Data-Efficient Bone Segmentation Using Feature Pyramid- Based SegFormer
Abstract
:1. Introduction
- We propose a segmentation method that improves the image feature extraction performance of SegFormer to extract more precise image features even on small datasets. Compared to the conventional SegFormer, our method can extract complicated shapes.
- We increase the feature map’s resolution and convolution layers for each hierarchical encoder to extract higher-quality image features. In addition, the FPN structure with attention effectively uses position information in the image for decoding.
- We evaluated the performance of the proposed model using about 270 spine, 420 hand and wrist, and 180 femur images. We achieved a high degree of accuracy compared to previous studies.
2. Related Work
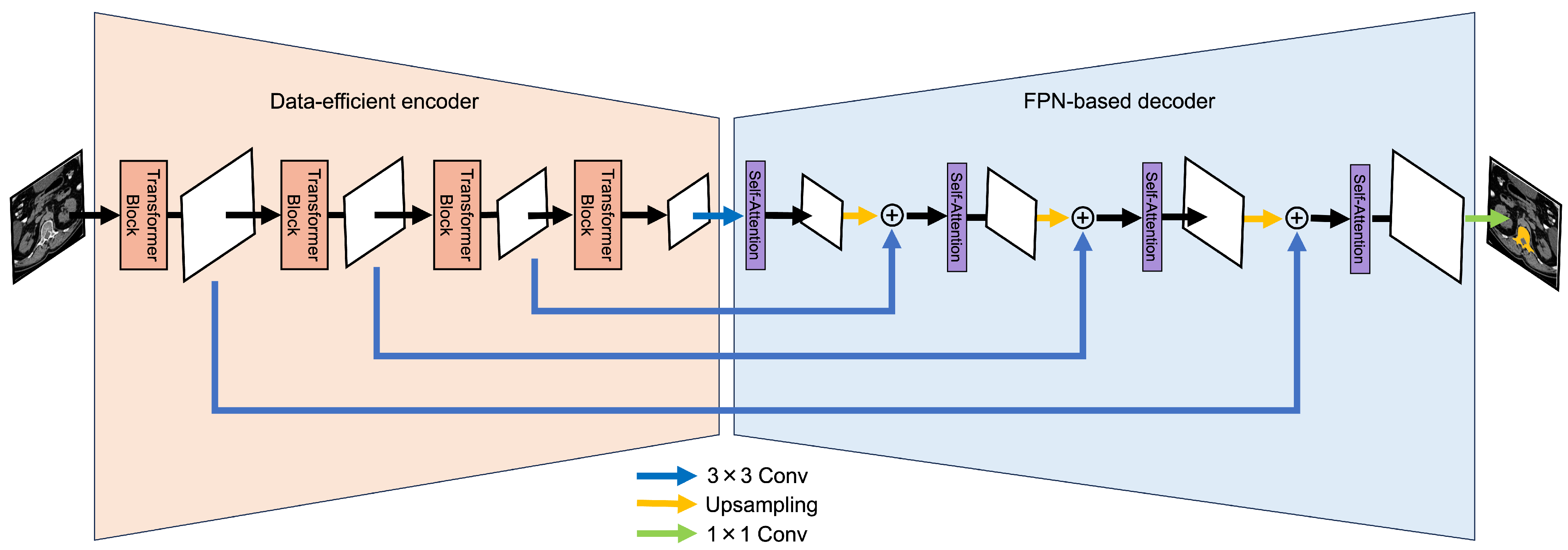
3. Proposed Method
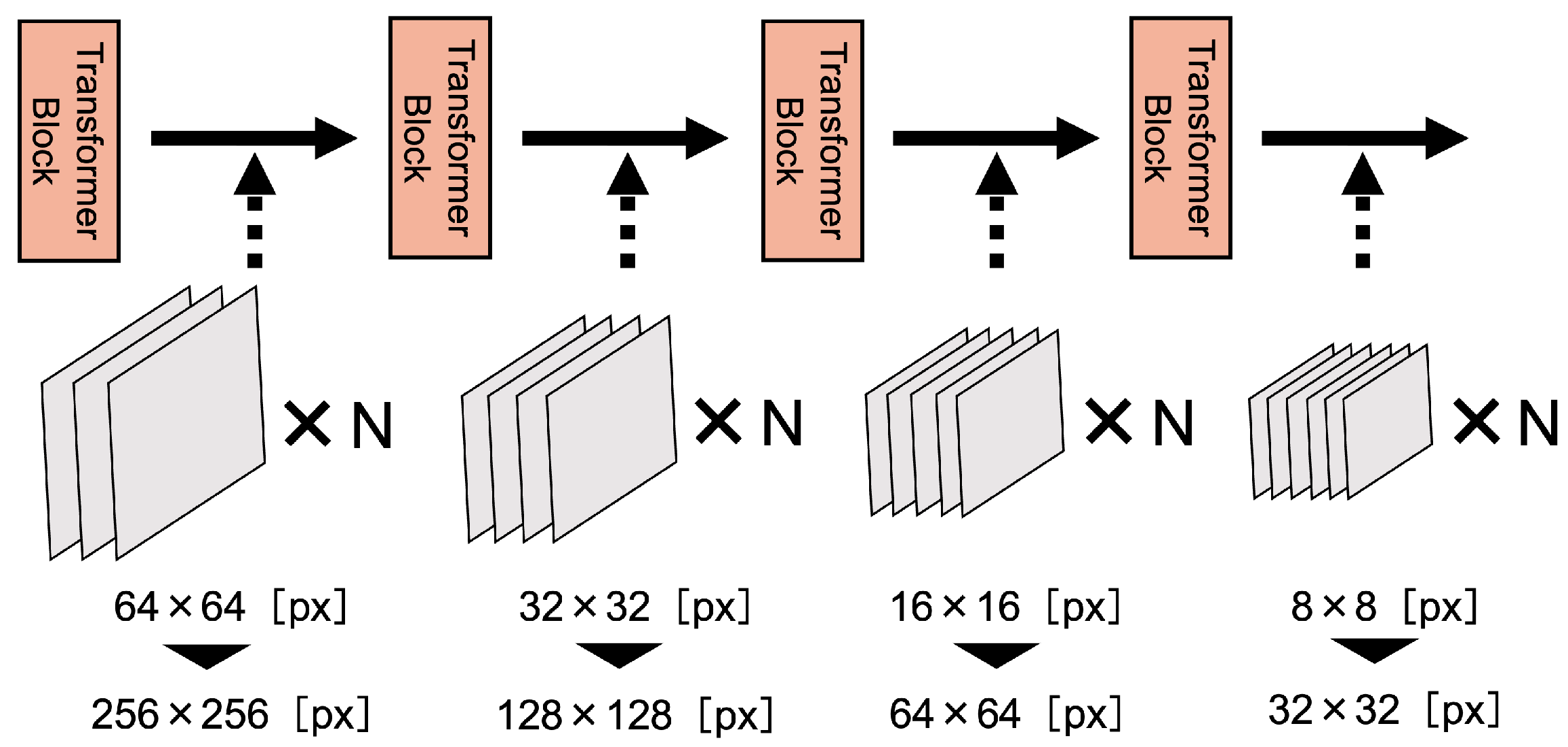
3.1. Data-Efficient Model
3.2. FPN-Based Model
4. Experimental Evaluations and Discussion
4.1. Datasets
4.2. Experimental Settings
4.3. Statistical Validation
4.4. Results with the Spine Dataset
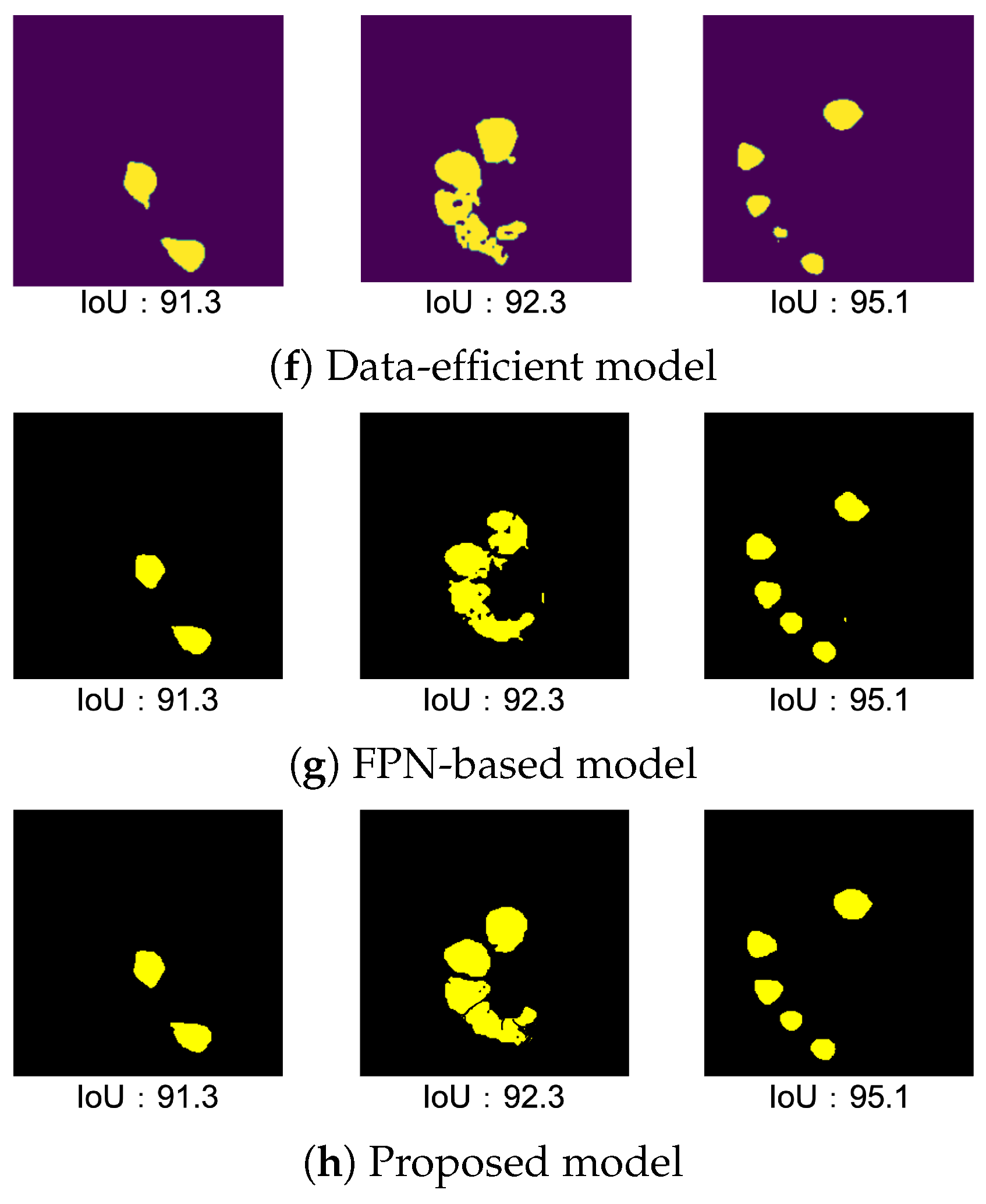
4.4.1. Result of Data-Efficient Model
4.4.2. Result of FPN-Based Model
4.4.3. Result of Proposed Model
4.4.4. Result of Validation and Test
4.5. Results of the Hand and Wrist Dataset
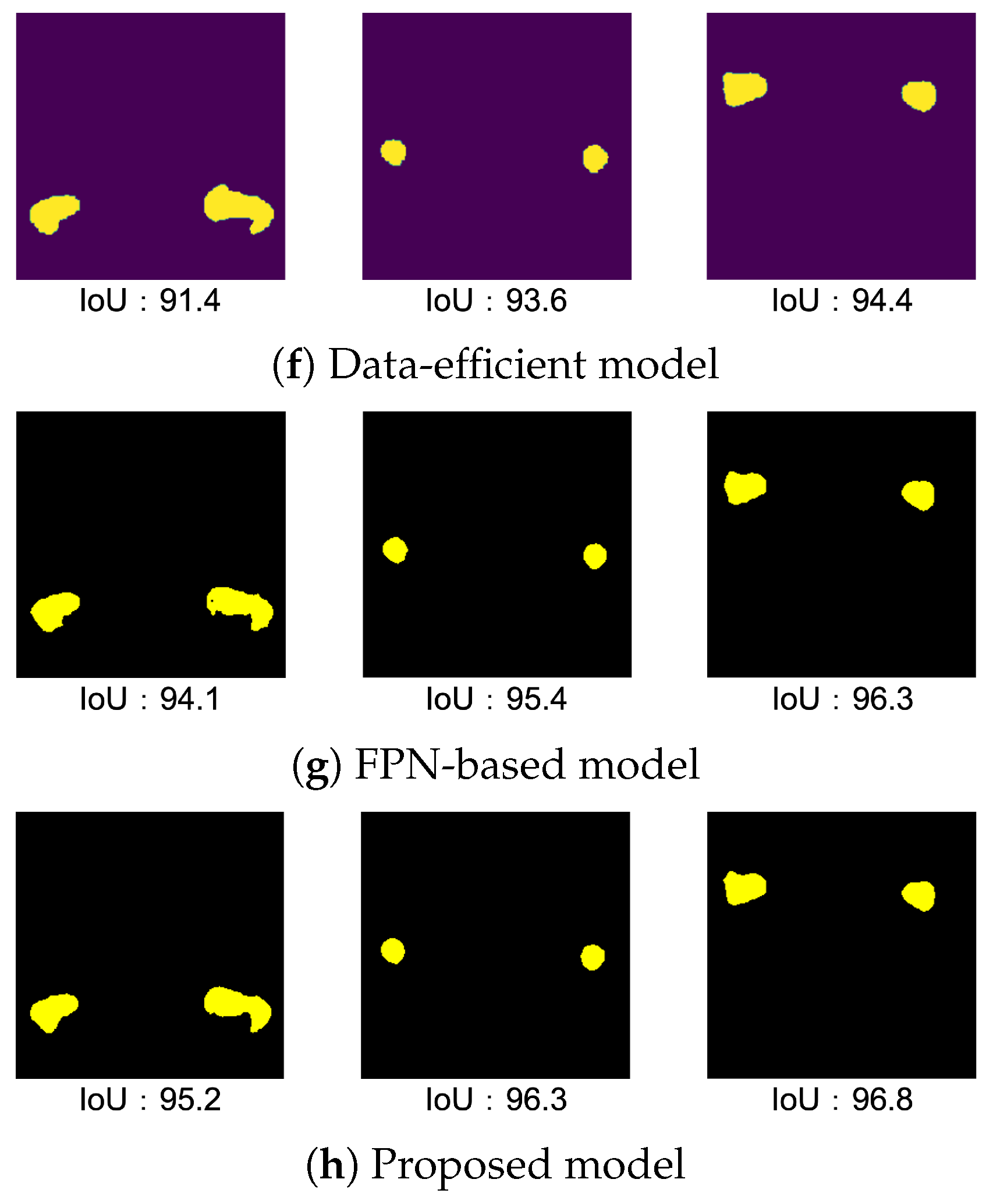
4.5.1. Result of Data-Efficient Model
4.5.2. Result of FPN-Based Model
4.5.3. Result of Proposed Model
4.5.4. Result of Validation and Test
4.6. Results of the Femur Dataset
4.6.1. Result of Data-Efficient Model
4.6.2. Result of FPN-Based Model
4.6.3. Result of Proposed Model
4.6.4. Result of Validation and Test
5. Conclusions
6. Future Works
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A






| Stage | Output Size | Layer Name | Parameter |
|---|---|---|---|
| Stage 1 | Overlapping Patch Embedding | ||
| Transformer Encoder | |||
| Stage 2 | Overlapping Patch Embedding | ||
| Transformer Encoder | |||
| Stage 3 | Overlapping Patch Embedding | ||
| Transformer Encoder | |||
| Stage 4 | Overlapping Patch Embedding | ||
| Transformer Encoder | |||
References
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Petit, O.; Thome, N.; Rambour, C.; Soler, L. U-Net Transformer: Self and Cross Attention for Medical Image Segmentation. arXiv 2021, arXiv:2103.06104. [Google Scholar]
- Fabian, I.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar]
- Zhan, X.; Liu, J.; Long, H.; Zhu, J.; Tang, H.; Gou, F.; Wu, J. An Intelligent Auxiliary Framework for Bone Malignant Tumor Lesion Segmentation in Medical Image Analysis. Diagnostics 2023, 13, 223. [Google Scholar] [CrossRef]
- Rich, J.M.; Bhardwaj, L.N.; Shah, A.; Gangal, K.; Rapaka, M.S.; Oberai, A.A.; Fields, B.K.K.; Matcuk, G.R., Jr.; Duddalwar, V.A. Deep learning image segmentation approaches for malignant bone lesions: A systematic review and meta-analysis. Front. Radiol. 2021, 3, 1241651. [Google Scholar] [CrossRef]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Tateishi, R.; Sato, M.; Hashizume, N.; Kamada, A.; Nakano, H.; Kabeya, Y.; Yonezawa, S.; Irie, R.; Tsujikawa, H.; et al. Deep learning and digital pathology powers prediction of HCC development in steatotic liver disease. Hepatology 2024. [Google Scholar] [CrossRef]
- Bai, Y.; Yu, Q.; Yun, B.; Jin, D.; Xia, Y.; Wang, Y. FS-MedSAM2: Exploring the Potential of SAM2 for Few-Shot Medical Image Segmentation without Fine-tuning. arXiv 2024. [Google Scholar] [CrossRef]
- Al-Asali, M.; Alqutaibi, A.Y.; Al-Sarem, M.; Saeed, F. Deep learning-based approach for 3D bone segmentation and prediction of missing tooth region for dental implant planning. Sci. Rep. 2024, 14, 13888. [Google Scholar] [CrossRef]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Hu, Z.; Yang, H.; Lou, T. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Palrmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017.
- Strudel, R.; Garcia, R.; Laptev, I.; Schmid, C. Segmenter: Transformer for Semantic Segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 7262–7272. [Google Scholar]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A Hybrid Transformer Architecture for Medical Image Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention (MICCAI), Strasbourg, France, 27 September–1 October 2021; pp. 61–71. [Google Scholar]
- Huang, X.; Deng, Z.; Li, D.; Yuan, X. MISSFormer: An Effective Medical Image Segmentation Transformer. arXiv 2021, arXiv:2109.07162. [Google Scholar] [CrossRef]
- Shi, J.; Wang, Y.; Yu, Z.; Li, G.; Hong, X.; Wang, F.; Gong, Y. Exploiting Multi-Scale Parallel Self-Attention and Local Variation via Dual-Branch Transformer-CNN Structure for Face Super-Resolution. IEEE Trans. Multimed. 2024, 26, 2608–2620. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, H.; Lou, T. Dual attention-guided feature pyramid network for instance segmentation of group pigs. Comput. Electron. Agric. 2021, 186, 106140. [Google Scholar] [CrossRef]
- Lee, S.; Islam, K.A.; Koganti, S.C.; Yaganti, V.; Mamillapalli, S.R.; Vitalos, H.; Williamson, D.F. SN-FPN: Self-Attention Nested Feature Pyramid Network for Digital Pathology Image Segmentation. IEEE Access 2024, 12, 92764–92773. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 11–17 October 2021; pp. 548–558. [Google Scholar]
- Klein, A.; Warszawski, J.; Hillenga, J.; Maier-Hein, K.H. Automatic bone segmentation in whole-body CT images. Int. J. Comput. Assist. Radiol. Surg. 2018, 14, 21–29. [Google Scholar] [CrossRef]
- Minnema, J.; Eijnatten, M.V.; Kouw, W.; Diblen, F.; Mendrik, A.; Wolff, J. CT image segmentation of bone for medical additive manufacturing using a convolutional neural network. Comput. Biol. Med. 2018, 103, 130–139. [Google Scholar] [CrossRef]
- Memiş, A.; Varlı, S.; Bilgili, F. Semantic segmentation of the multiform proximal femur and femoral head bones with the deep convolutional neural networks in low quality MRI sections acquired in different MRI protocols. Comput. Med. Imaging Graph. 2020, 81, 101715. [Google Scholar] [CrossRef]
- Eggermont, P.; Wal, G.V.D.; Westhoff, P.; Laar, A.; Jong, M.D.; Rozema, T.; Kroon, H.M.; Ayu, O.; Derikx, L.; Dijkstra, S.; et al. Patient-specific finite element computer models improve fracture risk assessments in cancer patients with femoral bone metastases compared to clinical guidelines. Bone 2019, 130, 115101. [Google Scholar] [CrossRef]
- Kar, M.K.; Nath, M.K.; Neog, D.R. A Review on Progress in Semantic Image Segmentation and Its Application to Medical Images. SN Comput. Sci. 2021, 2, 397. [Google Scholar] [CrossRef]
- Bowen, C.; Ishan, M.; Alexander, S.G.; Alexander, K.; Rohit, G. Masked-attention Mask Transformer for Universal Image Segmentation. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 1280–1289. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The cancer imaging archive (tcia): Maintaining and operating a public information repository. J. Digit. Imaging 2023, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Mazher, M.; Qayyum, A.; Puig, D.; Abdel-Nasser, M. Effective Approaches to Fetal Brain Segmentation in MRI and Gestational Age Estimation by Utilizing a Multiview Deep Inception Residual Network and Radiomics. Entropy 2022, 24, 1708. [Google Scholar] [CrossRef] [PubMed]

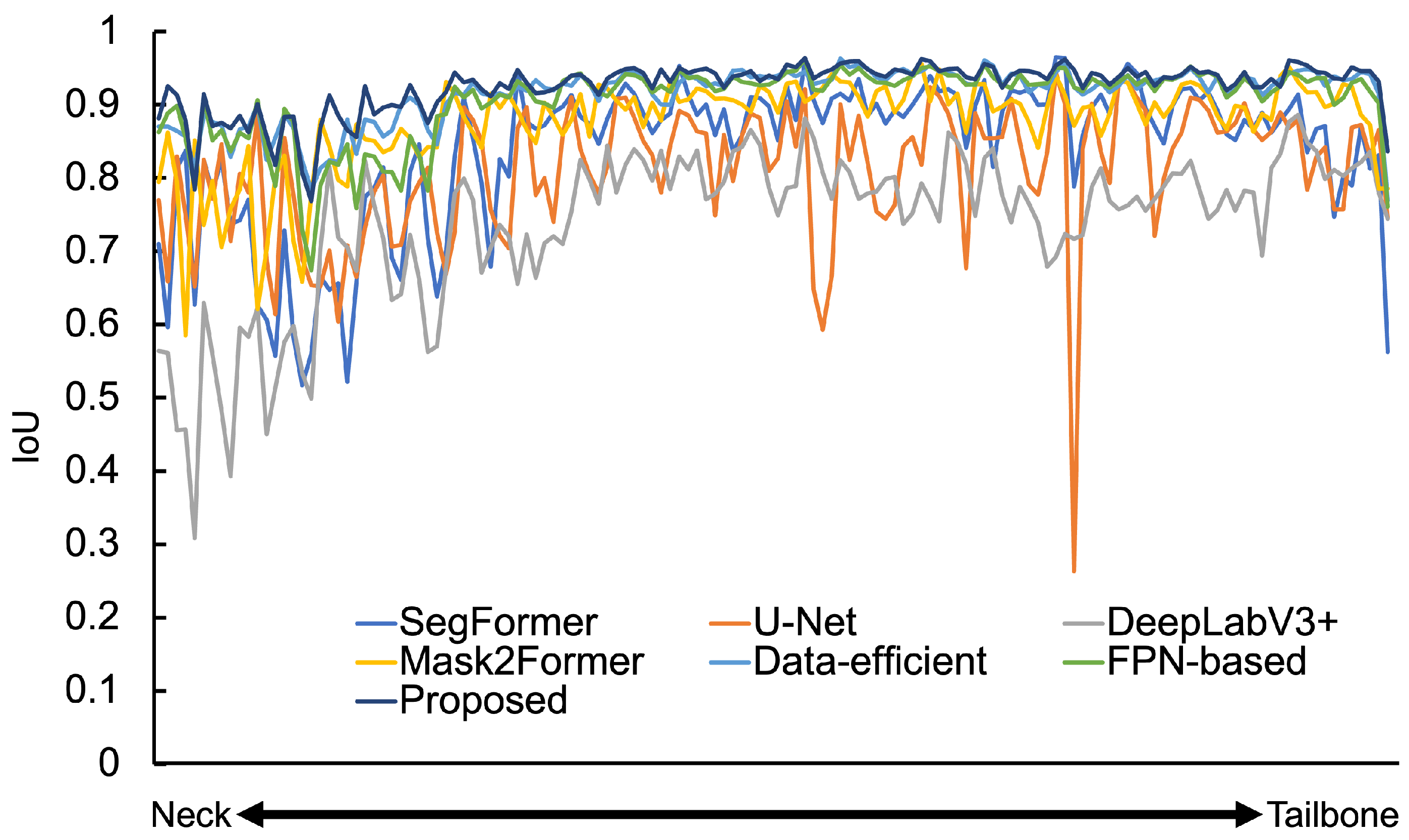
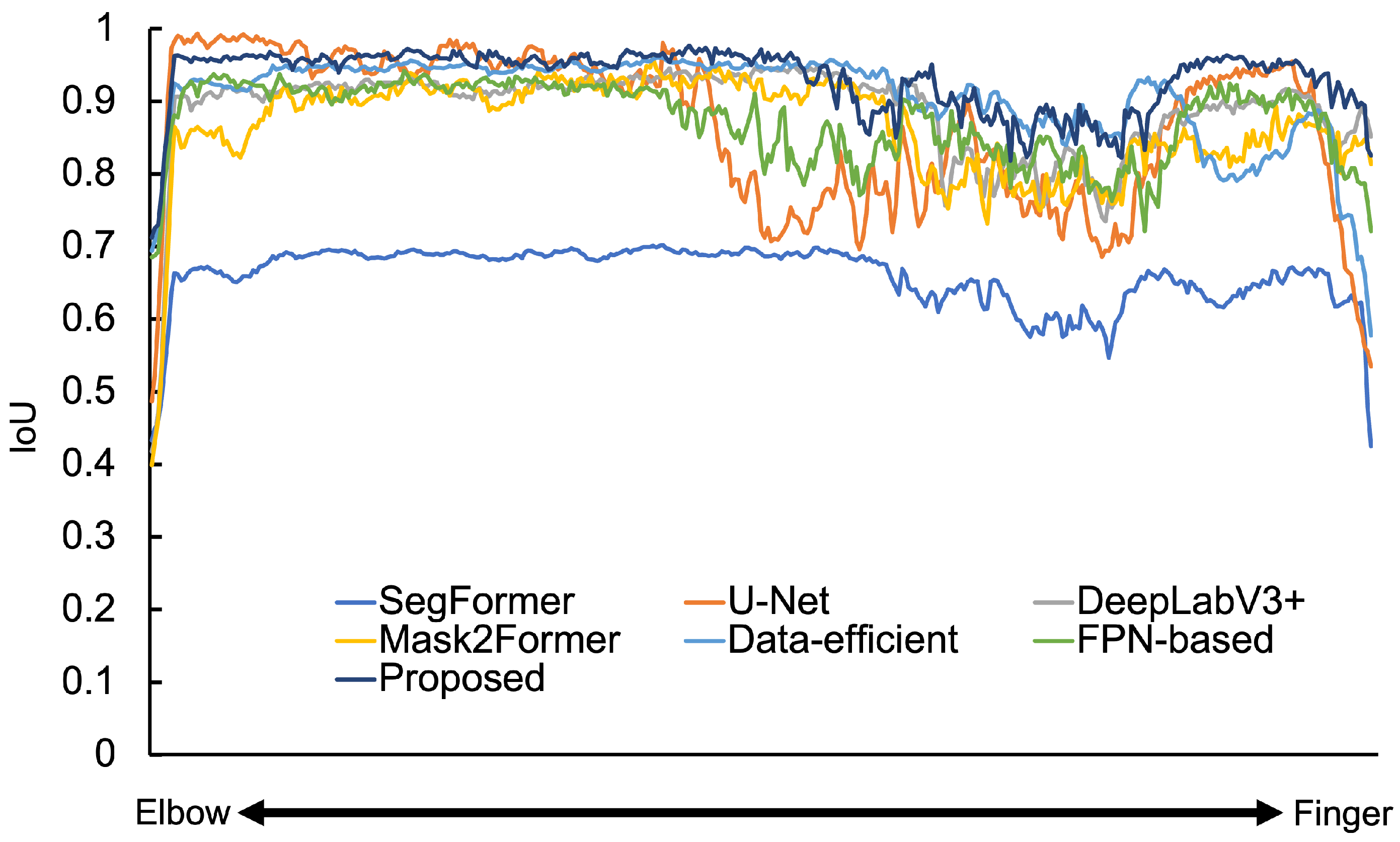

| Number of Convolution Layers | Spine IoU | Hand and Wrist IoU | Femur IoU |
|---|---|---|---|
| 1 (SegFormer) | 82.7 | 63.9 | 88.5 |
| 2 | 85.9 | 68.5 | 90.4 |
| 3 | 85.1 | 78.5 | 90.1 |
| 4 | 85.2 | 73.5 | 89.2 |
| 5 | 84.9 | 73.2 | 89.3 |
| Resolution of Feature Map (First Transformer Block) | Spine IoU | Hand and Wrist IoU | Femur IoU |
|---|---|---|---|
| 64 × 64 [px] (SegFormer) | 82.7 | 63.9 | 88.5 |
| 128 × 128 [px] | 86.2 | 79.5 | 90.2 |
| 256 × 256 [px] | 89.8 | 84.7 | 91.1 |
| 512 × 512 [px] | 89.1 | 84.1 | 90.7 |
| Methods | Spine IoU | Hand and Wrist IoU | Femur IoU |
|---|---|---|---|
| SegFormer | 82.7 | 63.9 | 88.5 |
| U-Net | 76.5 | 84.3 | 84.8 |
| DeepLabV3+ | 74.1 | 88.9 | 93.9 |
| Mask2Former | 89.1 | 87.6 | 95.3 |
| Data-efficient model | 91.2 | 90.7 | 92.8 |
| FPN-based model | 90.7 | 88.5 | 95.4 |
| Proposed Model | 93.1 | 93.2 | 96.3 |
| Methods | Spine DSC | Hand and Wrist DSC | Femur DSC |
|---|---|---|---|
| SegFormer | 87.3 | 73.8 | 92.1 |
| U-Net | 89.4 | 90.7 | 89.4 |
| DeepLabV3+ | 84.5 | 93.6 | 96.8 |
| Mask2Former | 93.3 | 92.6 | 97.6 |
| Data-efficient model | 95.1 | 94.9 | 96.2 |
| FPN-based model | 94.6 | 92.3 | 97.6 |
| Proposed Model | 96.6 | 96.4 | 98.1 |
| IoU by Attention Mechanism | Spine IoU | Hand and Wrist IoU | Femur IoU |
|---|---|---|---|
| SegFormer | 82.7 | 63.9 | 88.5 |
| FPN-based model without Attention mechanism | 88.1 | 86.2 | 93.9 |
| FPN-based model | 90.7 | 88.5 | 95.4 |
| Methods | Average Spine IoU | Standard Deviation |
|---|---|---|
| SegFormer | 83.3 | 0.34 |
| U-Net | 77.0 | 0.46 |
| DeepLabV3+ | 74.4 | 0.34 |
| Mask2Former | 88.9 | 0.71 |
| Proposed Model | 93.3 | 0.47 |
| Methods | Average Hand and Wrist IoU | Standard Deviation |
|---|---|---|
| SegFormer | 64.3 | 0.40 |
| U-Net | 83.6 | 0.70 |
| DeepLabV3+ | 88.3 | 0.55 |
| Mask2Former | 87.0 | 0.60 |
| Proposed Model | 92.9 | 0.25 |
| Methods | Average Femur IoU | Standard Deviation |
|---|---|---|
| SegFormer | 88.6 | 0.40 |
| U-Net | 84.9 | 0.48 |
| DeepLabV3+ | 93.1 | 0.55 |
| Mask2Former | 95.4 | 0.37 |
| Proposed Model | 96.3 | 0.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masuda, N.; Ono, K.; Tawara, D.; Matsuura, Y.; Sakabe, K. Data-Efficient Bone Segmentation Using Feature Pyramid- Based SegFormer. Sensors 2025, 25, 81. https://doi.org/10.3390/s25010081
Masuda N, Ono K, Tawara D, Matsuura Y, Sakabe K. Data-Efficient Bone Segmentation Using Feature Pyramid- Based SegFormer. Sensors. 2025; 25(1):81. https://doi.org/10.3390/s25010081
Chicago/Turabian StyleMasuda, Naohiro, Keiko Ono, Daisuke Tawara, Yusuke Matsuura, and Kentaro Sakabe. 2025. "Data-Efficient Bone Segmentation Using Feature Pyramid- Based SegFormer" Sensors 25, no. 1: 81. https://doi.org/10.3390/s25010081
APA StyleMasuda, N., Ono, K., Tawara, D., Matsuura, Y., & Sakabe, K. (2025). Data-Efficient Bone Segmentation Using Feature Pyramid- Based SegFormer. Sensors, 25(1), 81. https://doi.org/10.3390/s25010081






