, with higher entropy indicating a greater range of pixel intensities. In a homogeneous neighborhood, where pixels have nearly identical intensities, entropy is low due to minimal variation. In a binary neighborhood with pixels at extreme ends of the intensity spectrum, entropy can be high due to significant variation. Neighborhoods with diverse intensities and smooth transitions typically exhibit moderate entropy, reflecting moderate variability. Fuzzy entropy in a sliding local region of size n × n is defined as follows:
Summation Range: The sum is over all pixel coordinates (i,j) in the neighborhood W, where W is a 3 × 3 window centered at (x,y), i.e., (i,j) ranges over (x − 1,y − 1) to (x + 1,y + 1).
is calculated as follows:
Here, represents the histogram count of brightness values (x,y) in the neighborhood W, indicating the frequency of elements matching the brightness at coordinates (x,y). Equation (5) shows that homogeneous regions have the highest fuzzy entropy, while regions with brightness values at opposite extremes have the lowest.
Step 8: Fuzzy Entropy-Based Local Contrast Transformation
Fuzzy entropy is used to determine the extent of local contrast transformation:
where
s > 0.
Step 9: Fuzzy Standard Deviation
The standard deviation (σ\sigma σ) measures the dispersion of brightness values around their mean in a neighborhood. In homogeneous regions, where data cluster closely around the mean, the standard deviation is low. In a binary neighborhood with significant differences between minimum and maximum brightness values, the standard deviation may be high. Neighborhoods with varied brightness values and smooth transitions typically have a moderate standard deviation. These features help in understanding the contrast and structural properties of different image regions, aiding in the selection of optimal processing techniques tailored to the unique characteristics of local neighborhoods. The standard deviation of brightness values in a sliding neighborhood W is computed as follows:
where
is the fuzzy standard deviation computed in the local neighborhood of (
x,
y), and
is the fuzzy arithmetic mean of brightness values in W as follows:
Here, N and M are the dimensions of the image. In homogeneous neighborhoods, Equation (8) yields zero, increasing with greater heterogeneity.
Step 10: Standard Deviation-Based Local Contrast Transformation
The fuzzy standard deviation of brightness data is used to determine the degree of local contrast change:
—adaptive enhancement exponent at pixel (x,y).
, —minimum and maximum allowable enhancement exponents.
σ(x,y)—fuzzy standard deviation of intensity in a local k × k neighborhood centered at (x,y).
s > 0—scaling exponent controlling the sensitivity to local contrast.
Step 11: Increasing Local Contrast Measures
A nonlinear transformation is applied to enhance local contrast according to a specific rule:
where R = 1 is the maximum feasible local contrast, C(
x,
y) is the local contrast of the original image at coordinates (
x,
y), and C*(
x,
y) is the enhanced local contrast. C
min and C
max represent the minimum and maximum local contrast values in the original image, respectively. C* approximates the mathematical expectation of local contrast values as the arithmetic mean, with A
0 and B
0 as constant bias coefficients, and
α as the exponent.
Step 12: Defuzzification
The modified image regions are reconstructed using the enhanced local contrast values. Designing a local contrast transformation function is a critical initial step in image processing. Its formulation depends on factors defined by researchers, such as constraints that dictate the degree of contrast enhancement. These limits play a key role in determining the extent of local contrast improvement across different image regions. The selection of the contrast transform function’s parameters relies on the researcher’s expertise and understanding of local statistical features.
3.2. Generation of Transformed Datasets Using Local Contrast Characteristics
The image enhancement pipeline begins with a common FIS applied to the input images, which involves Steps 1–4: Normalization, Fuzzification, Fuzzification Refinement, and Local Contrast Quantification.
To demonstrate Steps 1–3, consider a 2 × 2 grayscale image with 8-bit pixel intensities (0–255), representing a small region of a medical image.
Table 1 shows the pixel values and their transformations through Normalization, Fuzzification, and Fuzzification Refinement.
Step 1: Normalization
Pixel intensities are scaled from [0, 255] to [0, 1] using the following:
For a 2 × 2 image with intensities [100, 150, 200, 50], the normalized values are [0.392, 0.588, 0.784, 0.196].
Step 2: Fuzzification
The normalized intensities are mapped to membership values using the following sigmoidal function:
where (a = 10), (b = 0.5). For example, for
= 0.392, the following applies:
This assigns a membership degree indicating the pixel’s association with a high-intensity set. Similar calculations are performed for other pixels.
Step 3: Fuzzification Refinement
Fuzzification Refinement adjusts membership values to optimize for medical image ambiguities. We apply a mathematical algorithm to fine-tune the membership function by introducing a contrast-enhancing factor
, set to 1.2 for CT images to emphasize edge transitions:
For , the refined membership is . This reduces the membership value slightly, emphasizing contrast in ambiguous regions.
The example shows how normalization standardizes intensities, fuzzification assigns membership degrees to model ambiguity, and refinement enhances contrast for downstream tasks. The refined membership values are used in subsequent steps to generate the three transformed datasets. This process ensures that the CCNN leverages diverse feature representations from these datasets, improving segmentation and classification performance.
This FIS-enhanced image serves as the base for generating three transformed datasets, each produced by applying one of three distinct local contrast characteristics: histogram spread function, fuzzy entropy, and fuzzy standard deviation. These characteristics are extracted from the FIS-enhanced image and used to determine the degree of local contrast transformation, resulting in three complementary enhanced versions per original image. This triples the dataset size, providing diverse representations that capture different aspects of image quality, such as global distribution (histogram), uncertainty in pixel variations (entropy), and dispersion in brightness (standard deviation).
The extraction and application process for each characteristic is as follows:
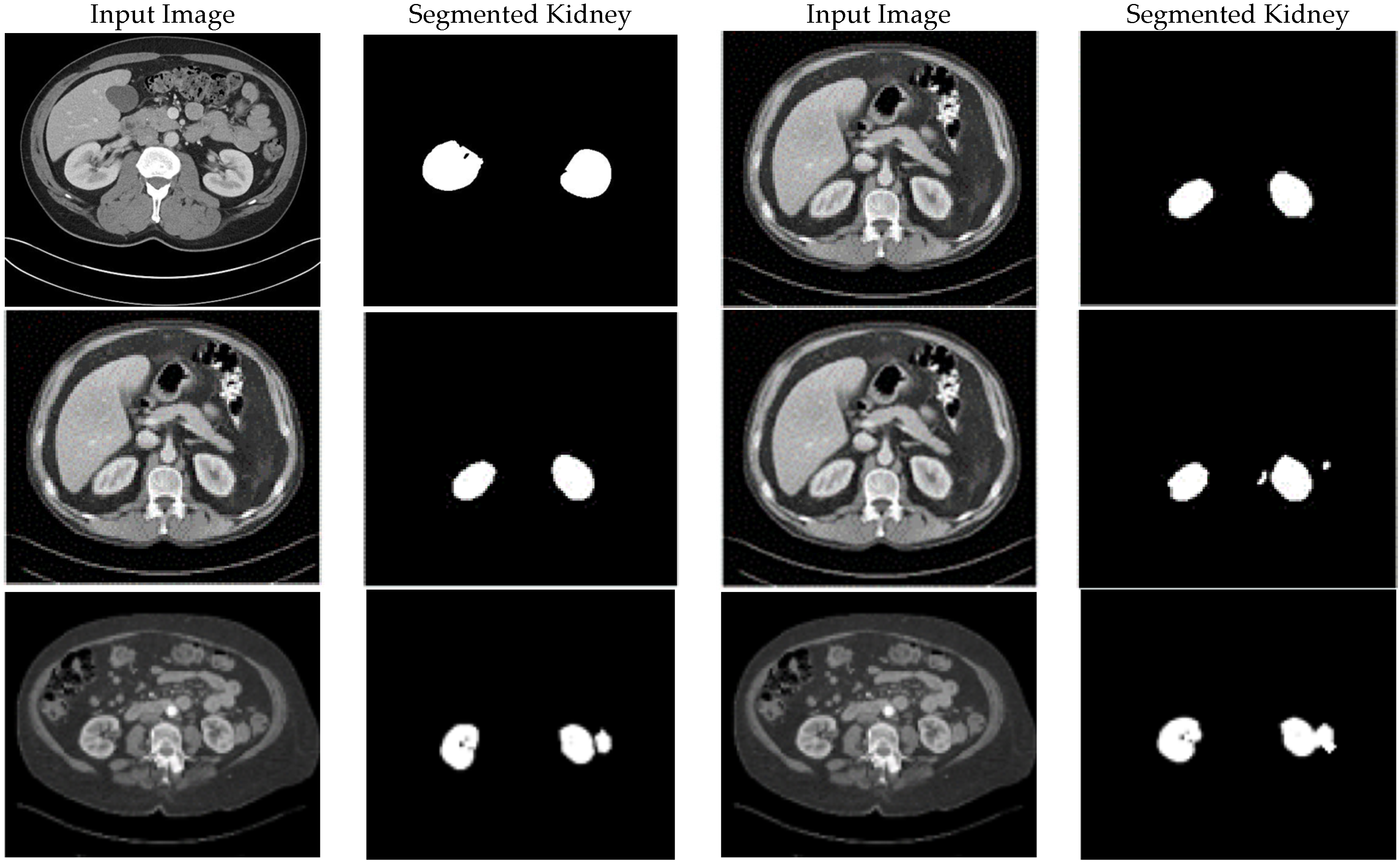
Histogram Spread Function (Steps 5–6): This characteristic quantifies the distribution of pixel brightness values using the Cumulative Distribution Function (CDF) in Equation (4), computed over sliding neighborhoods W centered at each pixel (x,y). It measures the proportion of pixels at or below a brightness threshold, with fmin and fmax as the minimum and maximum brightness in W, and hmax as the maximum histogram bin value. In homogeneous regions, the CDF is minimal and near-linear; in binary regions, it is maximal with steps; and in varied regions, it shows gradual progression. This is extracted by computing the histogram length function in Equation (4), which determines the transformation degree, where s > 0 is an empirically tuned exponent (typically s = 1.5 for medical images to balance enhancement). The transformation is applied nonlinearly in Step 11 using Equation (10) to increase local contrast C(x,y), followed by defuzzification in Step 12 to reconstruct the image. The resulting dataset emphasizes global contrast adjustments, making it suitable for enhancing overall visibility in low-contrast areas like kidney tumors in CT.
Fuzzy Entropy (Steps 7–8): This characteristic measures the uncertainty or variation in pixel membership degrees within neighborhoods, using Equation (5) for fuzzy entropy, where is the fuzzified membership from the FIS-enhanced image, normalized by the histogram count h(f(x,y)) in Equation (6). Extraction involves sliding window computation, yielding high entropy in homogeneous regions (low variation) and low entropy in binary extremes (high variation). The transformation degree (Equation (7), s > 0, typically s = 2 for noise-sensitive MRI) is used in Step 11 to enhance contrast selectively in uncertain areas. Defuzzification produces a dataset focused on reducing ambiguity and noise, highlighting subtle intensity variations.
Fuzzy Standard Deviation (Steps 9–10): This quantifies the dispersion of fuzzified brightness values around the mean, using Equations (8) and (9). Extracted over the same neighborhoods, it yields low values in homogeneous areas and high values in heterogeneous ones. The transformation using Equation (10), s > 0, typically s = 1.2 for X-ray opacity detection guides Step 11’s nonlinear enhancement, emphasizing edge preservation. Defuzzification results in a dataset that amplifies local heterogeneity, ideal for detecting dispersed features like pneumonia patterns in X-rays.
These three datasets differ as follows:
Image Content and Features: All start from the same FIS-enhanced base; hence, core content remains identical, but features are enhanced differently. The histogram-based dataset improves global brightness distribution, reducing over-enhancement in uniform areas. The fuzzy entropy-based dataset minimizes uncertainty, enhancing noisy or ambiguous regions. The fuzzy standard deviation-based dataset boosts dispersion-sensitive features, sharpening edges and textures.
Quantitative Differences: As shown in
Section 4.2, they vary in perceived quality. Feature-wise, histogram emphasizes uniform histograms, entropy high-variation areas, and std dev local deviations.
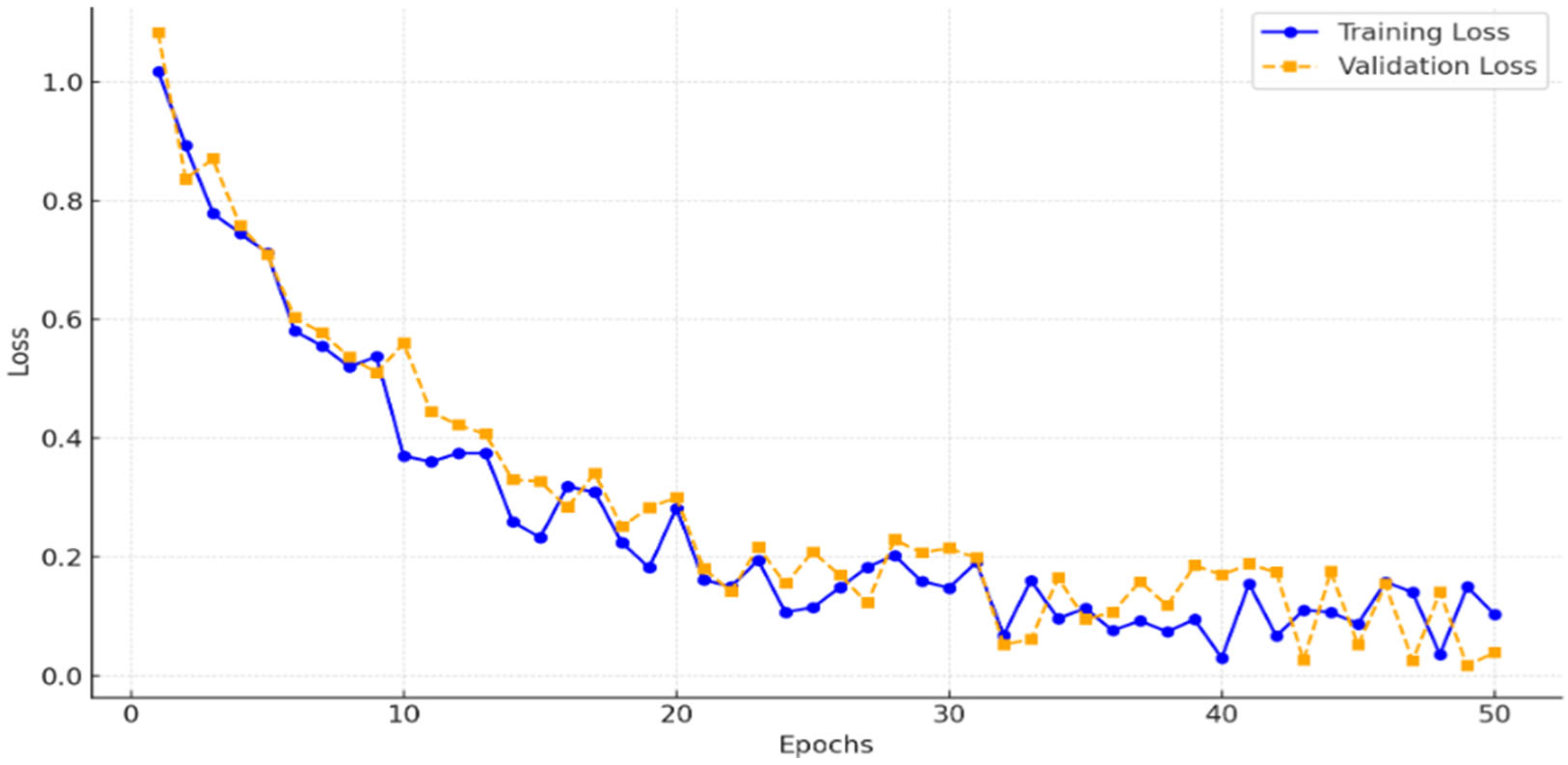
Utilization in CCNN: The CCNN processes these datasets in parallel streams, extracting complementary features (e.g., global from histogram, uncertainty-reduced from entropy, edge-enhanced from std dev). Feature maps are concatenated (64 × 64 × 768) before deeper layers, enabling robust learning by leveraging diversity, as evidenced by improved Dice coefficients.