A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning
Abstract
1. Introduction
1.1. Characteristics and Challenges of Hyperspectral Remote-Sensing Images
- (1)
- Curse of Dimensionality
- (2)
- Strong Inter-band Correlation
- (3)
- Nonlinear Data Structures
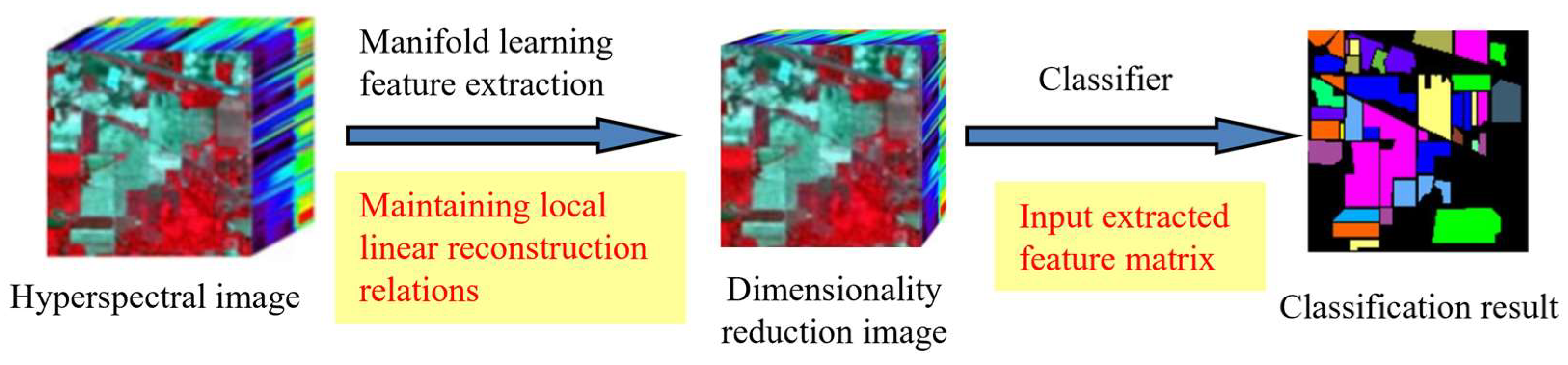
1.2. Hyperspectral Manifold Learning Dimensionality Reduction and Related Parameter Mathematical Expression
1.3. Manifold Expression in Hyperspectral Imagery Dimensionality Reduction
2. Materials and Methods
2.1. Study Area
2.2. Data Description
2.3. Methods
2.3.1. Linear Manifold Learning Methods
- (1)
- Principal Components Analysis (PCA)
| Algorithm 1 Principal Component Analysis (PCA) |
| Input: A dataset X, intrinsic dimensionality d Output: Return low-dimensional coordinate matrix Step 1. Standardize the original dataset: Step 2. Calculate the covariance matrix: Step 3. Compute the eigenvalues and eigenvectors of the covariance matrix: Step 4. Order the eigenvalues, select the principal components, construct the projection matrix, and transform into the new space: |
- (2)
- Multidimensional Scaling (MDS)
| Algorithm 2 Multidimensional Scaling (MDS) |
| Input: A dataset X, intrinsic dimensionality d Output: Return low-dimensional coordinate matrix Step 1. Calculate a distance matrix based on the original high-dimensional data Step 2. Perform double centering on the distance matrix: Step 3. Conduct eigenvalue decomposition on the double-centered matrix: Step 4. Select the principal components and compute the configuration: |
- (3)
- Linear Discriminant Analysis (LDA)
| Algorithm 3 Linear Discriminant Analysis (LDA) |
| Input: A dataset X, intrinsic dimensionality d,data labels label Output: Return low-dimensional coordinate matrix Step 1. Calculate the within-class mean and the overall mean: and Step 2. Compute the within-class scatter matrix and the between-class scatter matrix: and Step 3. Calculate the projection vector: Step 4. Select the eigenvector corresponding to the largest eigenvalue and transform into the new space: |
2.3.2. Nonlinear Manifold Learning Methods
- (1)
- Isometric Mapping (Isomap)
| Algorithm 4 Isometric Mapping (Isomap) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k Output: Return low-dimensional coordinate matrix Step 1. Calculate the double-centered distance matrix: Step 2. Perform eigenvalue decomposition on the double-centered matrix B, obtaining eigenvalues and their corresponding eigenvectors Step 3. Select the eigenvectors corresponding to the largest eigenvalues, which form the basis of the low-dimensional space Step 4. Use the selected eigenvectors and the square roots of their corresponding eigenvalues to compute the coordinates of the data points in the low-dimensional space |
- (2)
- Locally Linear Embedding (LLE)
| Algorithm 5 Locally linear embedding (LLE) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k Output: Return low-dimensional coordinates matrix Step 1. Given dataset , for each point , find its nearest neighbors Step 2. Calculate weights based on the linear relationship Step 3. Find the low-dimensional representation |
- (3)
- Laplacian Eigenmaps (LE)
| Algorithm 6 Laplacian Eigenmaps (LE) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k, the Gaussian kernel function bandwidth t Output: Return low-dimensional coordinate matrix Step 1. Based on the given dataset, construct an adjacency graph Step 2. Compute the Laplacian matrix: Step 3. Solve for the eigenvalues: Step 4. Select the eigenvectors corresponding to the smallest non-zero eigenvalues of as the coordinates of the data points in the low-dimensional space |
- (4)
- Hessian Locally Linear Embedding (HLLE)
| Algorithm 7 Hessian locally linear embedding (HLLE) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k Output: Return low-dimensional coordinate matrix Step 1. First, for each point in the dataset, find its nearest neighbors and construct a nearest neighbor graph Step 2. For each data point and its nearest neighbors, estimate the Hessian matrix to reflect the curvature of the local geometric structure Step 3. By combining the local Hessian estimates of each point, construct a global Hessian matrix Step 4. The space embedding involves eigenvalue decomposition and selection of the eigenvectors corresponding to the smallest non-zero eigenvalues: |
- (5)
- Local Tangent Space Alignment (LTSA)
| Algorithm 8 Local Tangent Space Alignment (LTSA) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k Output: Return low-dimensional coordinate matrix Step 1. For each point in the dataset, find its nearest neighbors and construct a nearest neighbor graph Step 2. For each point and its nearest neighbors, calculate the basis of the local tangent space through PCA of the local neighborhood Step 3. By rotating and translating each local tangent space, find a global reference frame that aligns all local tangent spaces within this frame as closely as possible Step 4. On the basis of the aligned local tangent spaces, reconstruct the global low-dimensional coordinates to preserve the geometric structure of local neighborhoods |
- (6)
- Maximum Variance Unfolding (MVU)
| Algorithm 9 Maximum Variance Unfolding (MVU) |
| Input: A dataset X, intrinsic dimensionality d, the neighborhood k, the lambda λ Output: Return low-dimensional coordinate matrix Step 1. For each point in the dataset, identify its nearest neighbors and construct a nearest neighbor graph Step 2. Calculate and store the Euclidean distances between the data point and its nearest neighbors Step 3. Maximize the variance in the low-dimensional space through semidefinite programming (SDP): Step 4. Use a semidefinite programming (SDP) solver to solve this optimization problem and extract the low-dimensional embedding from the solution of SDP, selecting the eigenvectors corresponding to the largest few eigenvalues as the coordinates in the low-dimensional space |
3. Results
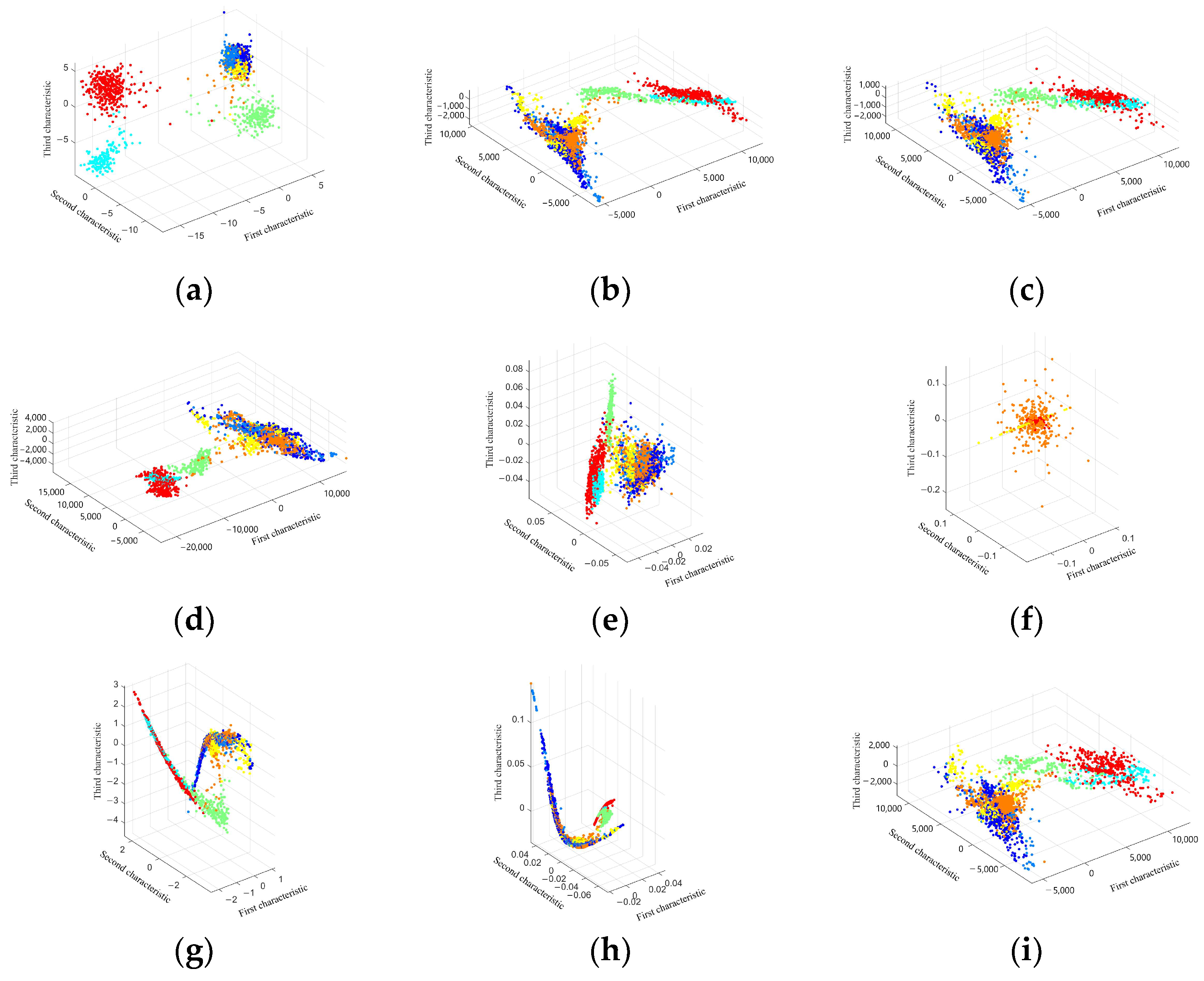
3.1. Visualization of Low-Dimensional Embedding of Hyperspectral Images
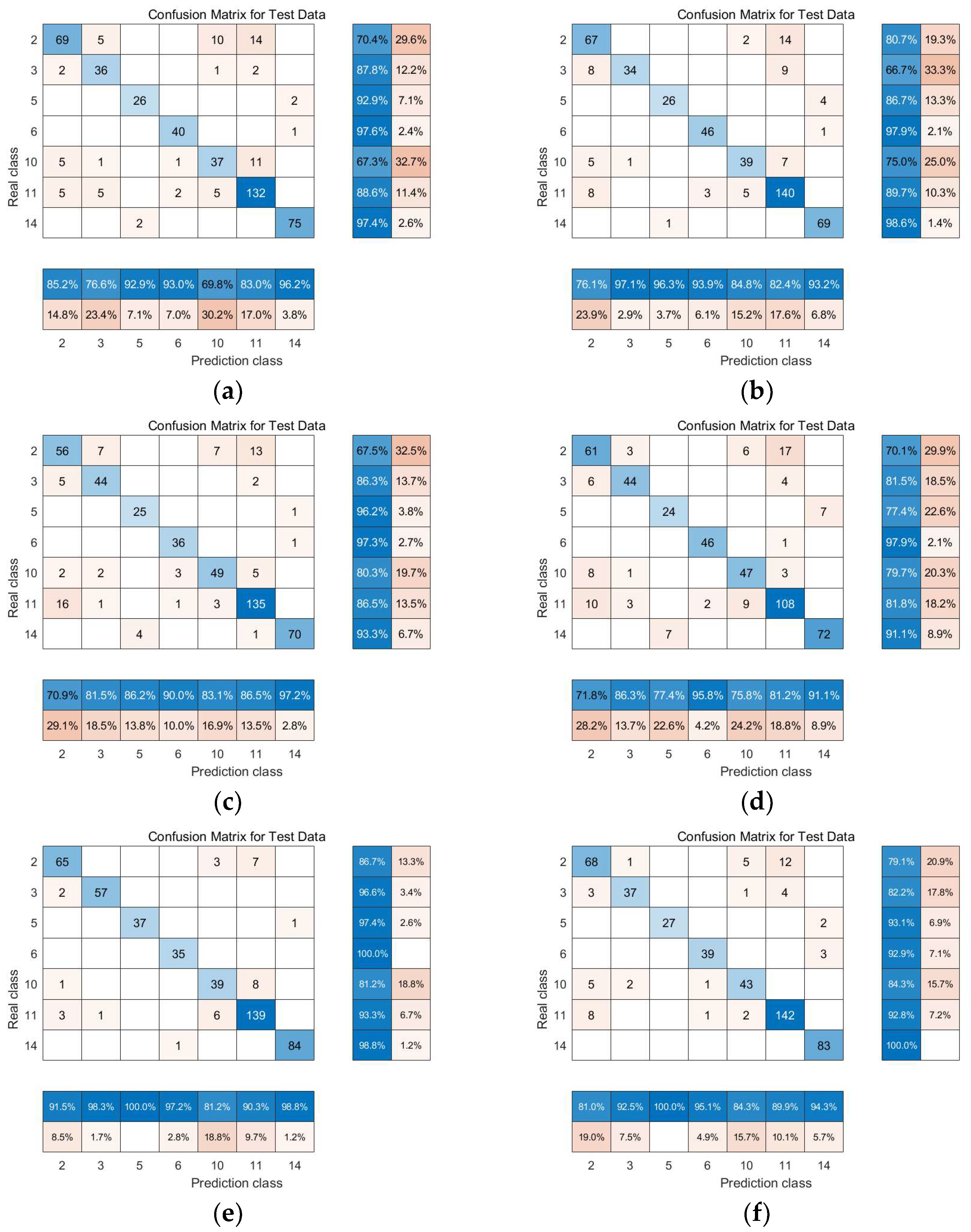
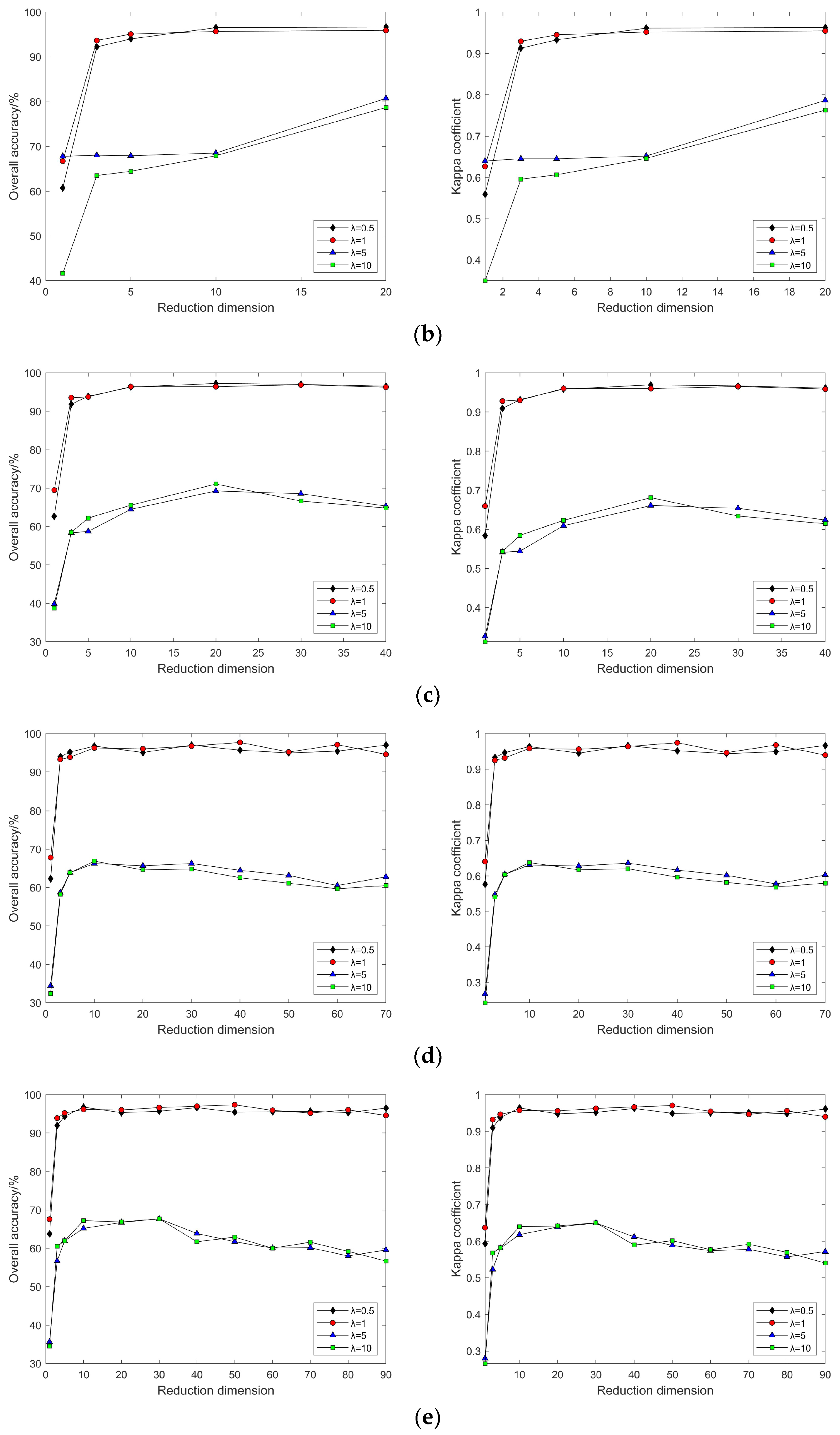
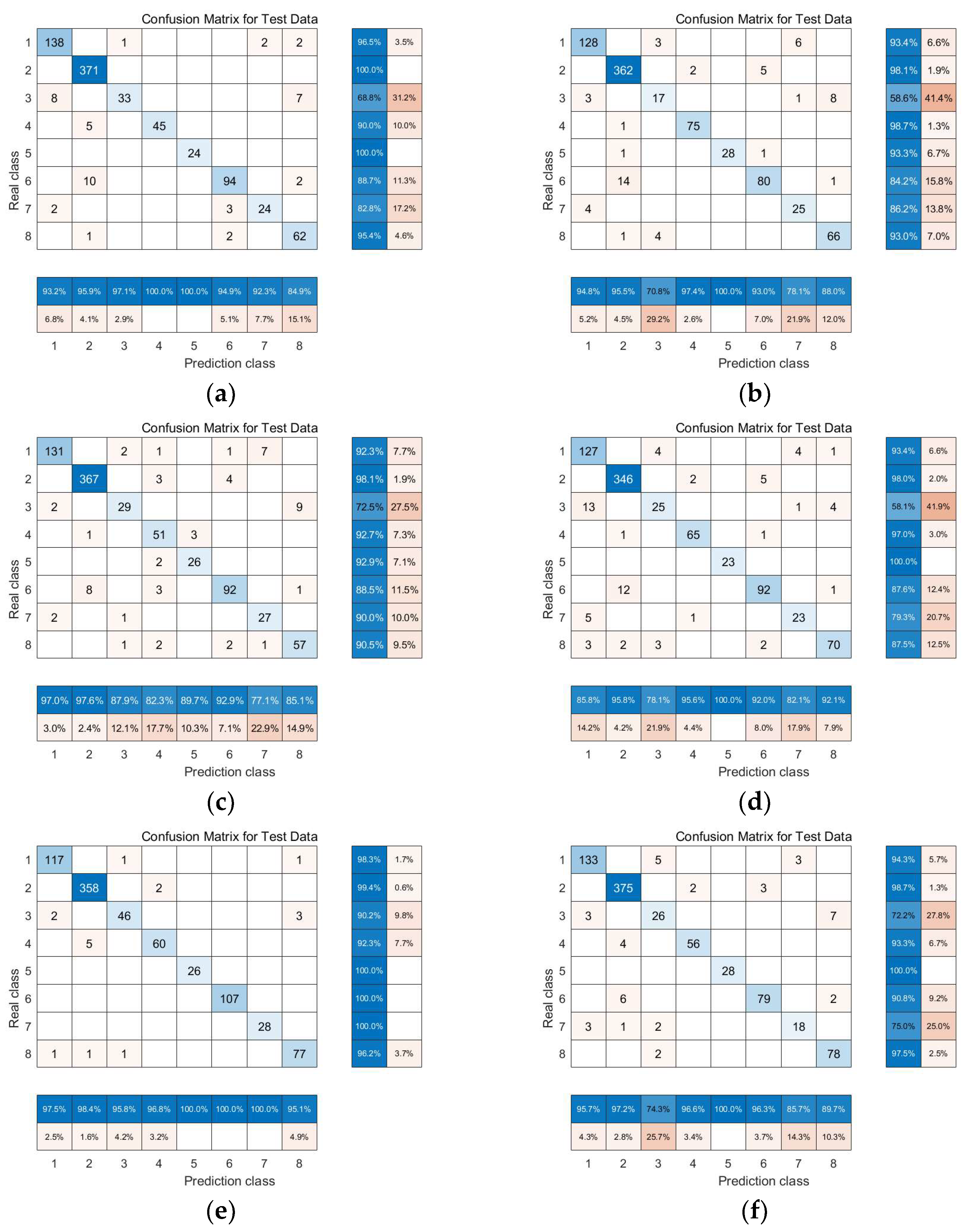
3.2. Experimental Results and Comparative Analysis Based on the Indian Pines Dataset
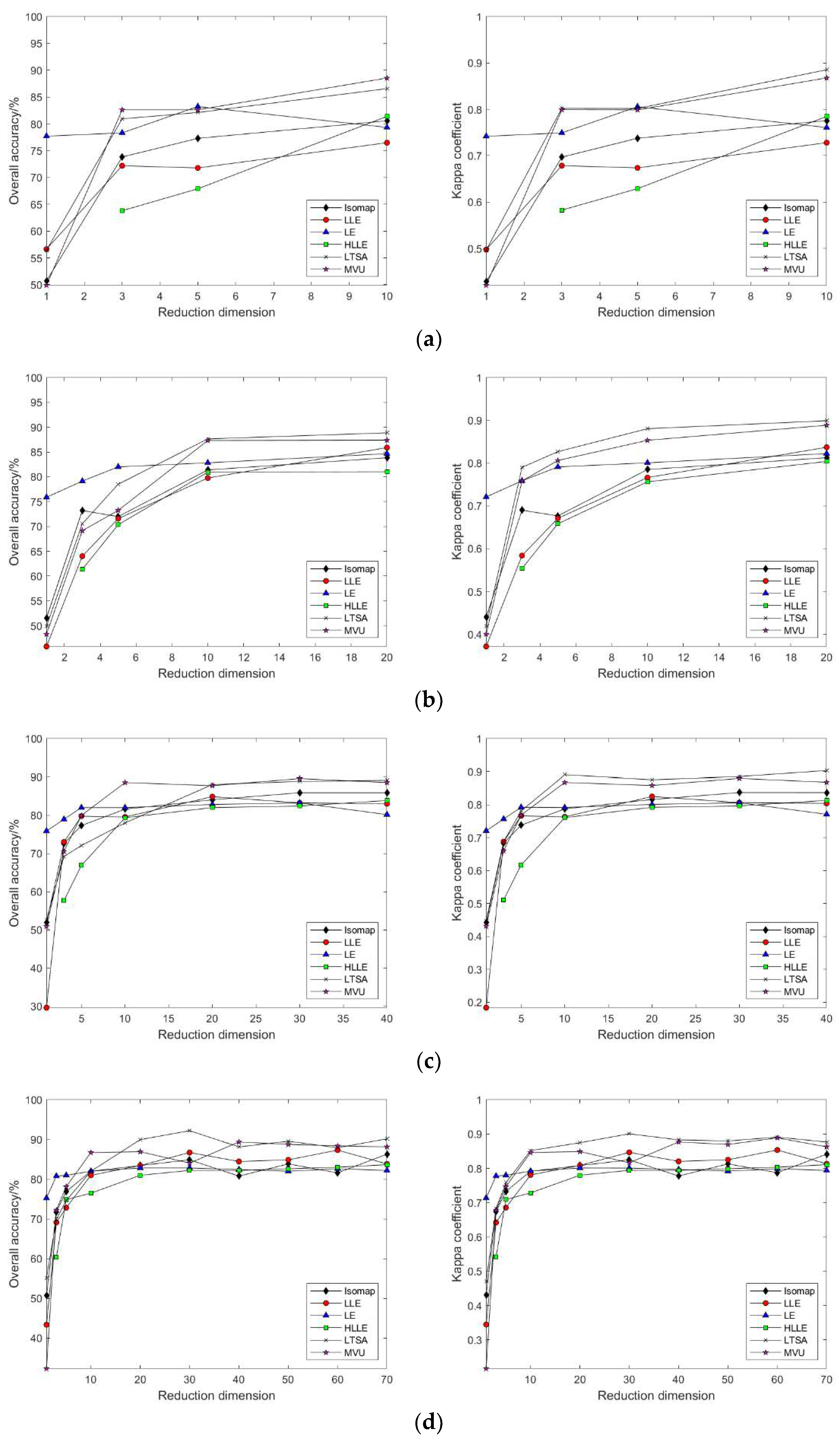
3.2.1. Linear Manifold Learning Dimensionality Reduction in Hyperspectral Imagery
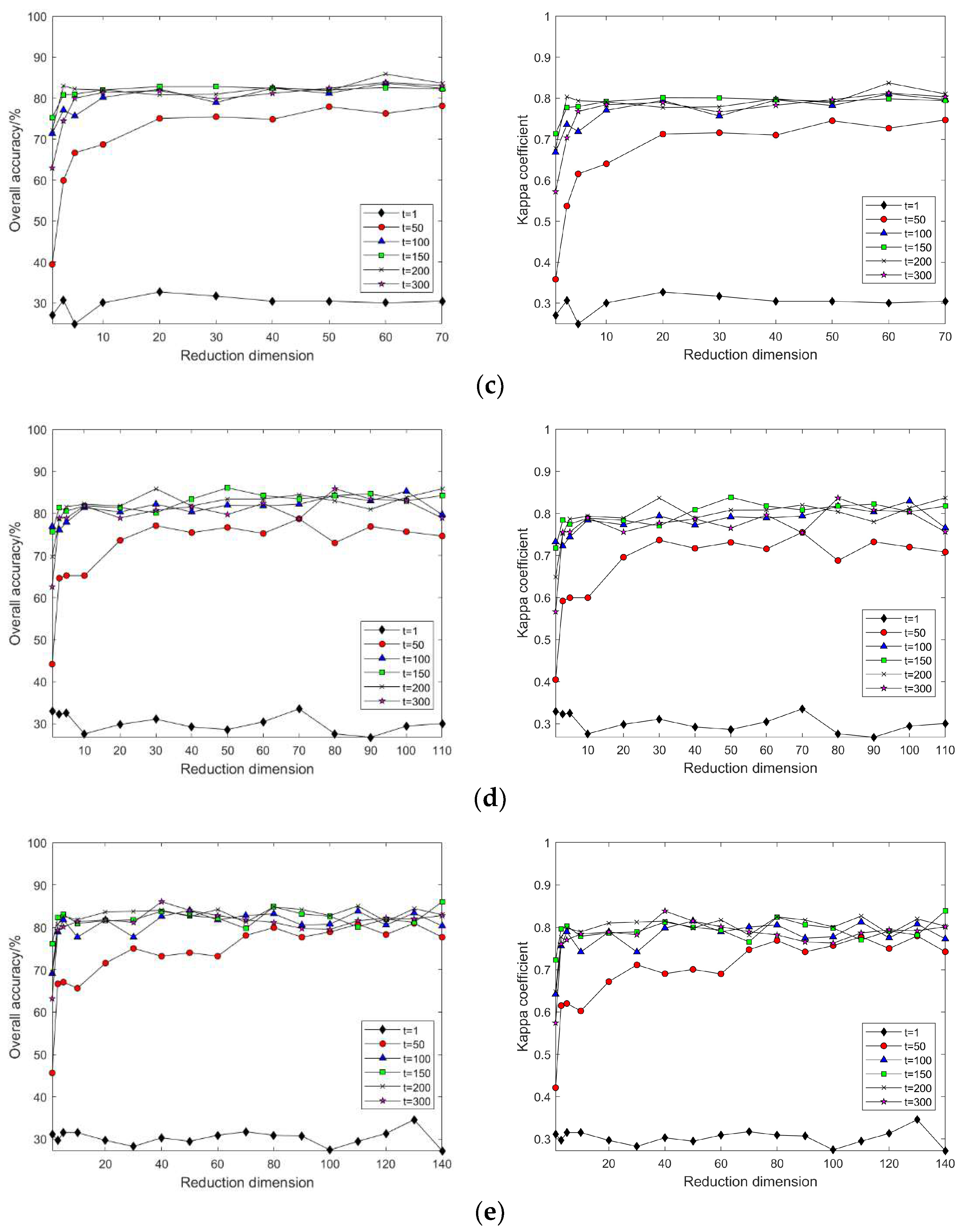
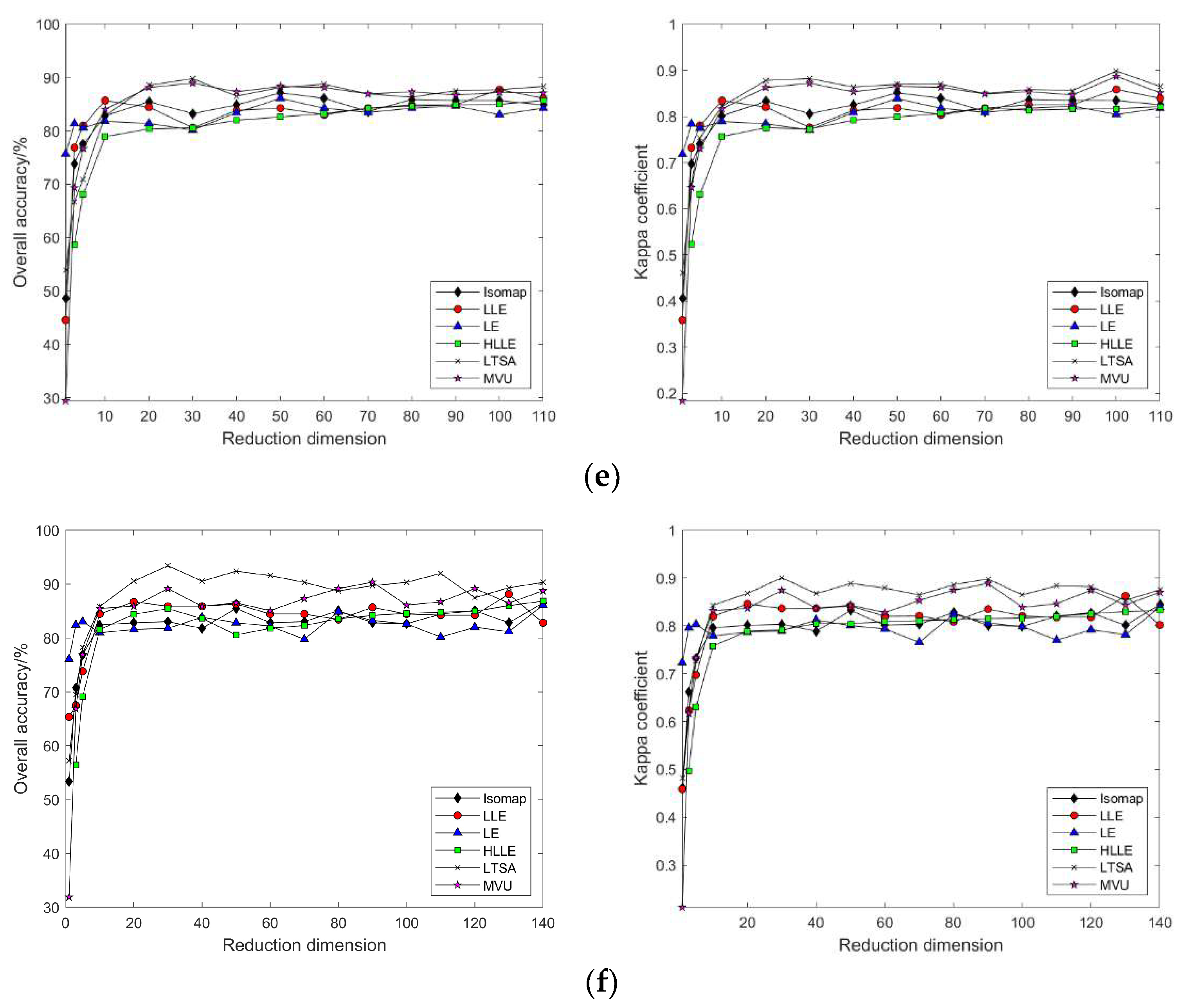
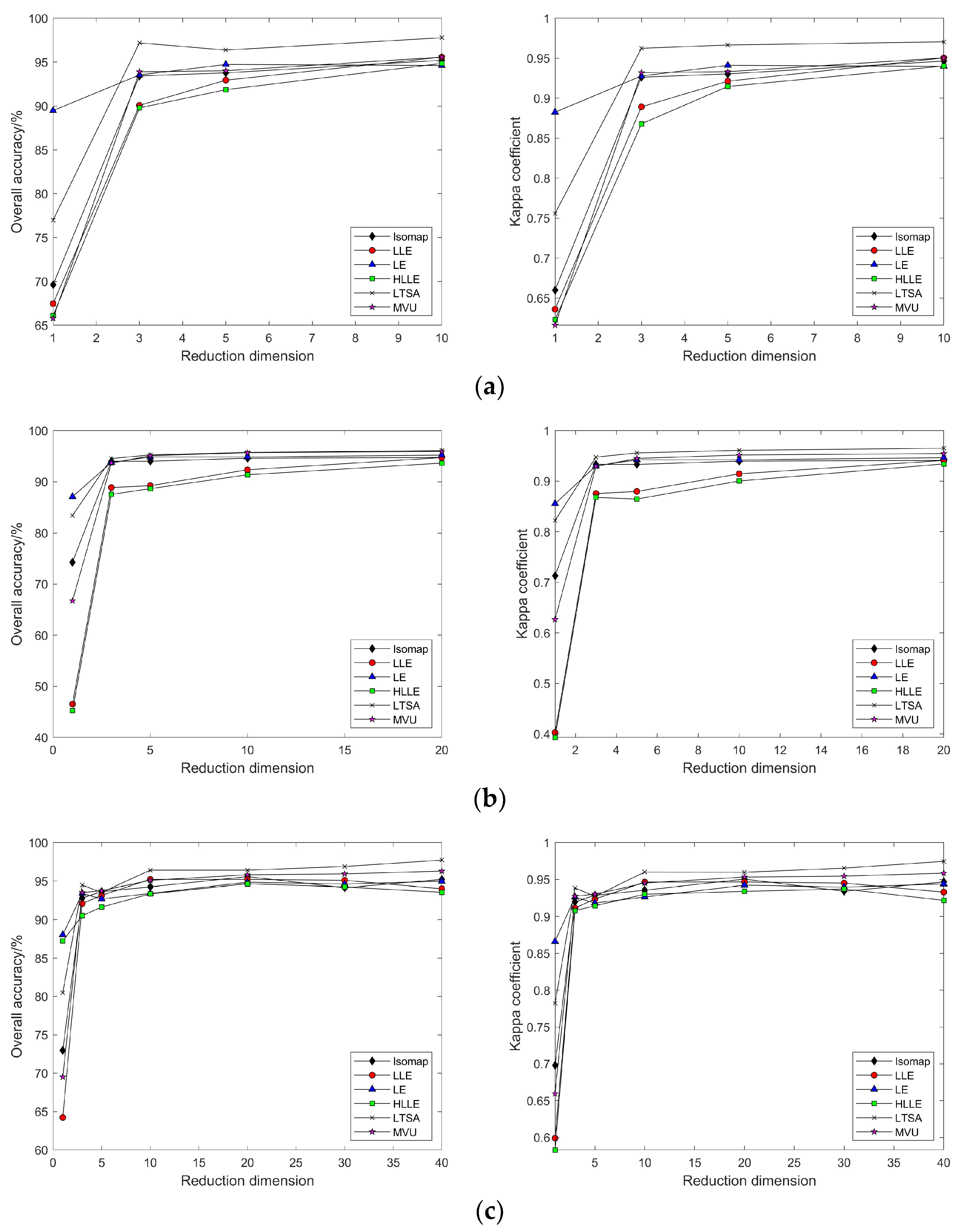
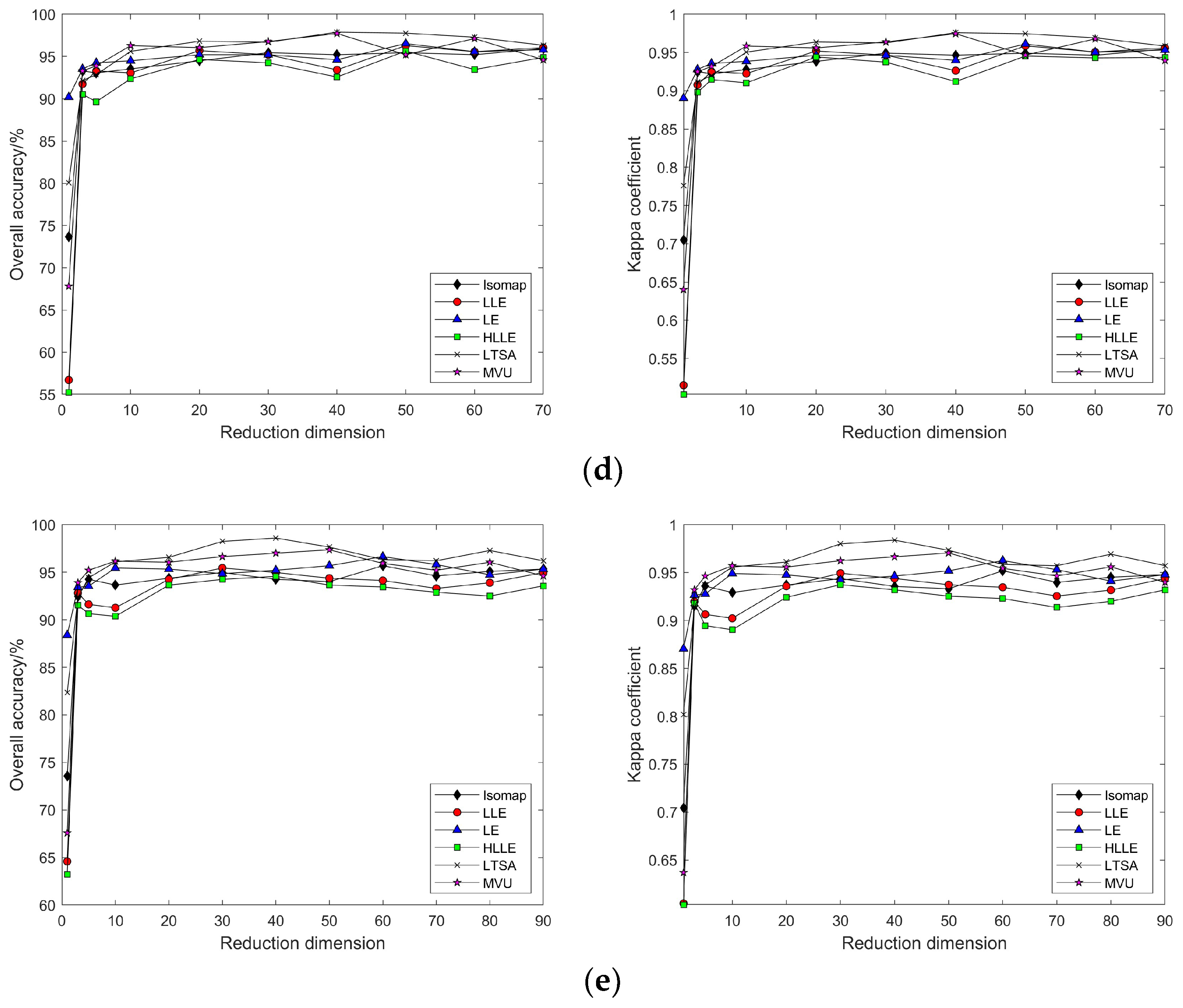
3.2.2. Nonlinear Manifold Learning Dimensionality Reduction in Hyperspectral Imagery
- Comparison of classification accuracy in nonlinear manifold learning
- 2.
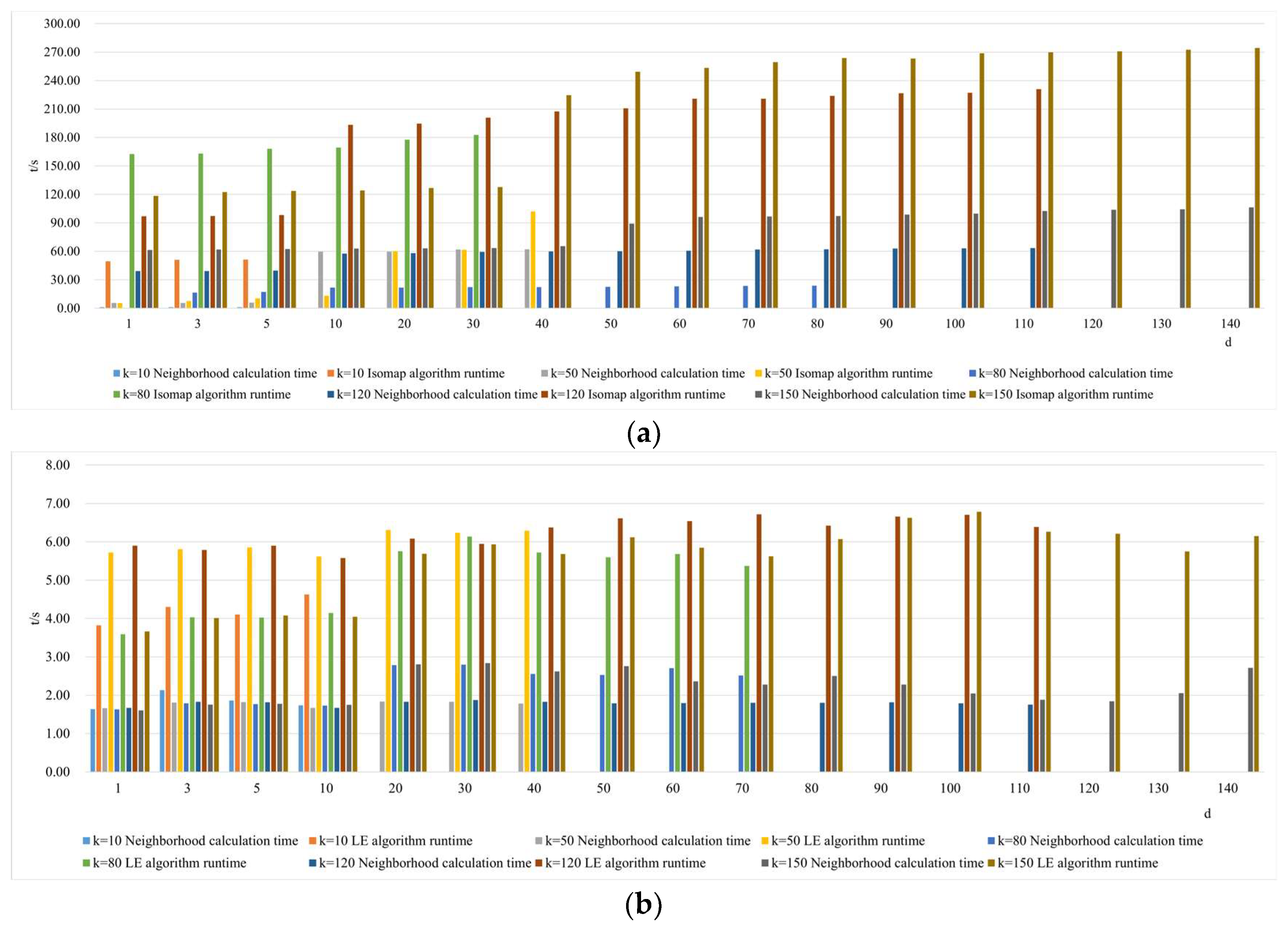
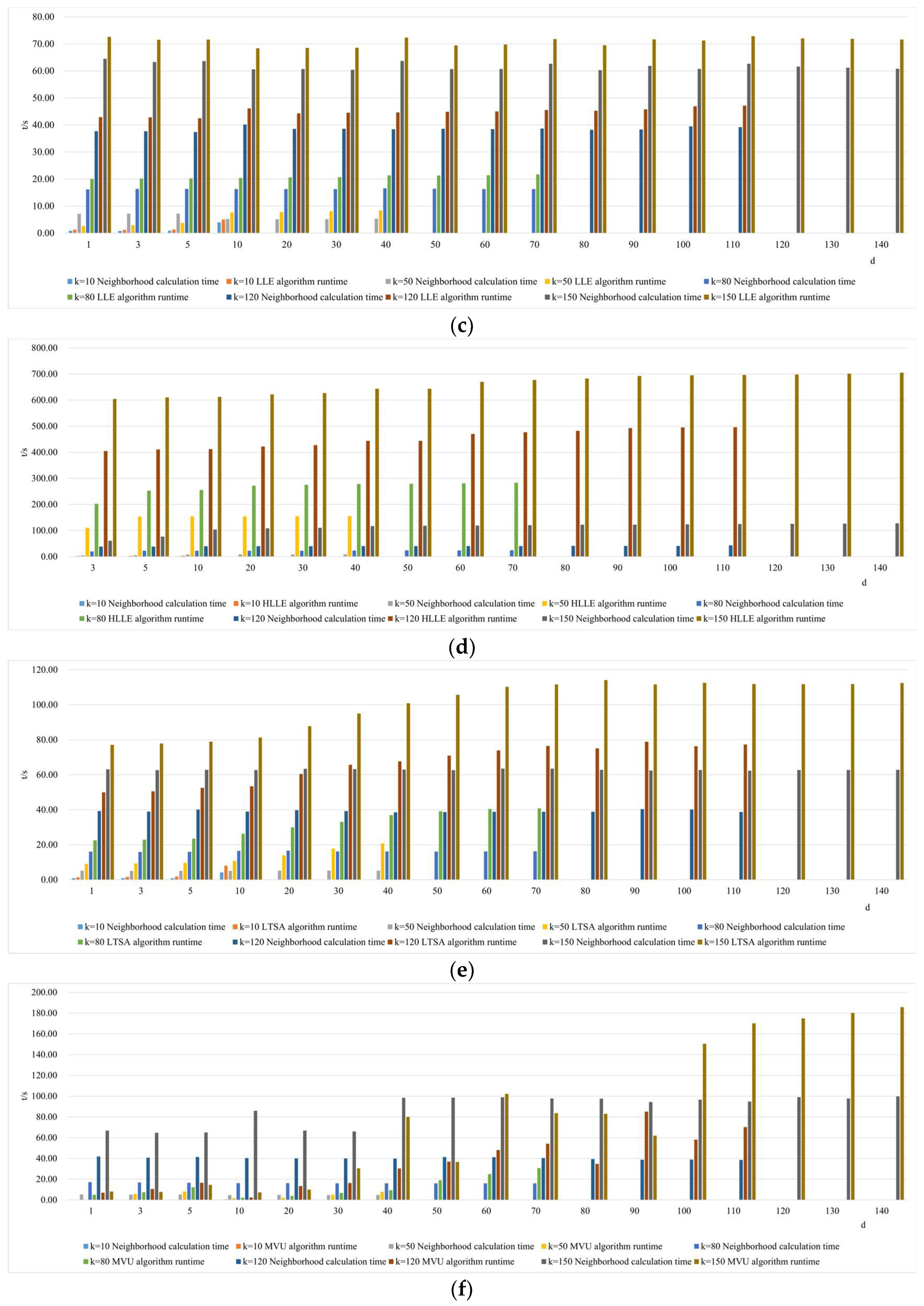
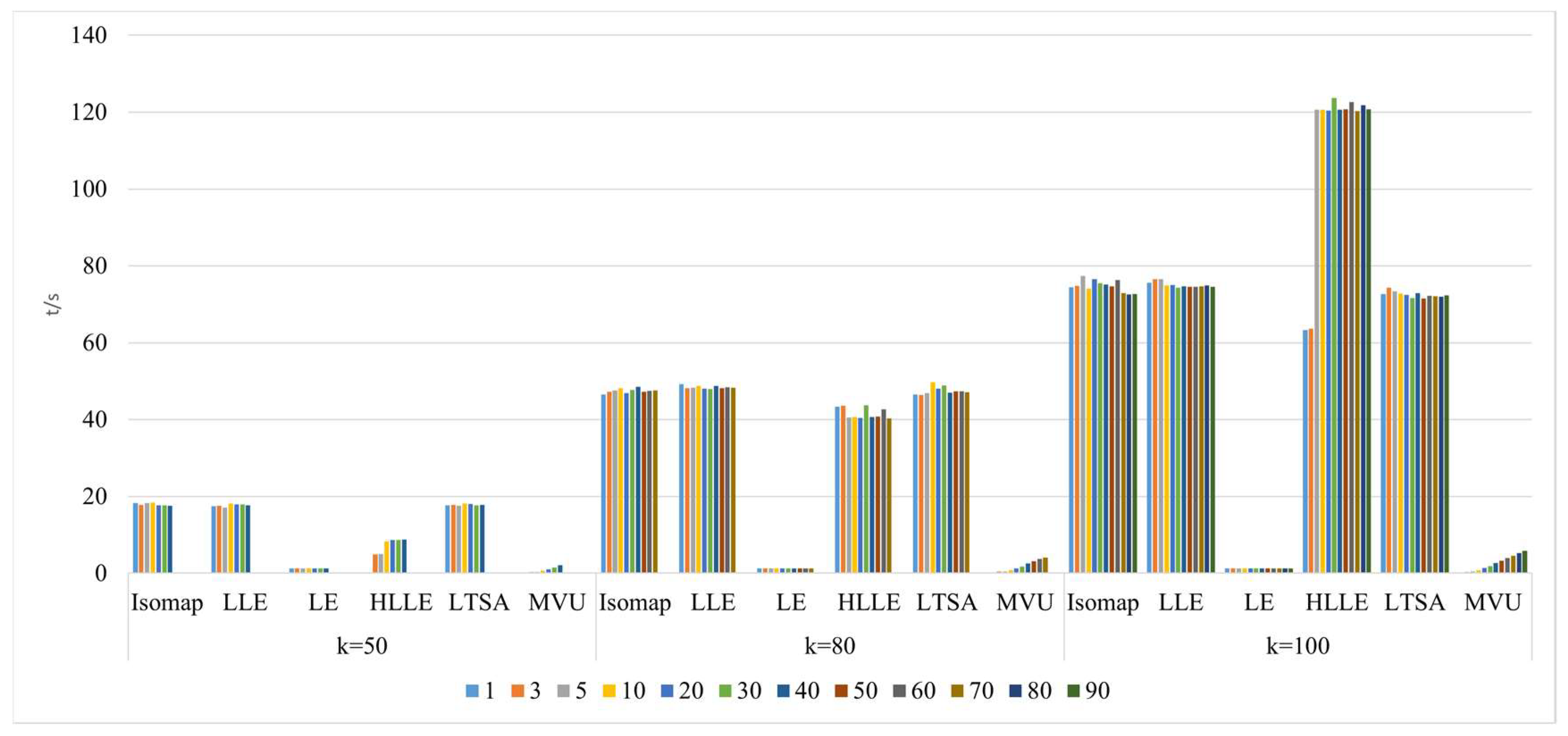
- Comparison of neighborhood computation time in nonlinear manifold learning
- 3.
- Comparison of algorithm runtime in nonlinear manifold learning
- 4.
- Comparison of neighborhood and algorithm runtime in nonlinear manifold learning
3.3. Experimental Results and Comparative Analysis Based on the Pavia University Dataset
3.3.1. Linear Manifold Learning Dimensionality Reduction in Hyperspectral Imagery
3.3.2. Nonlinear Manifold Learning Dimensionality Reduction in Hyperspectral Imagery
- Comparison of classification accuracy in nonlinear manifold learning
- 2.
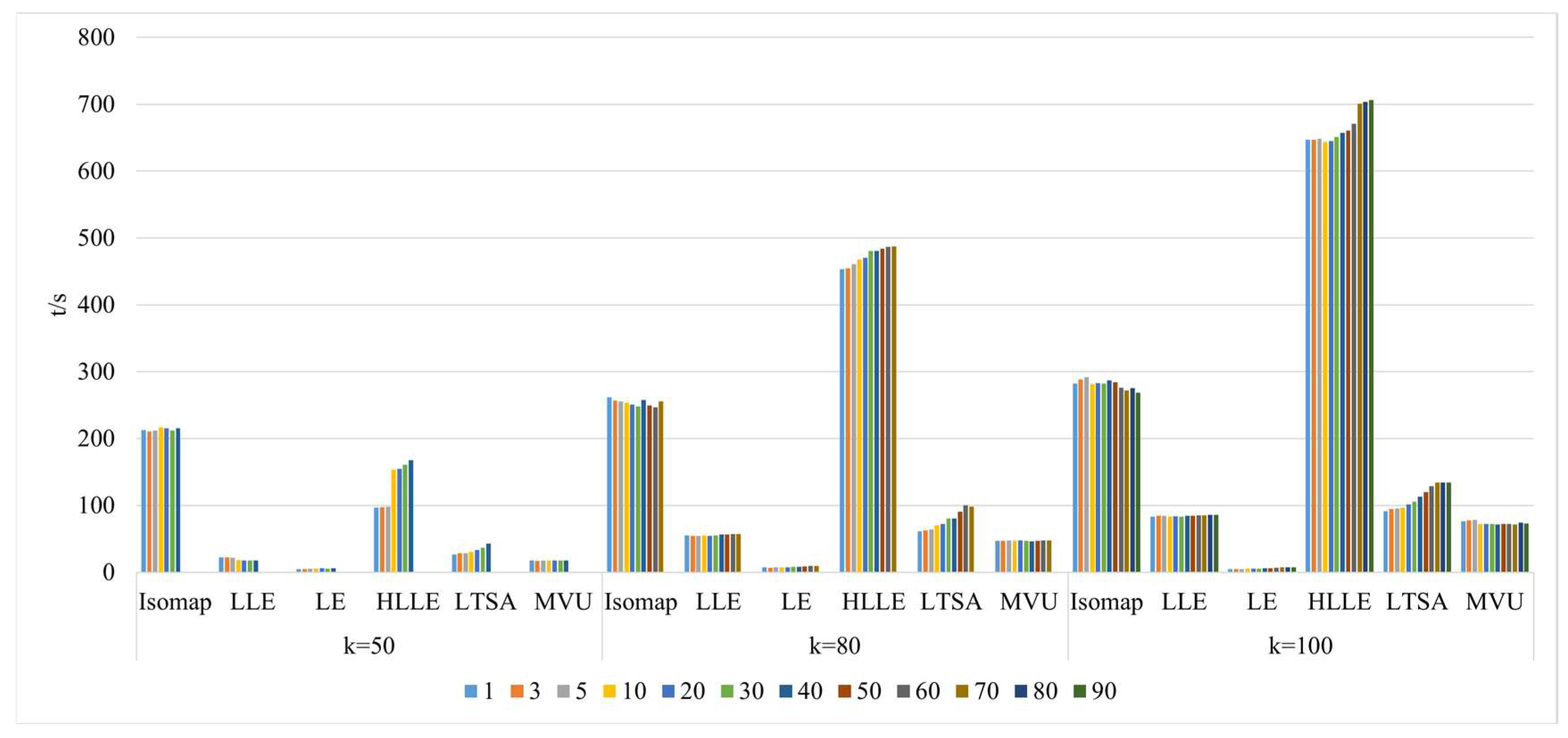
- Comparison of neighborhood computation time in nonlinear manifold learning
- 3.
- Comparison of algorithm runtime in nonlinear manifold learning
4. Discussion
4.1. Discussion on the Visualization of Hyperspectral Image Dimensionality Reduction Using Manifold Learning Methods
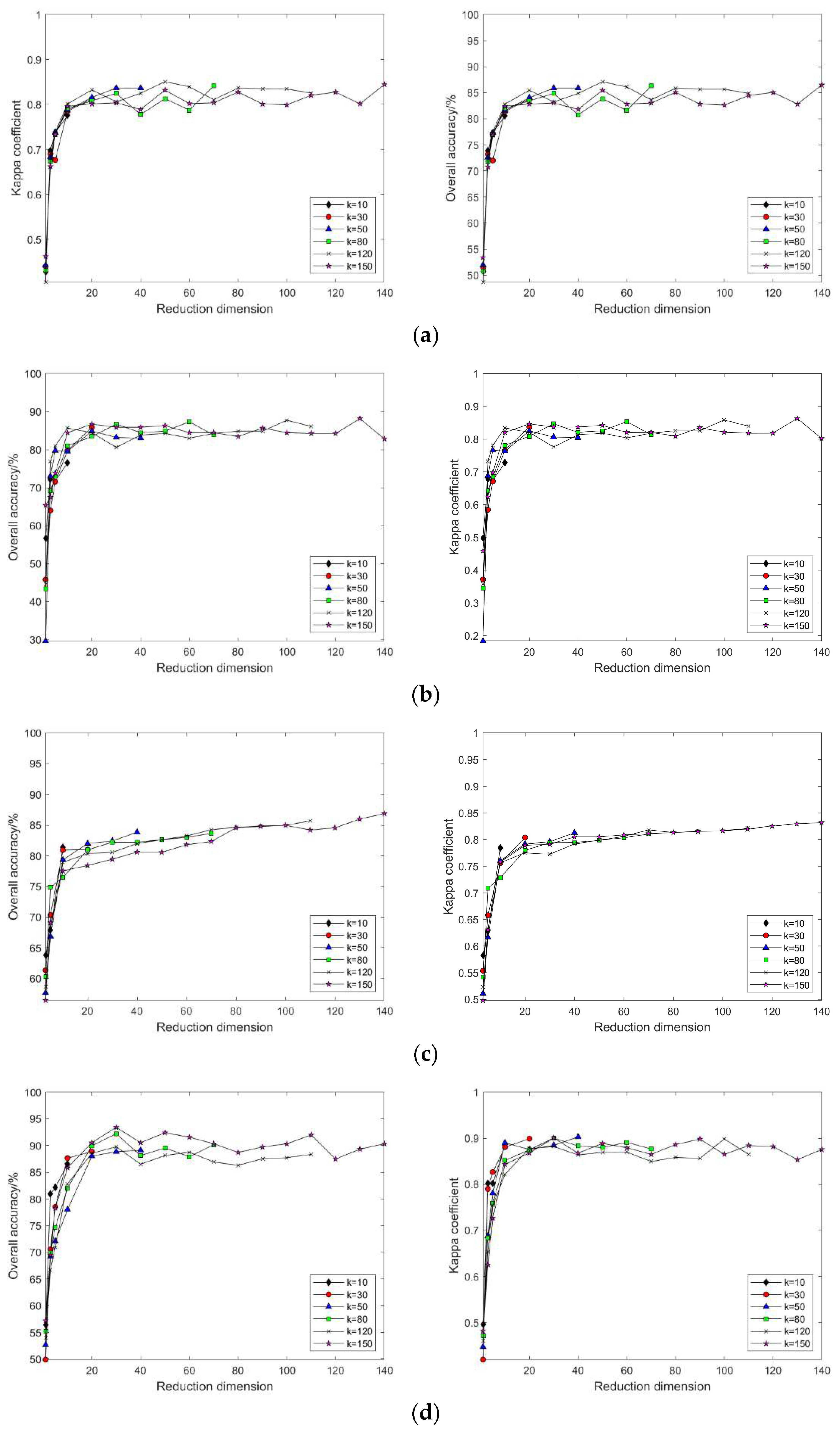
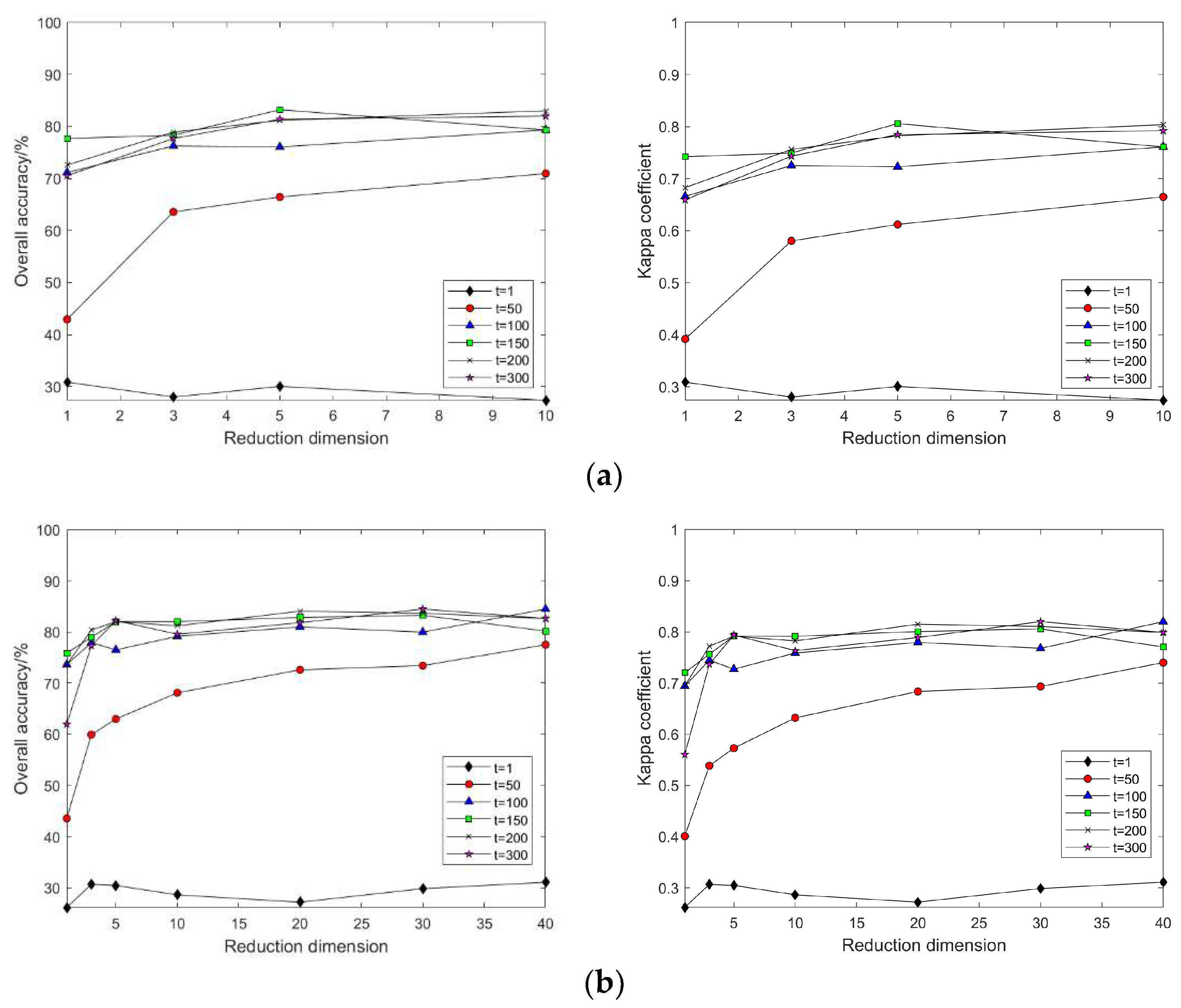
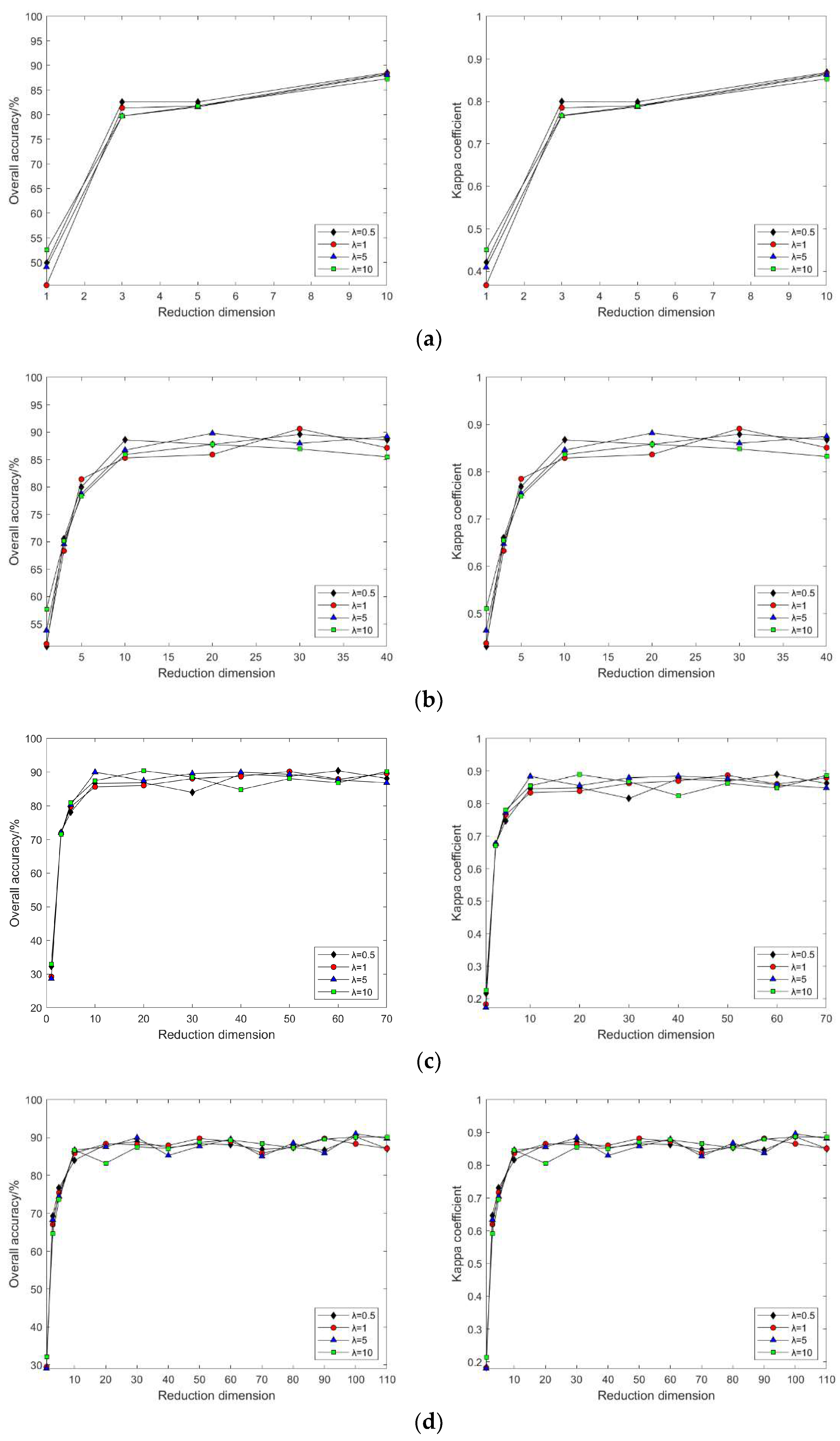
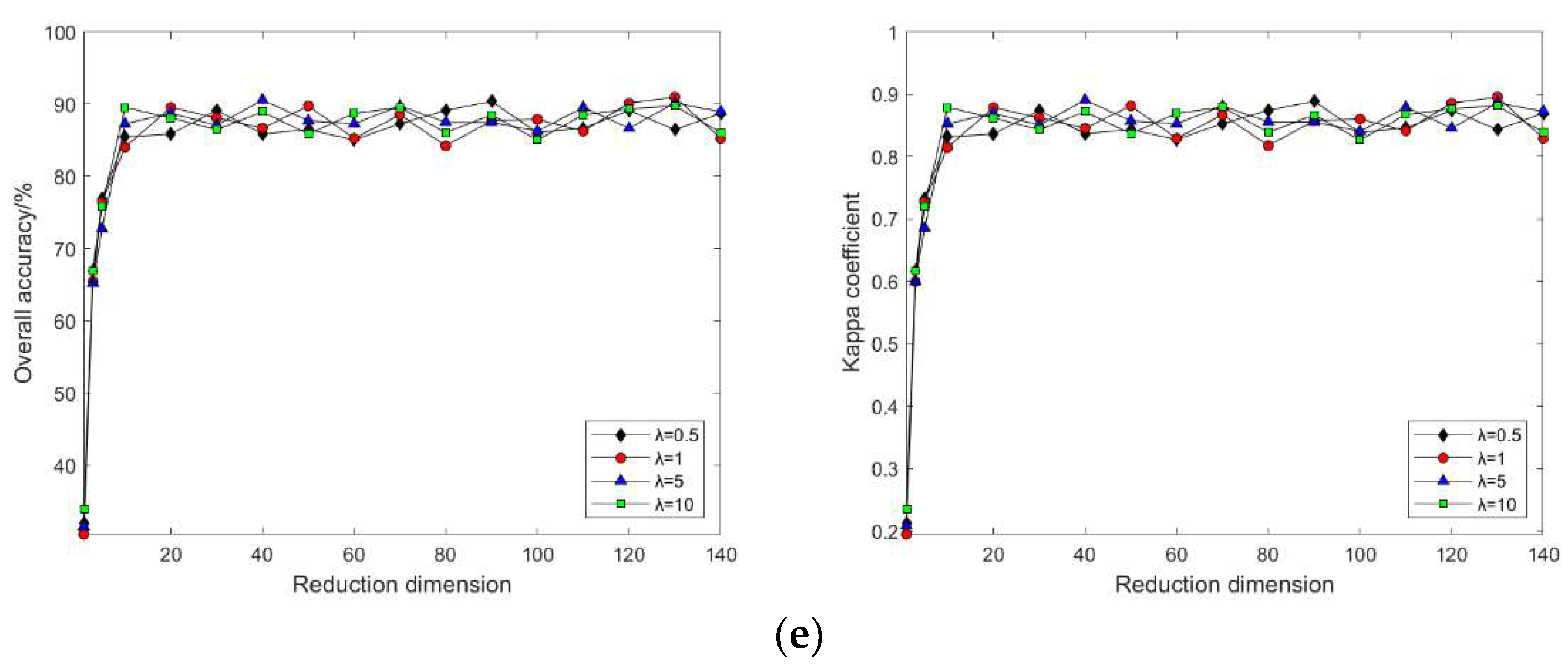
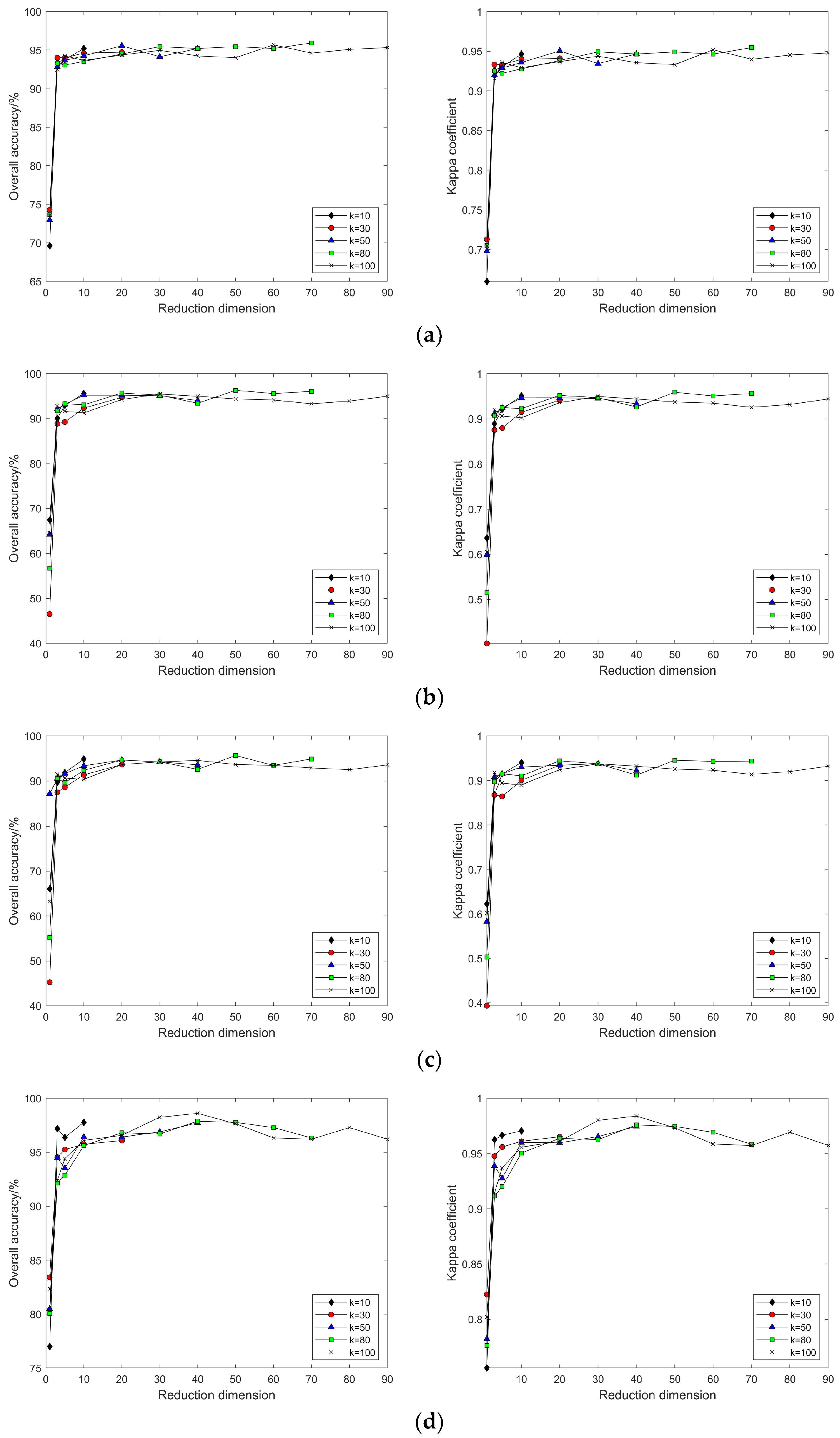
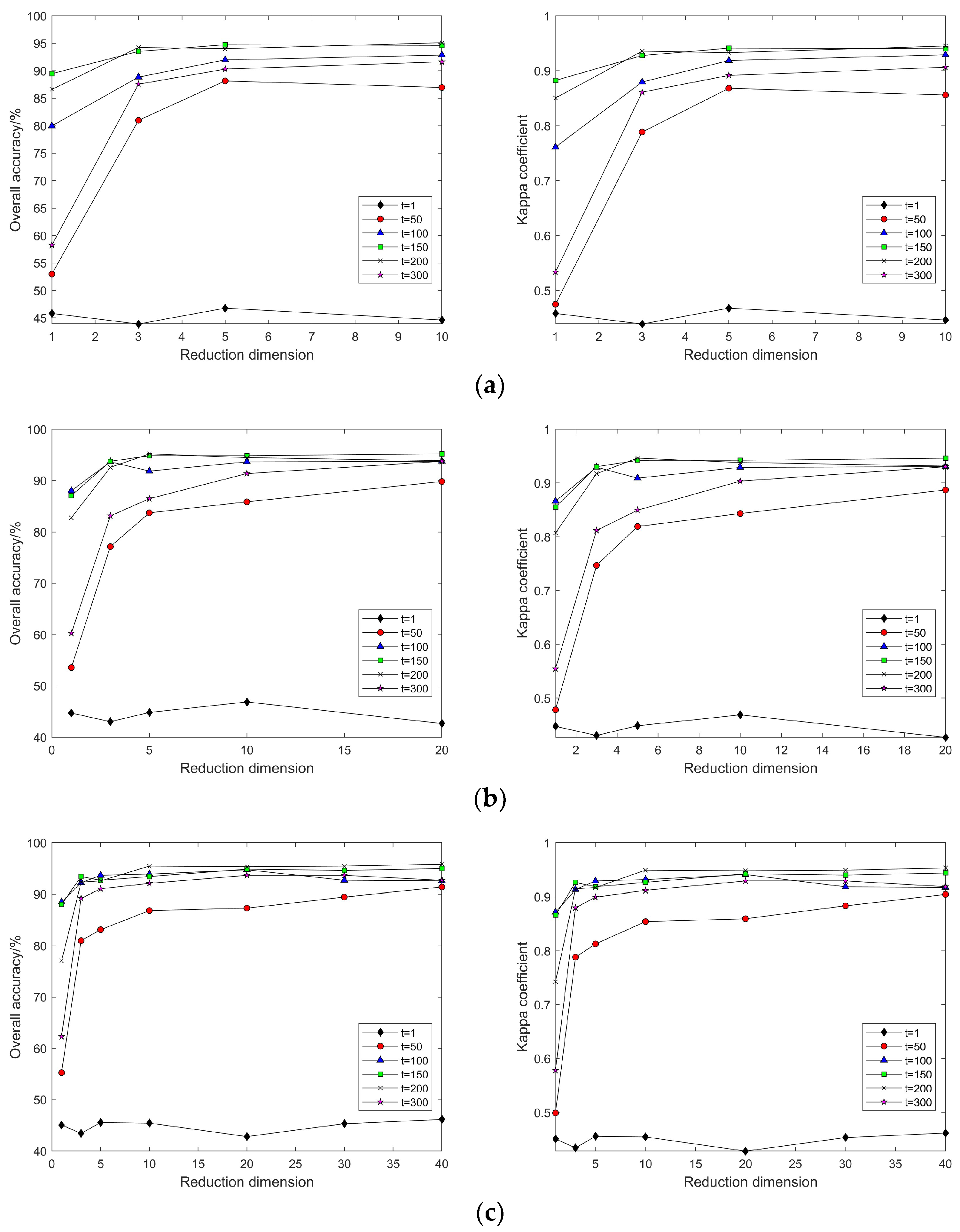
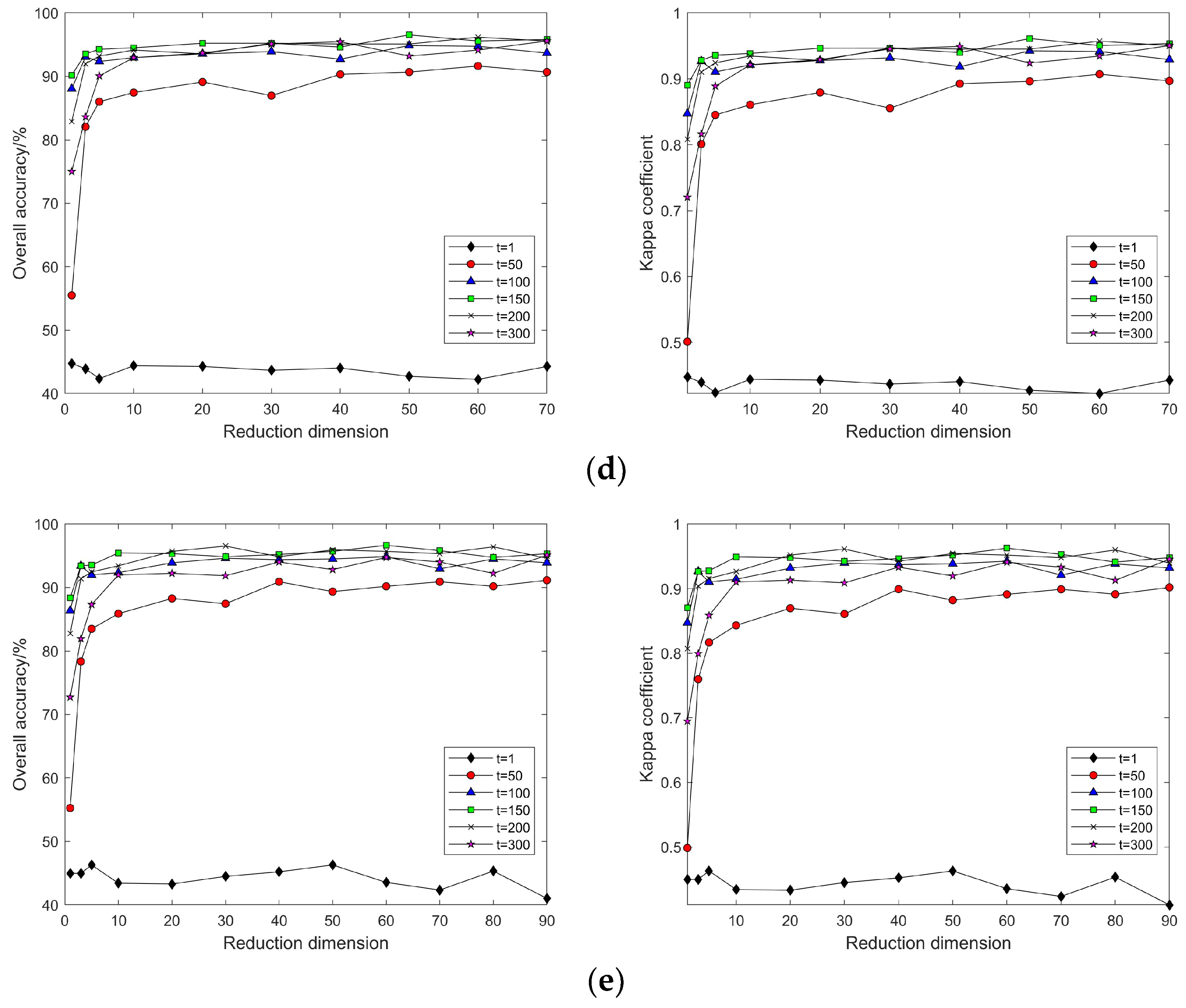
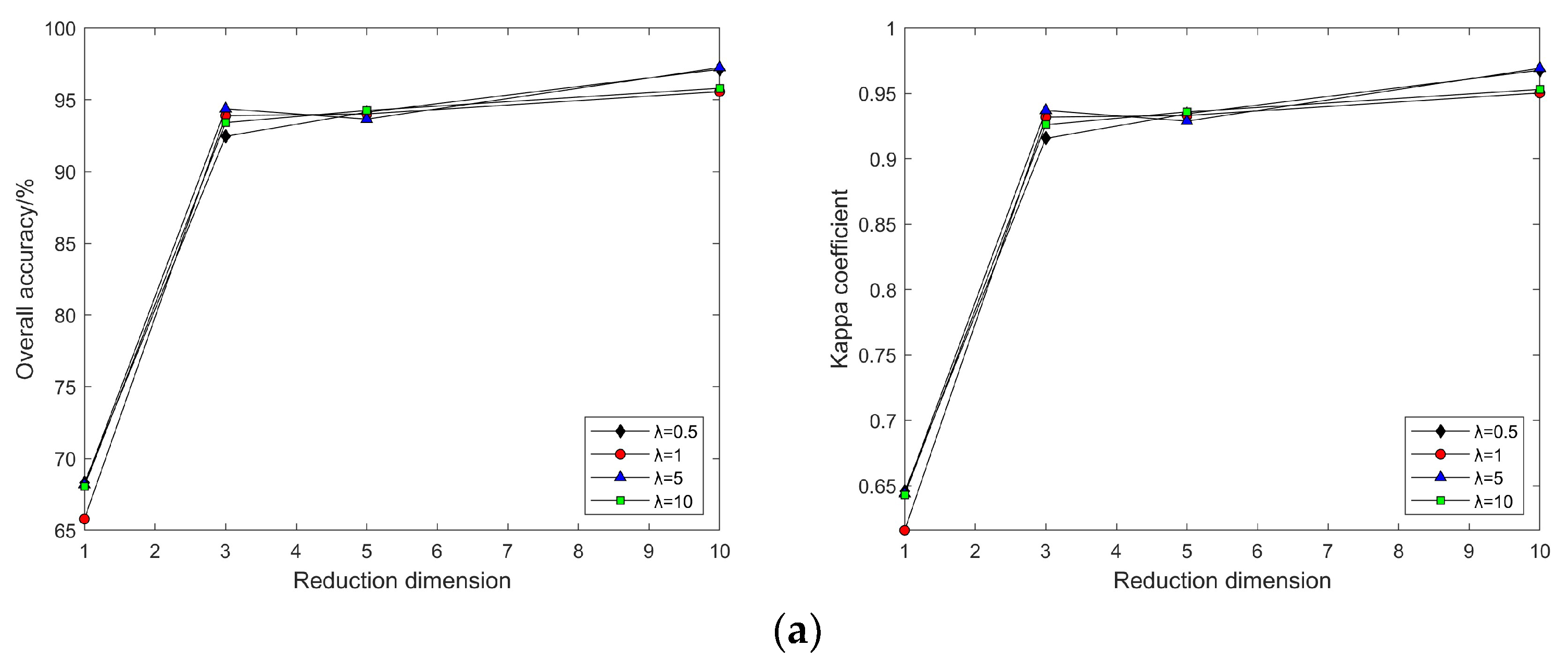
4.2. Impact of Different Parameters on the Results of Feature Extraction from Hyperspectral Images Using Manifold Learning
4.3. The Impact of Intrinsic Dimension d and Neighborhood k on Neighborhood Computation and Overall Algorithm Runtime
4.4. Discussion on Other Widely Researched Hyperspectral Image Feature Extraction Methods
4.5. Uncertainties, Limitations, and Future Direction
4.5.1. Uncertainties
4.5.2. Limitations
4.5.3. Future Direction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, P.; Xia, J.; Xue, C.; Tan, K.; Su, H.; Bao, R. Research Progress on Hyperspectral Remote Sensing Image Classification. J. Remote Sens. 2016, 20, 236–256. [Google Scholar]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.-S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep Learning in Remote Sensing: A Comprehensive Review and List of Resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Du, B. Deep Learning for Remote Sensing Data: A Technical Tutorial on the State of the Art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Tong, Q.X.; Zhang, B.; Zheng, L.F. Hyperspectral Remote Sensing: The Principle, Technology, and Application; Higher Education Press: Beijing, China, 2006. [Google Scholar]
- Chang, C.I. Hyperspectral Imaging: Techniques for Spectral Detection and Classification; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Landgrebe, D.A. Signal Theory Methods in Multispectral Remote Sensing; John Wiley & Sons: New York, NY, USA, 2003. [Google Scholar]
- Zhang, G.; Jia, X.; Hu, J. Superpixel-Based Graphical Model for Remote Sensing Image Mapping. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5861–5871. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spectral–Spatial Classification of Hyperspectral Images with a Superpixel-Based Discriminative Sparse Model. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4186–4201. [Google Scholar] [CrossRef]
- Yuan, H.; Tang, Y.Y. Learning with Hypergraph for Hyperspectral Image Feature Extraction. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1695–1699. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Tao, D.; Huang, X. A Modified Stochastic Neighbor Embedding for Multi-Feature Dimension Reduction of Remote Sensing Images. ISPRS J. Photogramm. Remote Sens. 2013, 83, 30–39. [Google Scholar] [CrossRef]
- Hughes, G. On the Mean Accuracy of Statistical Pattern Recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Yuan, H.; Li, L. Manifold-Based Sparse Representation for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7606–7618. [Google Scholar] [CrossRef]
- Luo, F.; Huang, H.; Liu, J.; Ma, Z. Fusion of Graph Embedding and Sparse Representation for Feature Extraction and Classification of Hyperspectral Imagery. Photogramm. Eng. Remote Sens. 2017, 83, 37–46. [Google Scholar] [CrossRef]
- Shao, Y.; Lan, J. A Spectral Unmixing Method by Maximum Margin Criterion and Derivative Weights to Address Spectral Variability in Hyperspectral Imagery. Remote Sens. 2019, 11, 1045. [Google Scholar] [CrossRef]
- Song, X.; Jiang, X.; Gao, J.; Cai, Z. Gaussian Process Graph-Based Discriminant Analysis for Hyperspectral Images Classification. Remote Sens. 2019, 11, 2288. [Google Scholar] [CrossRef]
- Xie, Z.; Xu, H.; Huang, Q.; Wang, P. Detection of Spinach Freshness Based on Hyperspectral Imaging and Deep Learning. Trans. Chin. Soc. Agric. Eng. 2019, 35, 277–284. [Google Scholar]
- Goodin, D.G.; Gao, J.; Henebry, G.M. The Effect of Solar Illumination Angle and Sensor View Angle on Observed Patterns of Spatial Structure in Tallgrass Prairie. IEEE Trans. Geosci. Remote Sens. 2004, 42, 154–165. [Google Scholar] [CrossRef]
- Sandmeier, S.R.; Middleton, E.M.; Deering, D.W.; Qin, W. The Potential of Hyperspectral Bidirectional Reflectance Distribution Function Data for Grass Canopy Characterization. J. Geophys. Res. Atmos. 1999, 104, 9547–9560. [Google Scholar] [CrossRef]
- Mobley, C.D. Light and Water: Radiative Transfer in Natural Waters; Academic Press: California, CA, USA, 1994. [Google Scholar]
- Keshava, N.; Mustard, J.F. Spectral Unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Roberts, D.A.; Smith, M.O.; Adams, J.B. Green Vegetation, Nonphotosynthetic Vegetation, and Soils in AVIRIS Data. Remote Sens. Environ. 1993, 44, 255–269. [Google Scholar] [CrossRef]
- Bachmann, C.M.; Ainsworth, T.L.; Fusina, R.A. Exploiting Manifold Geometry in Hyperspectral Imagery. IEEE Trans. Geosci. Remote Sens. 2005, 43, 441–454. [Google Scholar] [CrossRef]
- Kégl, B. Intrinsic Dimension Estimation Using Packing Numbers. 2002. Available online: https://dblp.uni-trier.de/db/conf/nips/nips2002.html (accessed on 5 February 2024).
- Bruske, J.; Sommer, G. Intrinsic Dimensionality Estimation with Optimally Topology Preserving Maps. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 572–575. [Google Scholar] [CrossRef]
- Camastra, F.; Vinciarelli, A. Estimating the Intrinsic Dimension of Data with a Fractal-Based Method. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 1404–1407. [Google Scholar] [CrossRef]
- Costa, J.A.; Hero, A.O. Manifold Learning Using Euclidean k-Nearest Neighbor Graphs. In Proceedings of the 2004 IEEE International Conference on Acoustics, Speech, and Signal Processing, Montreal, QC, Canada, 17–21 May 2004. [Google Scholar] [CrossRef]
- Costa, J.A.; Hero, A.O. Geodesic Entropic Graphs for Dimension and Entropy Estimation in Manifold Learning. IEEE Trans. Signal Process. 2004, 52, 2210–2221. [Google Scholar] [CrossRef]
- Fukunaga, K.; Olsen, D.R. An Algorithm for Finding Intrinsic Dimensionality of Data. IEEE Trans. Comput. 1971, C-20, 176–183. [Google Scholar] [CrossRef]
- Levina, E.; Bickel, P.J. Maximum Likelihood Estimation of Intrinsic Dimension. In Proceedings of the Advances in Neural Information Processing Systems 17 (NIPS 2004), Vancouver, BC, Canada, 13–18 December 2004; MIT Press: Cambridge, MA, USA, 2004. [Google Scholar]
- Pettis, K.W.; Bailey, T.A.; Jain, A.K.; Dubes, R.C. An Intrinsic Dimensionality Estimator from Near-Neighbor Information. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 25–37. [Google Scholar] [CrossRef]
- Verveer, P.J.; Duin, R.P.W. An Evaluation of Intrinsic Dimensionality Estimators. IEEE Trans. Pattern Anal. Mach. Intell. 1995, 17, 81–86. [Google Scholar] [CrossRef]
- Gu, Y.F.; Liu, Y.; Zhang, Y. A Selective KPCA Algorithm Based on High-Order Statistics for Anomaly Detection in Hyperspectral Imagery. IEEE Geosci. Remote Sens. Lett. 2008, 5, 43–47. [Google Scholar] [CrossRef]
- Xia, J.S.; Falco, N.; Benediktsson, J.A.; Du, P.J.; Chanussot, J. Hyperspectral Image Classification with Rotation Random Forest via KPCA. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 1601–1609. [Google Scholar] [CrossRef]
- Tenenbaum, J.B.; Silva, V.D.; Langford, J.C. A Global Geometric Framework for Nonlinear Dimensionality Reduction. Science 2000, 290, 2319–2323. [Google Scholar] [CrossRef]
- Roweis, S.; Saul, L. Nonlinear Dimensionality Reduction by Locally Linear Embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef]
- Belkin, M.; Niyogi, P. Laplacian Eigenmaps for Dimensionality Reduction and Data Representation. Neural Comput. 2003, 15, 1373–1396. [Google Scholar] [CrossRef]
- Donoho, D.; Grimes, C. Hessian Locally Linear Embedding: New Tools for Nonlinear Dimensionality Reduction. Proc. Natl. Acad. Sci. USA 2003, 100, 5591–5596. [Google Scholar] [CrossRef]
- Zhang, Z.; Zha, H. Principal Manifolds and Nonlinear Dimensionality Reduction via Tangent Space Alignment. SIAM J. Sci. Comput. 2004, 26, 313–338. [Google Scholar] [CrossRef]
- Weinberger, K.Q.; Saul, L.K. An Introduction to Nonlinear Dimensionality Reduction by Maximum Variance Unfolding. In Proceedings of the Twenty-First National Conference on Artificial Intelligence (AAAI-06), Boston, MA, USA, 16–20 July 2006; pp. 1683–1686. [Google Scholar]
- Weinberger, K.Q.; Saul, L.K. Unsupervised Learning of Image Manifolds by Semidefinite Programming. In Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR-04), Washington, DC, USA, 27 June–2 July 2004; pp. 988–995. [Google Scholar] [CrossRef]
- Lee, J.A.; Verleysen, M. Nonlinear Dimensionality Reduction; Springer: New York, NY, USA, 2007. [Google Scholar]
- Venna, J. Dimensionality Reduction for Visual Exploration of Similarity Structures; Helsinki University of Technology: Helsinki, Finland, 2007. [Google Scholar]
- Zhang, T.; Yang, J.; Zhao, D.; Ge, X.; Liu, C. Linear Local Tangent Space Alignment and Application to Face Recognition. Neurocomputing 2007, 70, 1547–1553. [Google Scholar] [CrossRef]
- Li, W.; Prasad, S.; Fowler, J.E.; Bruce, L.M. Locality-Preserving Dimensionality Reduction and Classification for Hyperspectral Image Analysis. IEEE Trans. Geosci. Remote Sens. 2011, 50, 1185–1198. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Tao, D.; Huang, X. On Combining Multiple Features for Hyperspectral Remote Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2011, 50, 879–893. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Tao, D.; Huang, X. Tensor Discriminative Locality Alignment for Hyperspectral Image Spectral–Spatial Feature Extraction. IEEE Trans. Geosci. Remote Sens. 2012, 51, 242–256. [Google Scholar] [CrossRef]






| Category | Classification | Color | Samples |
|---|---|---|---|
| 1 | Alfalfa | 46 | |
| 2 | Corn-notill | 1428 | |
| 3 | Corn-mintill | 830 | |
| 4 | Corn | 237 | |
| 5 | Grass-pasture | 483 | |
| 6 | Grass-tree | 730 | |
| 7 | Grass-pasture-mowed | 28 | |
| 8 | Hay-windrowed | 478 | |
| 9 | Oats | 20 | |
| 10 | Soybean-notill | 972 | |
| 11 | Soybean-mintill | 2455 | |
| 12 | Soybean-clean | 593 | |
| 13 | Wheat | 205 | |
| 14 | Woods | 1265 | |
| 15 | Buildings-Grass-Tress-Drives | 386 | |
| 16 | Stone-Steel-Towers | 93 |
| Category | Classification | Color | Samples |
|---|---|---|---|
| 1 | Asphalt | 6631 | |
| 2 | Meadows | 18,649 | |
| 3 | Gravel | 2099 | |
| 4 | Trees | 3064 | |
| 5 | Painted metal sheets | 1345 | |
| 6 | Bare Soil | 5029 | |
| 7 | Bitumen | 1330 | |
| 8 | Self-Blocking Bricks | 3682 | |
| 9 | Stone-Steel-Towers | 947 |
| Indian Pines Dataset | Pavia University Dataset | ||||||
|---|---|---|---|---|---|---|---|
| Category | Classification | Total Number of Samples | Number of Samples Selected | Category | Classification | Total Number of Samples | Number of Samples Selected |
| 2 | Corn-notill | 1428 | 428 | 1 | Asphalt | 6631 | 663 |
| 3 | Corn-mintill | 830 | 249 | 2 | Meadows | 18,649 | 1865 |
| 5 | Grass-pasture | 483 | 145 | 3 | Gravel | 2099 | 210 |
| 6 | Grass-tree | 730 | 219 | 4 | Trees | 3064 | 306 |
| 10 | Soybean-notill | 972 | 292 | 5 | Painted metal sheets | 1345 | 135 |
| 11 | Soybean-mintill | 2455 | 737 | 6 | Bare Soil | 5029 | 503 |
| 14 | Woods | 1265 | 380 | 7 | Bitumen | 1330 | 133 |
| —— | —— | —— | —— | 8 | Self-Blocking Bricks | 3682 | 368 |
| Total | —— | 8163 | 2449 | Total | —— | 41,829 | 4183 |
| Indian Pines Dataset | Pavia University Dataset | ||||||
|---|---|---|---|---|---|---|---|
| Category | Classification | Training Set | Validation Set | Category | Classification | Training Set | Validation Set |
| 2 | Corn-notill | 342 | 86 | 1 | Asphalt | 530 | 133 |
| 3 | Corn-mintill | 199 | 50 | 2 | Meadows | 1492 | 373 |
| 5 | Grass-pasture | 116 | 29 | 3 | Gravel | 168 | 42 |
| 6 | Grass-tree | 175 | 44 | 4 | Trees | 245 | 61 |
| 10 | Soybean-notill | 234 | 58 | 5 | Painted metal sheets | 108 | 27 |
| 11 | Soybean-mintill | 590 | 147 | 6 | Bare Soil | 402 | 101 |
| 14 | Woods | 304 | 76 | 7 | Bitumen | 106 | 27 |
| —— | —— | —— | —— | 8 | Self-Blocking Bricks | 295 | 74 |
| Total | —— | 1959 | 490 | Total | —— | 3346 | 837 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, W.; Zhang, X.; Yang, G.; Chen, Y.; Wang, L.; Xu, H. A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning. Sensors 2024, 24, 2089. https://doi.org/10.3390/s24072089
Song W, Zhang X, Yang G, Chen Y, Wang L, Xu H. A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning. Sensors. 2024; 24(7):2089. https://doi.org/10.3390/s24072089
Chicago/Turabian StyleSong, Wenhui, Xin Zhang, Guozhu Yang, Yijin Chen, Lianchao Wang, and Hanghang Xu. 2024. "A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning" Sensors 24, no. 7: 2089. https://doi.org/10.3390/s24072089
APA StyleSong, W., Zhang, X., Yang, G., Chen, Y., Wang, L., & Xu, H. (2024). A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning. Sensors, 24(7), 2089. https://doi.org/10.3390/s24072089






