Capillary Flow Profile Analysis on Paper-Based Microfluidic Chips for Classifying Astringency Intensity
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Paper-Based Microfluidic Chips and Equipment for Flow Velocity Profile Measurement
2.2.1. Paper-Based Microfluidic Chips
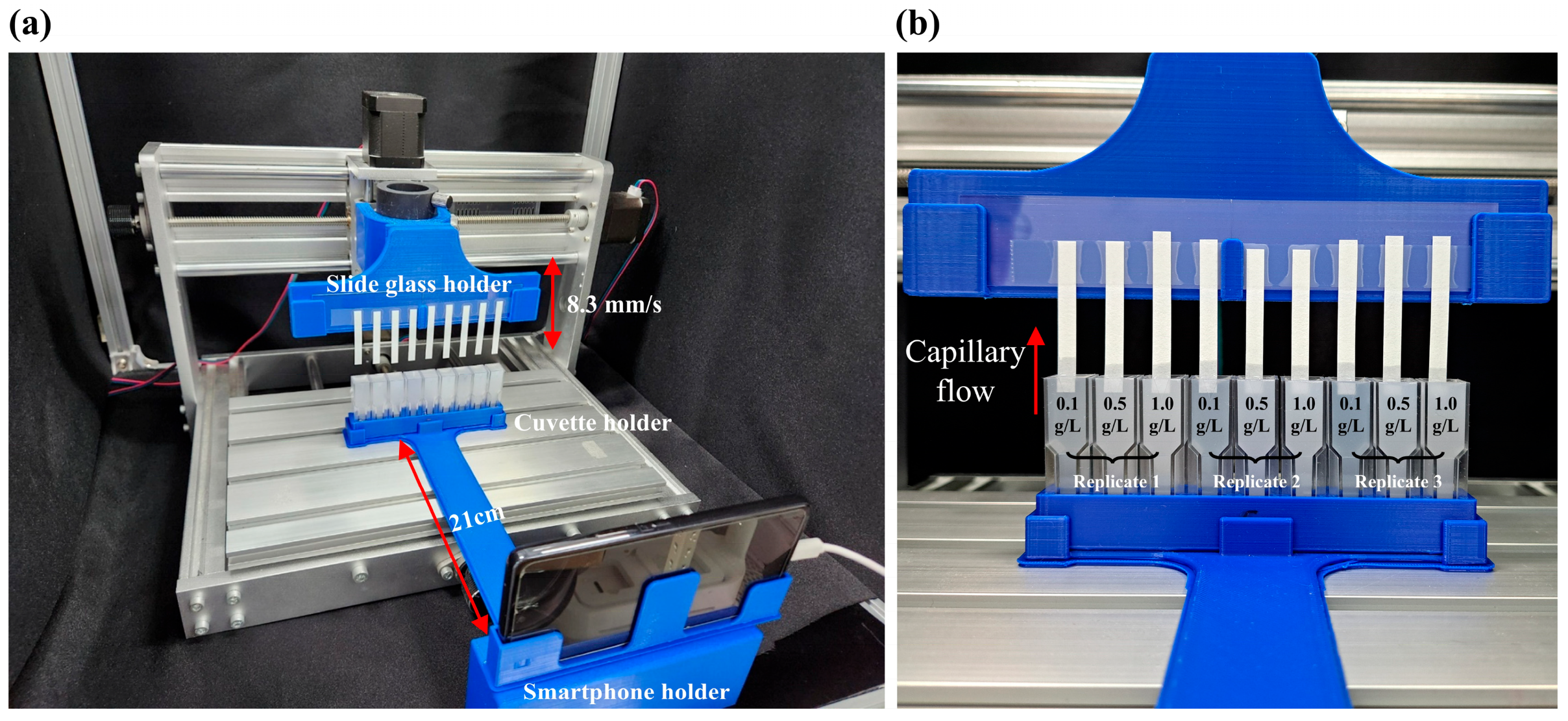
2.2.2. Motor-Controlled Sample Loading System
2.2.3. Devices and Conditions for Velocity Monitoring
2.3. Velocity Profile Measurement
2.4. Data Frame and Preprocessing
2.4.1. Raw Flow Profile Dataset
2.4.2. Model-Fitted Flow Profile Dataset
2.5. Machine Learning Models and Hyperparameter Tuning
3. Results and Discussion
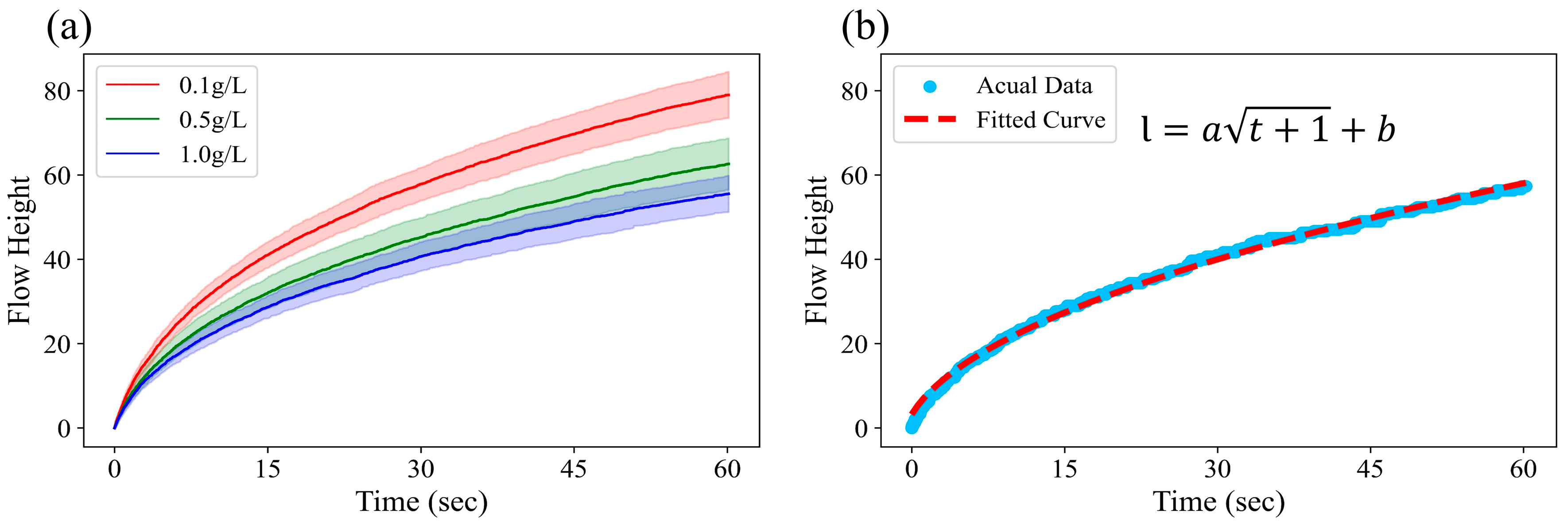
3.1. Flow Profiles on Paper-Based Microfluidic Chip and Theoretical Curve Fit
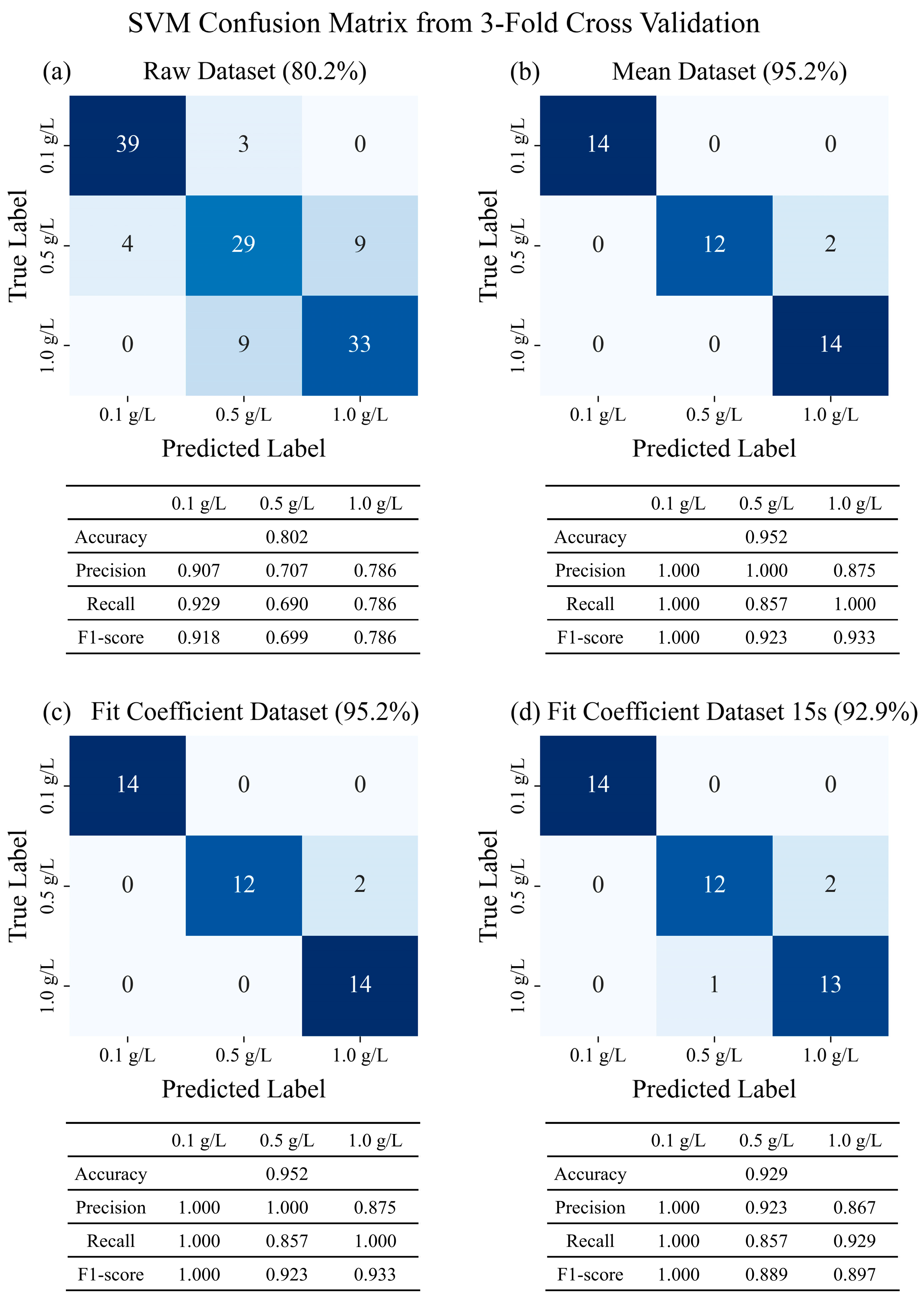
3.2. Astringency Intensity Classification Using Machine Learning
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, M.; Septier, C.; Brignot, H.; Martin, C.; Canon, F.; Feron, G. Astringency sensitivity to tannic acid: Effect of ageing and saliva. Molecules 2022, 27, 1617. [Google Scholar] [CrossRef]
- Ramos-Pineda, A.M.; Carpenter, G.H.; García-Estévez, I.; Escribano-Bailon, M.T. Influence of chemical species on polyphenol–protein interactions related to wine astringency. J. Agric. Food Chem. 2020, 68, 2948–2954. [Google Scholar] [CrossRef]
- Melis, M.; Yousaf, N.Y.; Mattes, M.Z.; Cabras, T.; Messana, I.; Crnjar, R.; Barbarossa, I.T.; Tepper, B.J. Sensory perception of and salivary protein response to astringency as a function of the 6-n-propylthioural (PROP) bitter-taste phenotype. Physiol. Behav. 2017, 173, 163–173. [Google Scholar] [CrossRef]
- Monteleone, E.; Condelli, N.; Dinnella, C.; Bertuccioli, M. Prediction of perceived astringency induced by phenolic compounds. Food Qual. Prefer. 2004, 15, 761–769. [Google Scholar] [CrossRef]
- Sarneckis, C.J.; Dambergs, R.G.; Jones, P.; Mercurio, M.; Herderich, M.J.; Smith, P.A. Quantification of condensed tannins by precipitation with methyl cellulose: Development and validation of an optimised tool for grape and wine analysis. Aust. J. Grape Wine Res. 2006, 12, 39–49. [Google Scholar] [CrossRef]
- Tamara, M.R.; Lelono, D.; Roto, R.; Triyana, K. All-solid-state astringent taste sensor using polypyrrole-carbon black composite as ion-electron transducer. Sens. Actuators A Phys. 2023, 351, 114170. [Google Scholar] [CrossRef]
- Yeom, J.; Choe, A.; Lim, S.; Lee, Y.; Na, S.; Ko, H. Soft and ion-conducting hydrogel artificial tongue for astringency perception. Sci. Adv. 2020, 6, eaba5785. [Google Scholar] [CrossRef]
- Costa, J.J.; Moreira, F.T.C.; Soares, S.; Brandão, E.; Mateus, N.; De Freitas, V.; Sales, M.G.F. Wine astringent compounds monitored by an electrochemical biosensor. Food Chem. 2022, 395, 133587. [Google Scholar] [CrossRef]
- Chung, S.; Jennings, C.M.; Yoon, J. Distance versus Capillary Flow Dynamics-Based Detection Methods on a Microfluidic Paper-Based Analytical Device (μPAD). Chem. Eur. J. 2019, 25, 13070. [Google Scholar] [CrossRef]
- Mabbott, S.; Fernandes, S.C.; Schechinger, M.; Cote, G.L.; Faulds, K.; Mace, C.R.; Graham, D. Detection of cardiovascular disease associated miR-29a using paper-based microfluidics and surface enhanced Raman scattering. Analyst 2020, 145, 983–991. [Google Scholar] [CrossRef]
- Yang, X.; Bi, Z.; Li, J.; Wang, L.; Huang, H.; Li, Y. Paper-based colorimetric sensor array integrated with smartphone APP for the identification of tea polyphenol and Longjing tea. Biosens. Bioelectron. 2025, 278, 117391. [Google Scholar] [CrossRef]
- Charbaji, A.; Smith, W.; Anagnostopoulos, C.; Faghri, M. Zinculose: A new fibrous material with embedded zinc particles. Eng. Sci. Technol. Int. J. 2021, 24, 571–578. [Google Scholar] [CrossRef]
- Klug, K.E.; Reynolds, K.A.; Yoon, J. A capillary flow dynamics-based sensing modality for direct environmental pathogen monitoring. Chem. Eur. J. 2018, 24, 6025–6029. [Google Scholar] [CrossRef] [PubMed]
- Yahaya, M.L.; Zakaria, N.D.; Noordin, R.; Razak, K.A. Development of rapid gold nanoparticles based lateral flow assays for simultaneous detection of Shigella and Salmonella genera. Biotechnol. Appl. Biochem. 2021, 68, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Danchana, K.; Iwasaki, H.; Ochiai, K.; Namba, H.; Kaneta, T. Determination of glutamate using paper-based microfluidic devices with colorimetric detection for food samples. Microchem. J. 2022, 179, 107513. [Google Scholar] [CrossRef]
- Gonzalez, S.; Armenta, C.; Hoyos-Arbeláez, L.F.; Álvarez-Lueje, M.; Contreras, A. Development of the simultaneous colorimetric enzymatic detection of sucrose, fructose and glucose using a microfluidic paper-based analytical device. Talanta 2020, 208, 120384. [Google Scholar] [CrossRef]
- Kim, S.; Day, A.S.; Yoon, J.-Y. Machine learning classification of bacterial species using mix-and-match reagents on paper microfluidic chips and smartphone-based capillary flow analysis. Anal. Bioanal. Chem. 2022, 414, 3895–3904. [Google Scholar] [CrossRef]
- Chung, S.; Loh, A.; Jennings, C.M.; Sosnowski, K.; Ha, S.Y.; Yim, U.H.; Yoon, J.-Y. Capillary flow velocity profile analysis on paper-based microfluidic chips for screening oil types using machine learning. J. Hazard. Mater. 2023, 447, 130806. [Google Scholar] [CrossRef]
- Breshears, L.E.; Mata-Robles, S.; Tang, Y.; Baker, J.C.; Reynolds, K.A.; Yoon, J.-Y. Rapid, sensitive detection of PFOA with smartphone-based flow rate analysis utilizing competitive molecular interactions during capillary action. J. Hazard. Mater. 2023, 446, 130699. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006, 11, 164–170. [Google Scholar] [CrossRef]
- Brossard, N.; Bordeu, E.; Ibáñez, R.A.; Chen, J.; Osorio, F. Rheological study of tannin and protein interactions based on model systems. J. Texture Stud. 2020, 51, 585–592. [Google Scholar] [CrossRef]
- Franzl, M. Grbl-Streamer. 2023. Available online: https://github.com/michaelfranzl/grbl-streamer (accessed on 9 April 2025).
- Franzl, M. Gcode-Machine. 2023. Available online: https://github.com/michaelfranzl/gcode-machine (accessed on 9 April 2025).
- ISO 12232:2019; Photography—Digital Still Cameras—Determination of Exposure Index, ISO Speed Ratings, Standard Output Sensitivity, and Recommended Exposure Index. International Organization for Standardization: Geneva, Switzerland, 2019.
- Scikit-Learn, Machine Learning in Python. 2025. Available online: https://scikit-learn.org/stable/ (accessed on 11 April 2025).
- Martin, C.; Rollinat, A.; Tournier, C.; Peltier, C. A simple procedure to assess the reliability of the best estimate thresholds (BETs): A case study on the detection threshold of tannic acid astringency. Food Qual. Prefer. 2025, 127, 105437. [Google Scholar] [CrossRef]
- Ployon, S.; Morzel, M.; Belloir, C.; Bonnotte, A.; Bourillot, E.; Briand, L.; Lesniewska, E.; Lherminier, J.; Aybeke, E.; Canon, F. Mechanisms of astringency: Structural alteration of the oral mucosal pellicle by dietary tannins and protective effect of bPRPs. Food Chem. 2018, 253, 79–87. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, Y.; Ma, J.; Chen, Y.; Pan, X.; Nishinari, K.; Yang, N. New insight in characterization of red wine astringency using soft tribology method. J. Texture Stud. 2024, 55, e12820. [Google Scholar] [CrossRef]
- Obreque-Slier, E.; Peña-Neira, Á.; López-Solís, R. Interactions of enological tannins with the protein fraction of saliva and astringency perception are affected by pH. LWT-Food Sci. Technol. 2012, 45, 88–93. [Google Scholar] [CrossRef]
- Maleki, A.; Lafitte, G.; Kjøniksen, A.-L.; Thuresson, K.; Nyström, B. Effect of pH on the association behavior in aqueous solutions of pig gastric mucin. Carbohydr. Res. 2008, 343, 328–340. [Google Scholar] [CrossRef]
- Zhang, L.; Guan, Q.; Zhang, H.; Tang, L. Effect of metal ions on the interaction of condensed tannins with protein. Foods 2023, 12, 829. [Google Scholar] [CrossRef]


| Accuracy (Standard Deviation) | ||||||||
|---|---|---|---|---|---|---|---|---|
| SVM | LR | MLP | KNNs | RF | LDA | NB | DT | |
| Raw dataset | 0.802 (0.030) | 0.786 (0.019) | 0.770 (0.030) | 0.762 (0.039) | 0.794 (0.040) | 0.778 (0.040) | 0.746 (0.040) | 0.762 (0.067) |
| Mean dataset | 0.952 (0.067) | 0.905 (0.089) | 0.929 (0.058) | 0.905 (0.067) | 0.929 (0.058) | 0.952 (0.067) | 0.881 (0.089) | 0.929 (0.058) |
| Fit coefficient dataset | 0.952 (0.067) | 0.929 (0.058) | 0.952 (0.067) | 0.952 (0.067) | 0.952 (0.067) | 0.952 (0.067) | 0.952 (0.067) | 0.952 (0.067) |
| Fit coefficient dataset (15 s) | 0.929 (0.058) | 0.881 (0.089) | 0.881 (0.089) | 0.881 (0.089) | 0.881 (0.089) | 0.857 (0.058) | 0.881 (0.089) | 0.857 (0.058) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, D.; Bae, J.; Park, C.; Song, J.; Chung, S. Capillary Flow Profile Analysis on Paper-Based Microfluidic Chips for Classifying Astringency Intensity. Sensors 2025, 25, 5068. https://doi.org/10.3390/s25165068
Son D, Bae J, Park C, Song J, Chung S. Capillary Flow Profile Analysis on Paper-Based Microfluidic Chips for Classifying Astringency Intensity. Sensors. 2025; 25(16):5068. https://doi.org/10.3390/s25165068
Chicago/Turabian StyleSon, Daesik, Junseung Bae, Chanwoo Park, Jihoon Song, and Soo Chung. 2025. "Capillary Flow Profile Analysis on Paper-Based Microfluidic Chips for Classifying Astringency Intensity" Sensors 25, no. 16: 5068. https://doi.org/10.3390/s25165068
APA StyleSon, D., Bae, J., Park, C., Song, J., & Chung, S. (2025). Capillary Flow Profile Analysis on Paper-Based Microfluidic Chips for Classifying Astringency Intensity. Sensors, 25(16), 5068. https://doi.org/10.3390/s25165068







