FED-UNet++: An Improved Nested UNet for Hippocampus Segmentation in Alzheimer’s Disease Diagnosis
Abstract
1. Introduction
2. Related Works
2.1. Evolution of Deep Segmentation Networks
2.2. The Application of Deep Learning Methods in Medical Image Segmentation
2.3. CNN-Based Methods for Hippocampus Segmentation
3. Materials and Methods
3.1. Architecture of the Proposed Method
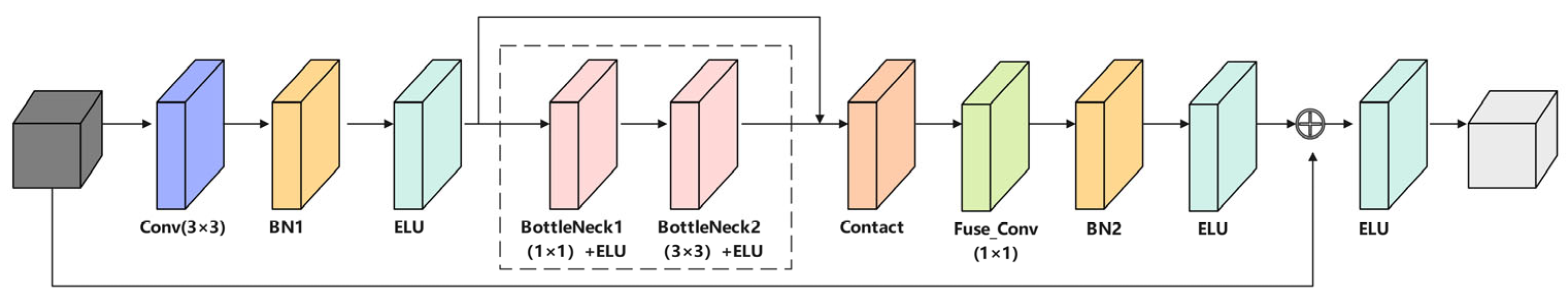
3.2. Residual Feature Reconstruction Block (FRBlock)
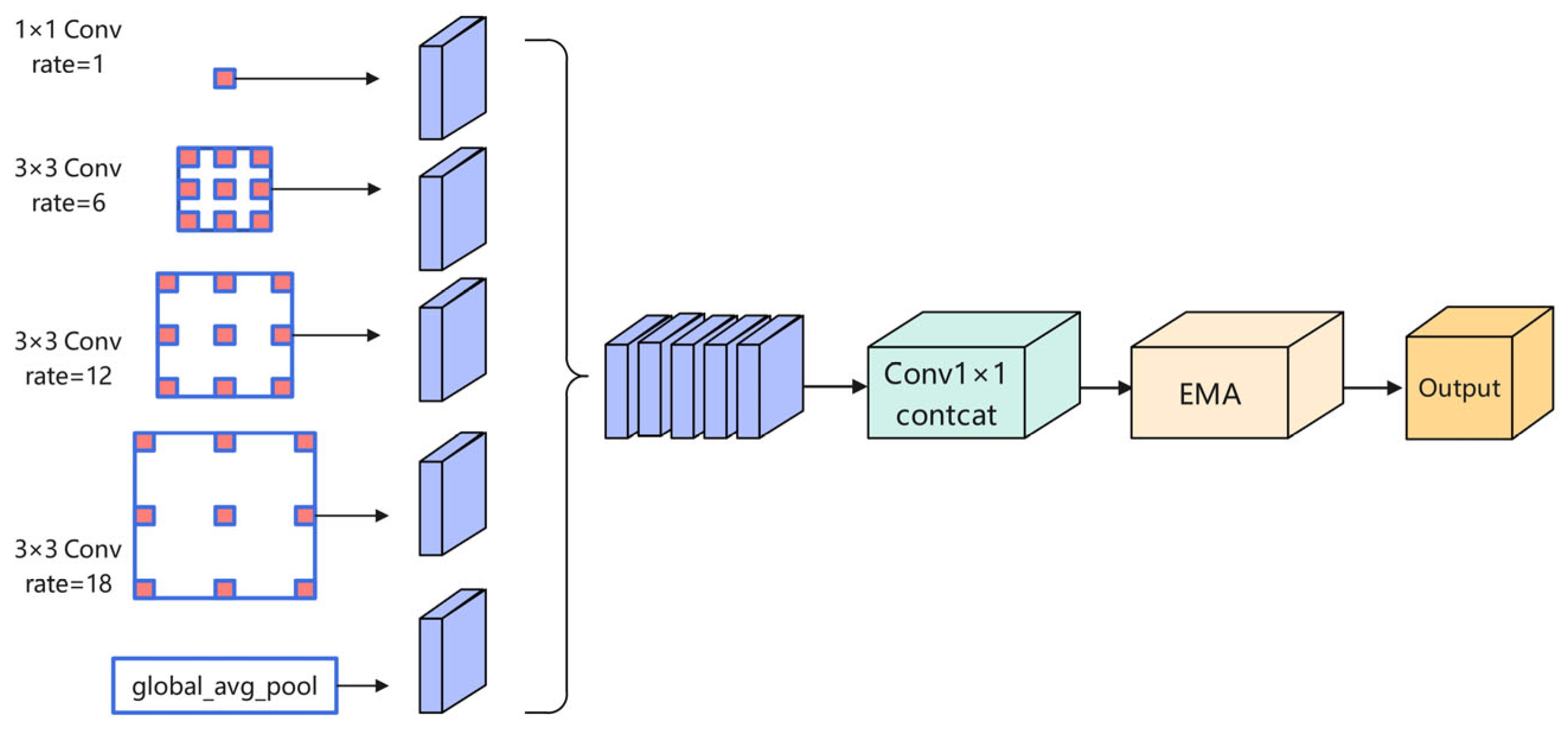
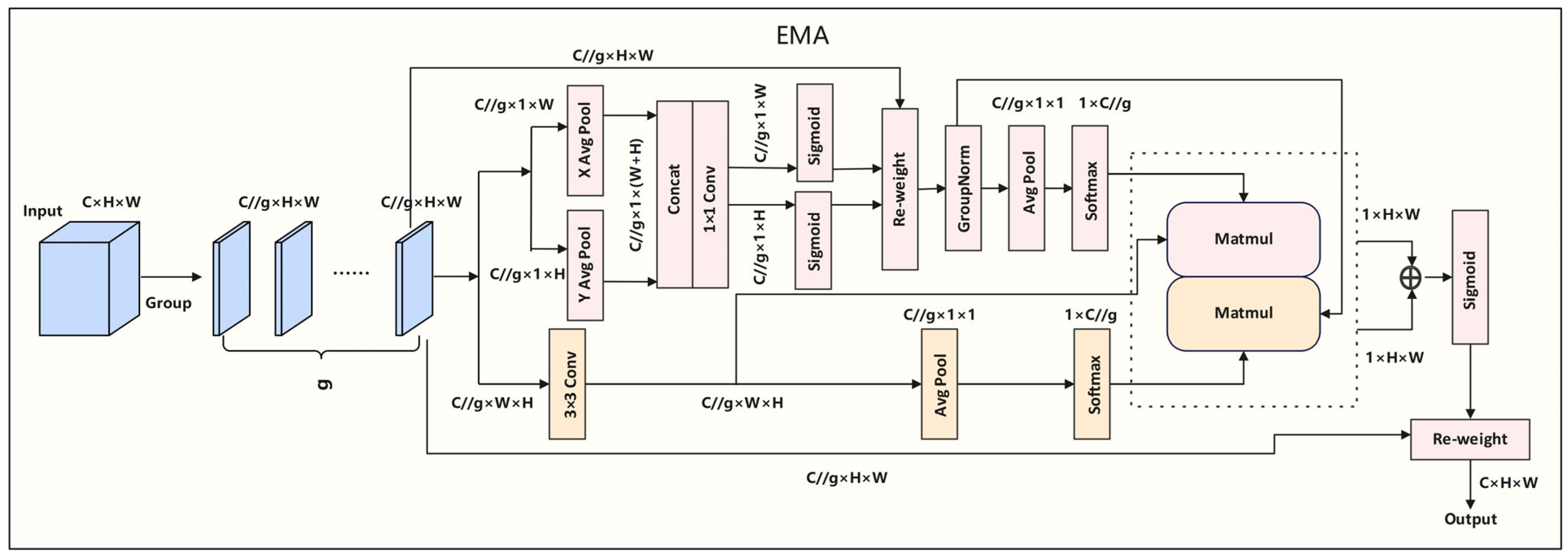
3.3. Efficient Attention Pyramid Module (EAP)
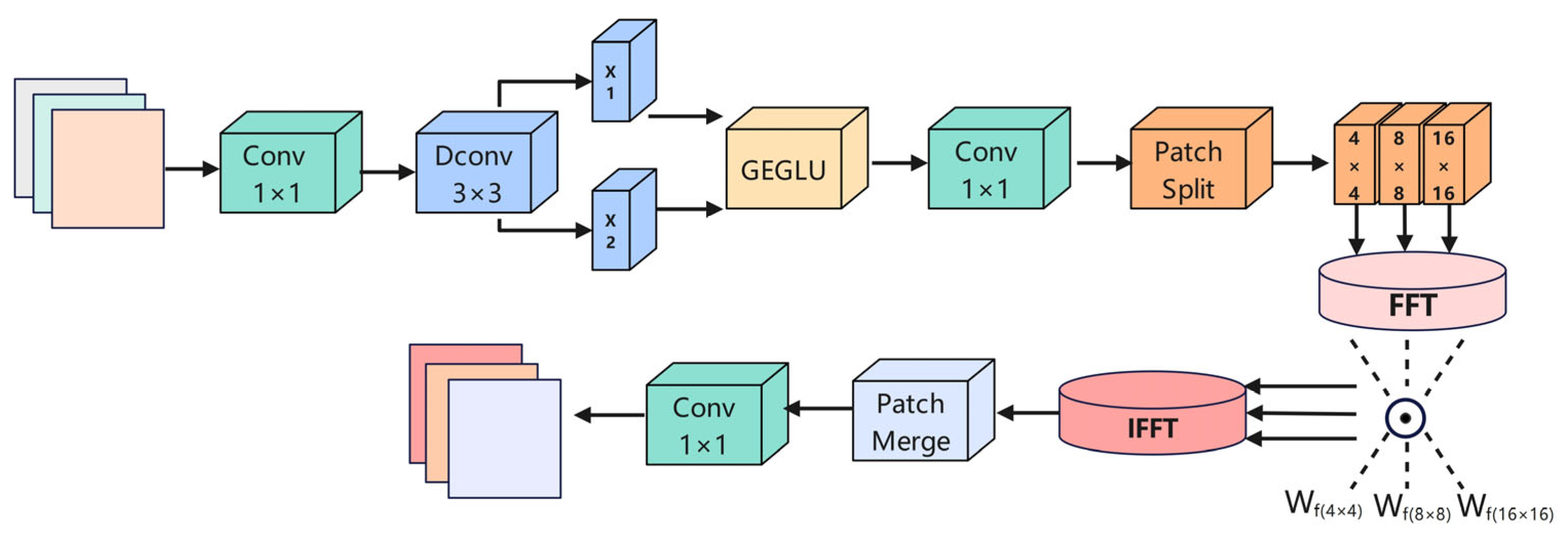
3.4. Dynamic Frequency Context Network (DFCN)
4. Experiments
4.1. Datasets
4.2. Experimental Settings and Parameters
4.3. Evaluation MetricsS
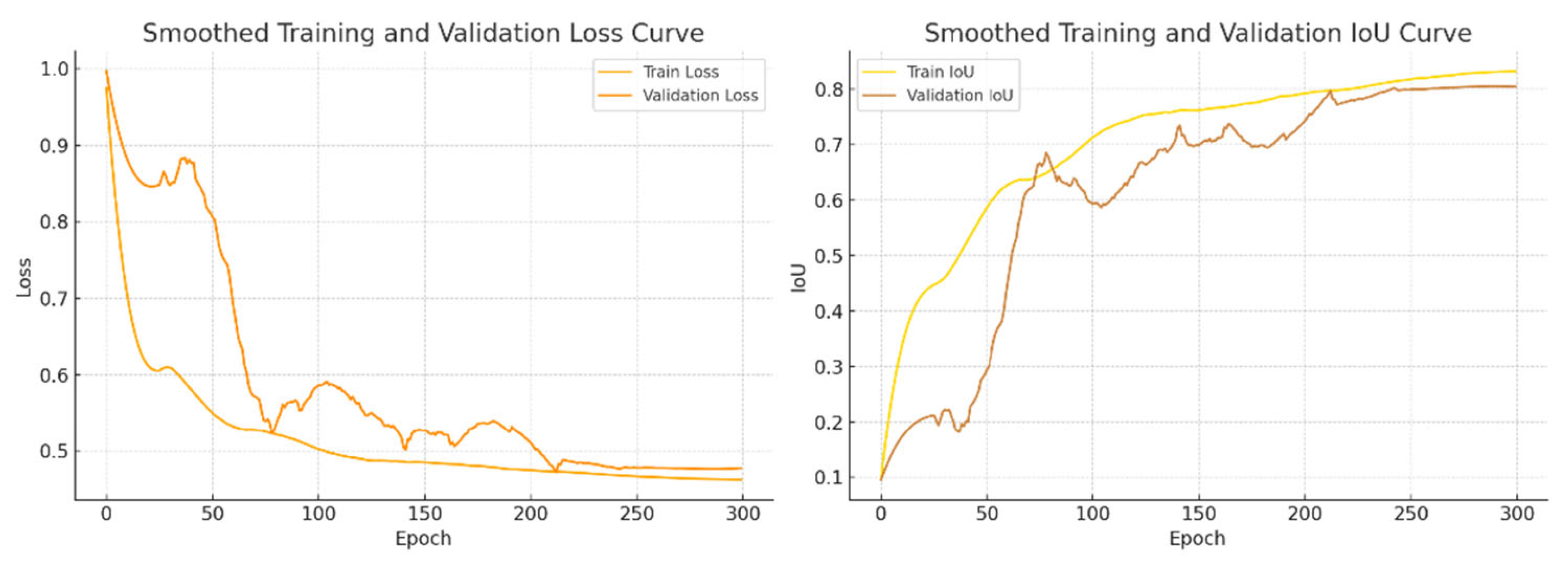
4.4. Results
4.4.1. Results and Analysis
4.4.2. Ablation Study of Key Modules
4.4.3. Robustness to Noise
4.4.4. Generalization Experiments
4.4.5. Model Efficiency Analysis
5. Conclusions
- Transitioning from 2D slices to 3D volumetric modeling could enable better representation of inter-slice continuity through 3D convolutions.
- Clinical validation using real-world and multi-center MRI datasets is needed to evaluate the model’s robustness under diverse imaging conditions.
- Diagnostic integration may allow for automated hippocampal volumetry and disease stage prediction, further extending the framework toward intelligent clinical decision support systems.
- Deployment optimization through pruning, quantization, and lightweight architectural design can facilitate real-time inference on edge devices and within practical medical environments.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, H.; Gamboa, H.; Schultz, T. Sensor-Based Human Activity and Behavior Research: Where Advanced Sensing and Recognition Technologies Meet. Sensors 2023, 23, 125. [Google Scholar] [CrossRef]
- Liu, H.; Gamboa, H.; Schultz, T. Human Activity Recognition, Monitoring, and Analysis Facilitated by Novel and Widespread Applications of Sensors. Sensors 2024, 24, 5250. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2024 Alzheimer’s disease facts and figures. Alzheimers Dement. 2024, 20, 3708–3821. [Google Scholar] [CrossRef]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R., Jr.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef]
- Mueller, S.G.; Weiner, M.W.; Thal, L.J.; Petersen, R.C.; Jack, C.R.; Jagust, W.; Trojanowski, J.Q.; Toga, A.W.; Beckett, L. Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005, 1, 55–66. [Google Scholar] [CrossRef]
- Cuingnet, R.; Gerardin, E.; Tessieras, J.; Auzias, G.; Lehéricy, S.; Habert, M.O.; Chupin, M.; Benali, H.; Colliot, O.; Alzheimer’s Disease Neuroimaging Initiative. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage 2011, 56, 766–781. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11045, pp. 3–11. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Computer Vision—ECCV 2018, Proceedings of the 15th European Conference, Munich, Germany, 8–14 September 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11211, pp. 3–19. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Ouyang, D.; Deng, Y.; Wu, Y.; Li, C.; Jin, L.; Sun, Y. Efficient Multi-Scale Attention Module with Cross-Spatial Learning. In Proceedings of the 2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–9 June 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Kong, L.; Dong, J.; Tang, J.; Yang, M.-H.; Pan, J. Efficient Visual State Space Model for Image Deblurring. arXiv 2025, arXiv:2405.14343. [Google Scholar] [CrossRef]
- Malekzadeh, S. MRI Hippocampus Segmentation. Kaggle. 2019. Available online: https://www.kaggle.com (accessed on 26 June 2025).
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A Large Annotated Medical Image Dataset for the Development and Evaluation of Segmentation Algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar] [CrossRef]
- Zhang, D.; Shen, D.; Alzheimer’s Disease Neuroimaging Initiative. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage 2012, 59, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, L.; Igel, C.; Pai, A.; Balas, I.; Anker, C.; Lillholm, M.; Nielsen, M.; Alzheimer’s Disease Neuroimaging Initiative; Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing. Differential diagnosis of mild cognitive impairment and Alzheimer’s disease using structural MRI cortical thickness, hippocampal shape, hippocampal texture, and volumetry. Neuroimage Clin. 2016, 13, 470–482. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), 2018, Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R. UNETR: Transformers for 3D medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), 2022, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like pure transformer for medical image segmentation. arXiv 2021, arXiv:2105.05537. [Google Scholar]
- Wang, J.; Li, X.; Ma, Z. Multi-Scale Three-Path Network (MSTP-Net): A New Architecture for Retinal Vessel Segmentation. Measurement 2025, 250, 117100. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Zhou, C.; Peng, H.; Zheng, Z.; Chen, J.; Ding, W. A Review of Cancer Data Fusion Methods Based on Deep Learning. Inf. Fusion 2024, 108, 102361. [Google Scholar] [CrossRef]
- Huang, K.W.; Yang, Y.R.; Huang, Z.H.; Liu, Y.Y.; Lee, S.H. Retinal Vascular Image Segmentation Using Improved UNet Based on Residual Module. Bioengineering 2023, 10, 722. [Google Scholar] [CrossRef]
- Xing, R. FreqU-FNet: Frequency-Aware U-Net for Imbalanced Medical Image Segmentation. arXiv 2025, arXiv:2505.17544. [Google Scholar] [CrossRef]
- Hayat, M.; Aramvith, S.; Bhattacharjee, S.; Ahmad, N. Attention GhostUNet++: Enhanced Segmentation of Adipose Tissue and Liver in CT Images. arXiv 2025, arXiv:2504.11491. [Google Scholar] [CrossRef]
- Hayat, M.; Gupta, M.; Suanpang, P.; Nanthaamornphong, A. Super-Resolution Methods for Endoscopic Imaging: A Review. In Proceedings of the 2024 12th International Conference on Internet of Everything, Microwave, Embedded, Communication and Networks (IEMECON), Jaipur, India, 22–24 February 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Liang, T.Y.; Zhao, H.; Wang, X.; Chen, Q.; Lin, Y.; Zhang, D. Integrating Vision Transformer with UNet++ for Hippocampus Segmentation in Alzheimer’s Disease. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15–19 July 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Helaly, H.A.; Badawy, M.; Haikal, A.Y. Toward Deep MRI Segmentation for Alzheimer’s Disease Detection. Neural Comput. Appl. 2022, 34, 1047–1063. [Google Scholar] [CrossRef]
- Sanjay, V.; Swarnalatha, P. Dominant Hippocampus Segmentation with Brain Atrophy Analysis-Based AD Subtype Classification Using KLW-RU-Net and T1FL. Alex. Eng. J. 2024, 104, 451–463. [Google Scholar] [CrossRef]
- Alzheimer’s Disease Neuroimaging Initiative. ADNI Data. 2023. Available online: https://adni.loni.usc.edu/ (accessed on 26 June 2025).

| BCE:Dice | IoU | Dice | Recall | Precision | HD95 |
|---|---|---|---|---|---|
| 0.7:0.3 | 74.83% | 84.31% ± 0.23% | 85.11% | 85.67% | 2.12 |
| 0.5:0.5 | 74.95% | 84.43% ± 0.21% | 85.31% | 85.81% | 2.06 |
| 0.3:0.7 | 74.74% | 84.25% ± 0.24% | 85.08% | 85.59% | 2.14 |
| Datasets | Method | IoU | Dice | Recall | Precision | Accuracy | HD95 |
|---|---|---|---|---|---|---|---|
| Kaggle dataset | U-Net | 66.60% | 79.20% | 77.01% | 82.33% | 99.85% | 6.1837 |
| U-Net++ | 70.51% | 81.62% ± 0.38% | 80.96% | 83.71% | 99.87% | 4.7924 | |
| SwinUNet | 60.75% | 73.56% | 73.03% | 75.78% | 99.82% | 5.8372 | |
| PSPNet | 51.70% | 68.15% | 72.40% | 64.39% | 99.75% | 7.2048 | |
| DeepLabv3+ | 69.89% | 82.29% | 89.70% | 75.99% | 99.86% | 3.5913 | |
| Ours | 74.95% | 84.43% ± 0.21% | 85.31% | 85.81% | 99.89% | 2.0611 |
| Datasets | Method | IoU | Dice | Recall | Precision | Accuracy | HD95 |
|---|---|---|---|---|---|---|---|
| Kaggle dataset | Baseline (U-Net++) | 70.51% | 81.62% ± 0.38% | 80.80% | 84.58% | 99.87% | 4.7924 |
| FR + U-Net++ | 72.10% | 82.74% ± 0.36% | 82.10% | 85.21% | 99.88% | 3.4372 | |
| EA + U-Net++ | 72.29% | 82.91% ± 0.33% | 84.37% | 83.40% | 99.88% | 3.0167 | |
| DF + U-Net++ | 72.45% | 83.02% ± 0.31% | 83.51% | 84.47% | 99.88% | 2.4452 | |
| FR + EA + U-Net++ | 73.18% | 83.88% ± 0.27% | 83.26% | 85.37% | 99.88% | 2.5823 | |
| FR + DF + U-Net++ | 73.08% | 84.01% ± 0.25% | 84.17% | 84.57% | 99.88% | 2.3714 | |
| EA + DF + U-Net++ | 73.37% | 84.17% ± 0.23% | 83.32% | 85.79% | 99.89% | 2.2259 | |
| FED-UNet++ | 74.95% | 84.43% ± 0.21% | 85.31% | 85.81% | 99.89% | 2.0611 |
| Original |  |  |  | 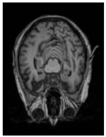 | 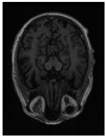 |
| GT |  |  |  |  |  |
| Baseline |  |  |  |  |  |
| FR+ |  |  |  |  |  |
| EA+ |  |  |  |  |  |
| DF+ |  |  |  |  |  |
| FR + EA+ |  |  |  |  |  |
| FR + DF+ |  |  |  |  |  |
| EA + DF+ |  |  |  |  |  |
| FED-UNet++ (FR + EA + DF+) |  |  |  |  |  |
| Original | GT | Representative Attention Activation Map from the EAP Module | Representative Frequency Response Visualization from the DFCN Module |
|---|---|---|---|
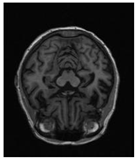 |  | 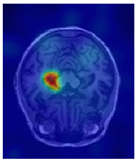 | 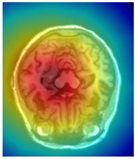 |
| Original | GT | Predict |
|---|---|---|
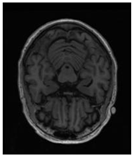 |  |  |
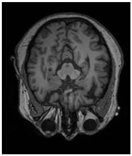 |  |  |
| Model: FED-UNet++ | Original | GT | Segmentation Result |
|---|---|---|---|
| Before noise addition | 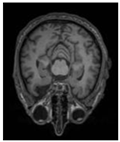 |  |  |
| After adding noise |  |  |  |
| Model: FED-UNet++ | IoU | Dice | Recall | Precision | HD95 |
|---|---|---|---|---|---|
| Before noise addition | 74.95% | 84.43% ± 0.21% | 85.31% | 85.81% | 2.0611 |
| After adding noise | 72.63% | 82.73 ± 0.33% | 83.25% | 82.57% | 2.8088 |
| Datasets | Method | IoU | Dice | Recall | Precision | Accuracy | HD95 |
|---|---|---|---|---|---|---|---|
| Task004_ Hippocamps | Baseline | 79.23% | 88.40% ± 0.41% | 87.81% | 88.74% | 98.40% | 4.7285 |
| UNet | 79.04% | 88.02% | 86.93% | 89.42% | 98.47% | 4.8192 | |
| PSPNet | 76.98% | 86.53% | 87.04% | 86.62% | 98.27% | 5.4796 | |
| DeepLabv3+ | 78.44% | 87.69% | 87.80% | 87.87% | 98.33% | 4.8951 | |
| SwinUNet | 77.65% | 87.51% | 86.02% | 88.55% | 98.26% | 5.0143 | |
| FR+ | 80.64% | 88.89% ± 0.33% | 88.67% | 89.74% | 98.51% | 3.8629 | |
| FR+EA+ | 81.78% | 89.55% ± 0.30% | 89.23% | 90.67% | 98.61% | 3.2074 | |
| Ours | 82.51% | 90.12% ± 0.27% | 89.86% | 90.94% | 98.65% | 2.8826 |
| Original |  |  | 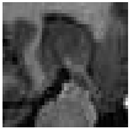 |  |
| GT |  |  |  |  |
| Baseline |  |  |  |  |
| UNet |  |  |  |  |
| PSPNet |  |  |  |  |
| DeepLabv3+ |  |  |  |  |
| SwinUNet |  |  |  |  |
| FR+ |  |  |  |  |
| FR+EA+ |  |  |  |  |
| Ours |  |  |  |  |
| Block | Kernel Config | Channels (In/Out) | Params (M) | FLOPs (G) | Memory (MB) |
|---|---|---|---|---|---|
| VGGBlock (U-Net++) | 3 × 3 + 3 × 3 | 64/64 | 0.0741 | 0.3041 | 6.08 |
| FRBlock | 3 × 3 + 1 × 1 + 3 × 3 + 1 × 1 | 64/64 | 0.0331 | 0.1358 | 8.50 |
| EAP | ASPP + EMA | 64/64 | 0.2500 | 0.9500 | 31.00 |
| DFCN | 1 × 1 + 3 × 3 + FFT + 1 × 1 | 64/64 | 0.0269 | 0.1101 | 23.50 |
| Model | Params (M) | FLOPs (G) | Inference Time (ms) | Peak VRAM | IoU (%) | Dice (%) |
|---|---|---|---|---|---|---|
| U-Net++ (baseline) | 9.16 | 34.90 | 5.33 | 157.38 MB | 70.51 | 81.62 ± 0.38 |
| FRBlock | 4.68 | 19.51 | 8.82 | 155.47 MB | 72.10 | 82.74 ± 0.36 |
| EAP | 18.12 | 37.21 | 6.24 | 192.02 MB | 72.29 | 82.91 ± 0.33 |
| DFCN | 5.90 | 20.35 | 9.55 | 168.10 MB | 72.45 | 83.02 ± 0.31 |
| FED-UNet++ | 18.82 | 42.75 | 11.91 | 198.37 MB | 74.95 | 84.43 ± 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Zhang, W.; Wang, S.; Yu, X.; Jing, B.; Sun, N.; Sun, T.; Wang, W. FED-UNet++: An Improved Nested UNet for Hippocampus Segmentation in Alzheimer’s Disease Diagnosis. Sensors 2025, 25, 5155. https://doi.org/10.3390/s25165155
Yang L, Zhang W, Wang S, Yu X, Jing B, Sun N, Sun T, Wang W. FED-UNet++: An Improved Nested UNet for Hippocampus Segmentation in Alzheimer’s Disease Diagnosis. Sensors. 2025; 25(16):5155. https://doi.org/10.3390/s25165155
Chicago/Turabian StyleYang, Liping, Wei Zhang, Shengyu Wang, Xiaoru Yu, Bin Jing, Nairui Sun, Tengchao Sun, and Wei Wang. 2025. "FED-UNet++: An Improved Nested UNet for Hippocampus Segmentation in Alzheimer’s Disease Diagnosis" Sensors 25, no. 16: 5155. https://doi.org/10.3390/s25165155
APA StyleYang, L., Zhang, W., Wang, S., Yu, X., Jing, B., Sun, N., Sun, T., & Wang, W. (2025). FED-UNet++: An Improved Nested UNet for Hippocampus Segmentation in Alzheimer’s Disease Diagnosis. Sensors, 25(16), 5155. https://doi.org/10.3390/s25165155








