A Handheld Laser-Scanning-Based Methodology for Monitoring Tree Growth in Chestnut Orchards
Abstract
1. Introduction
2. Materials and Methods
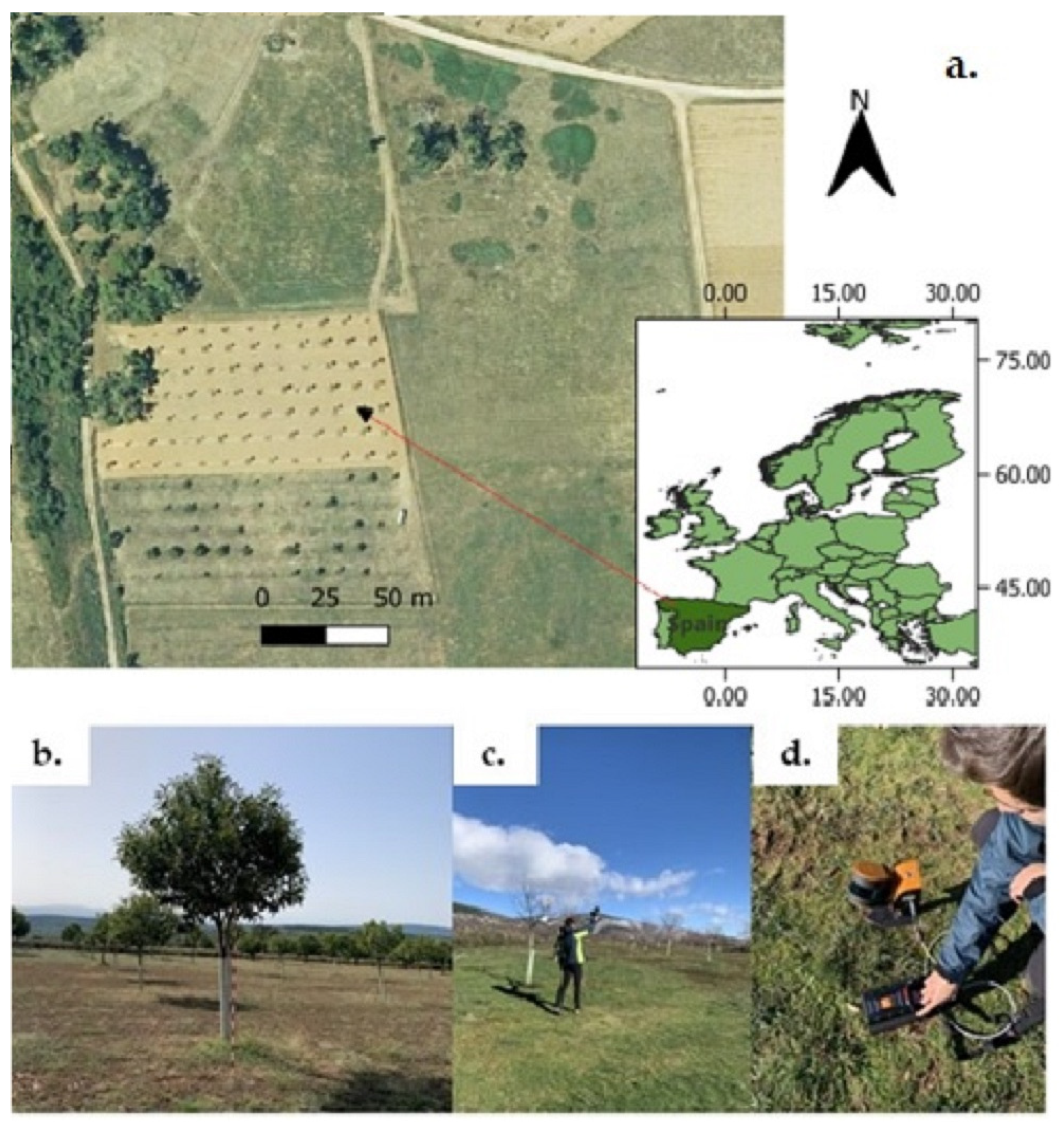
2.1. Study Site and MLS Data Acquisition
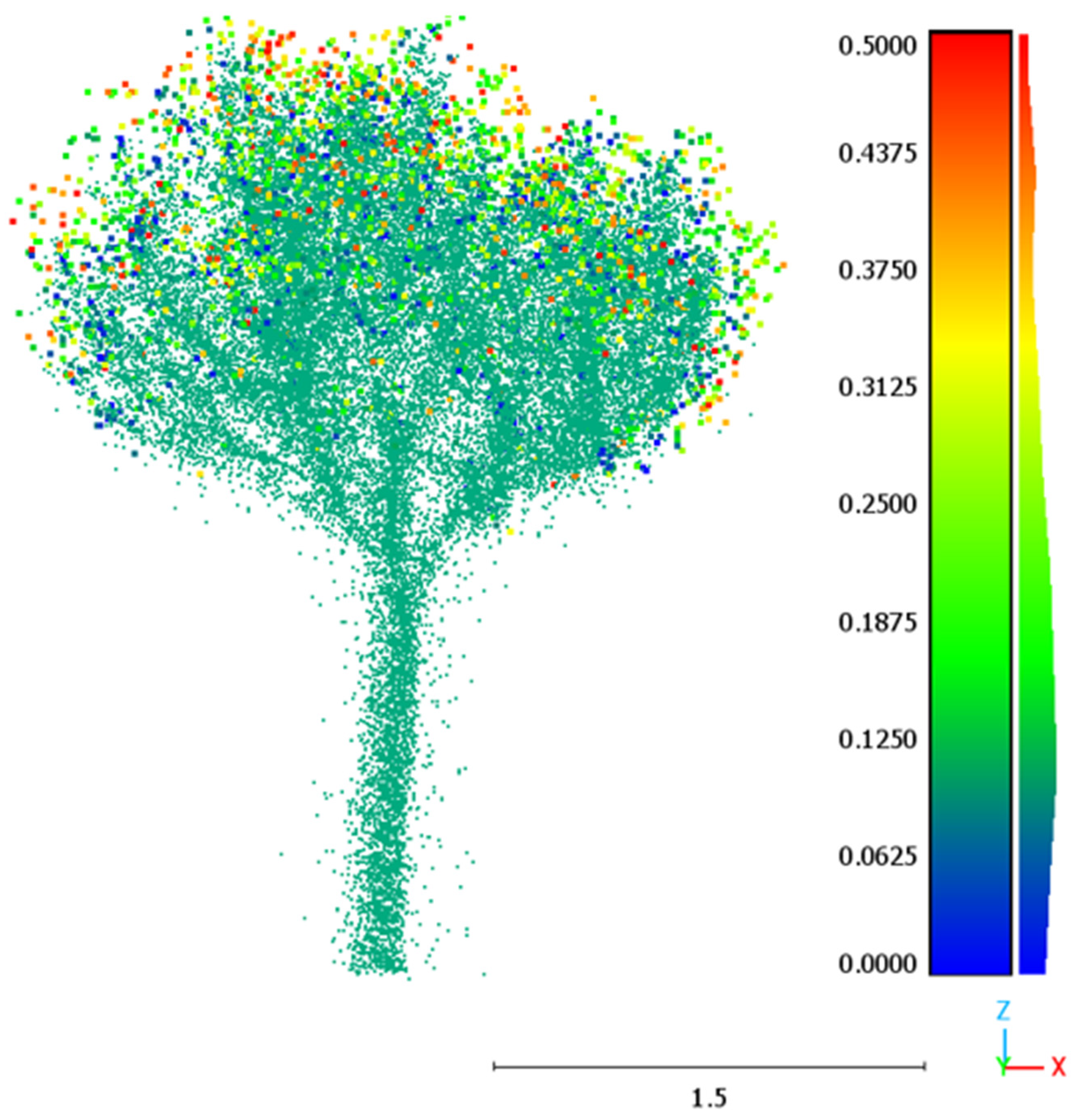
2.2. Tree Point Cloud Processing
- Transformation of the tree point clouds from Cartesian to polar coordinates with an origin in the center of mass of each tree. As explained above, this transformation allows to leverage the radial structure of the branches to estimate their length.
- Point cloud discretization in radial sections to calculate branch length. This discretization makes it easier to detect the ends of the branches.
- Comparison of branch ends measured on two different dates to estimate tree growth over time. In essence, the comparison is carried out by subtracting matrices whose elements are distances to the central point.
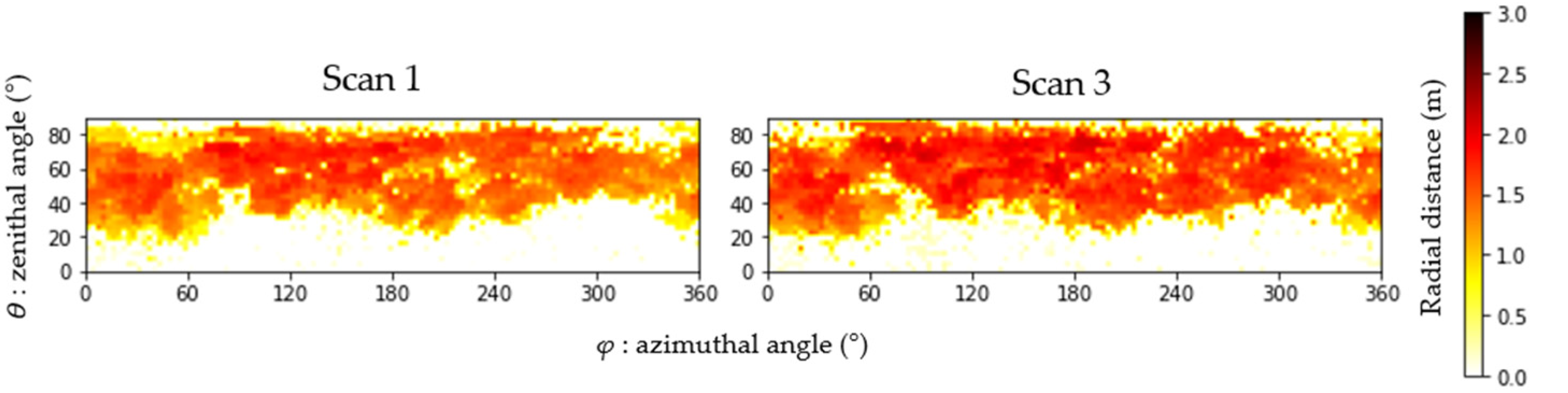
2.2.1. Radial Distance and Branch End Estimation
2.2.2. Growth Estimation
3. Results and Discussion
3.1. Validation
3.2. Growth Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guineacute:, R.P.F.; Costa, C.; Florenccedila, S.G.; Correia, P.M.R. A Review of the Use of Chestnut in Traditional and Innovative Food Products. J. Nuts 2022, 14, 1–8. [Google Scholar] [CrossRef]
- Fernández-Cruz, J.; Míguez-Soto, B.; Fernández-López, J. Origin of Traditional Sweet Chestnut (Castanea Sativa Mill.) Varieties from the Northwest of the Iberian Peninsula. Tree Genet. Genomes 2022, 18, 34. [Google Scholar] [CrossRef]
- Fernandes, P.; Colavolpe, M.B.; Serrazina, S.; Costa, R.L. European and American Chestnuts: An Overview of the Main Threats and Control Efforts. Front. Plant Sci. 2022, 13, 951844. [Google Scholar] [CrossRef] [PubMed]
- Aglietti, C.; Cappelli, A.; Andreani, A. From Chestnut Tree (Castanea sativa) to Flour and Foods: A Systematic Review of the Main Criticalities and Control Strategies towards the Relaunch of Chestnut Production Chain. Sustainability 2022, 14, 12181. [Google Scholar] [CrossRef]
- Rivera, G.; Porras, R.; Florencia, R.; Sánchez-Solís, J.P. LiDAR Applications in Precision Agriculture for Cultivating Crops: A Review of Recent Advances. Comput. Electron. Agric. 2023, 207, 107737. [Google Scholar] [CrossRef]
- Özdemir, S.; Akbulut, Z.; Karslı, F.; Acar, H. Automatic Extraction Of Trees By Using Multiple Return Properties of The Lidar Point Cloud. Int. J. Eng. Geosci. 2021, 6, 20–26. [Google Scholar] [CrossRef]
- Canaz Sevgen, S.; Karsli, F. Automatıc Ground Extractıon For Urban Areas From Aırborne Lıdar Data. Turk. J. Eng. 2020, 4, 113–122. [Google Scholar] [CrossRef]
- Ozdarici-ok, A.; Ok, A.O. Using Remote Sensing to Identify Individual Tree Species in Orchards: A Review. Sci. Hortic. 2023, 321, 112333. [Google Scholar] [CrossRef]
- Pádua, L.; Marques, P.; Martins, L.; Sousa, A.; Peres, E.; Sousa, J.J. Monitoring of Chestnut Trees Using Machine Learning Techniques Applied to UAV-Based Multispectral Data. Remote Sens. 2020, 12, 3032. [Google Scholar] [CrossRef]
- DİAZ, B.S.; Mata-Zayas, E.E.; Gama-Campillo, L.M.; Rincon-Ramirez, J.A.; Vidal-Garcia, F.; Rullan-Silva, C.D.; Sanchez-Gutierrez, F. Lidar Modeling to Determine the Height of Shade Canopy Tree in Cocoa Agrosystems as Available Habitat for Wildlife. Int. J. Eng. Geosci. 2022, 7, 283–293. [Google Scholar] [CrossRef]
- Liang, X.; Kukko, A.; Balenovic, I.; Saarinen, N.; Junttila, S.; Kankare, V.; Holopainen, M.; Mokros, M.; Surovy, P.; Kaartinen, H.; et al. Close-Range Remote Sensing of Forests: The State of the Art, Challenges, and Opportunities for Systems and Data Acquisitions. IEEE Geosci. Remote Sens. Mag. 2022, 10, 32–71. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International Benchmarking of Terrestrial Laser Scanning Approaches for Forest Inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Schindler, Z.; Morhart, C.; Sheppard, J.P.; Frey, J.; Seifert, T. In a Nutshell: Exploring Single Tree Parameters and above-Ground Carbon Sequestration Potential of Common Walnut (Juglans regia L.) in Agroforestry Systems. Agrofor. Syst. 2023, 97, 1007–1024. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Bailey, B.N.; Fu, K.; Shackel, K. Topological and Spatial Analysis of Within-Tree Fruiting Characteristics for Walnut Trees. Sci. Hortic. 2023, 318, 112127. [Google Scholar] [CrossRef]
- Schindler, Z.; Seifert, T.; Sheppard, J.P.; Morhart, C. Allometric Models for Above-Ground Biomass, Carbon and Nutrient Content of Wild Cherry (Prunus avium L.) Trees in Agroforestry Systems. Ann. For. Sci. 2023, 80, 28. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Escolà, A.; Isabel de Castro, A.; López-Granados, F.; Rosell-Polo, J.R.; Sebé, F.; Manuel Jiménez-Brenes, F.; Sanz, R.; Gregorio, E.; Peña, J.M. Mobile Terrestrial Laser Scanner vs. UAV Photogrammetry to Estimate Woody Crop Canopy Parameters—Part 2: Comparison for Different Crops and Training Systems. Comput. Electron. Agric. 2023, 212, 108083. [Google Scholar] [CrossRef]
- Alonso, L.; Picos, J.; Bastos, G.; Armesto, J. Detection of Very Small Tree Plantations and Tree-Level Characterization Using Open-Access Remote-Sensing Databases. Remote Sens. 2020, 12, 2276. [Google Scholar] [CrossRef]
- Arakawa, T.; Tanaka, T.S.T.; Kamio, S. Detection of On-Tree Chestnut Fruits Using Deep Learning and RGB UAV Imagery for Estimation of Yield and Fruit Load. Agron. J. 2023. [Google Scholar] [CrossRef]
- Balestra, M.; Tonelli, E.; Vitali, A.; Urbinati, C.; Frontoni, E.; Pierdicca, R. Geomatic Data Fusion for 3D Tree Modeling: The Case Study of Monumental Chestnut Trees. Remote Sens. 2023, 15, 2197. [Google Scholar] [CrossRef]
- Mokroš, M.; Mikita, T.; Singh, A.; Tomaštík, J.; Chudá, J.; Wężyk, P.; Kuželka, K.; Surový, P.; Klimánek, M.; Zięba-Kulawik, K.; et al. Novel Low-Cost Mobile Mapping Systems for Forest Inventories as Terrestrial Laser Scanning Alternatives. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102512. [Google Scholar] [CrossRef]
- Spadavecchia, C.; Belcore, E.; Grasso, N.; Piras, M. A fully automatic forest parameters extraction at single-tree level: A comparison of mls and tls applications. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2023, XLVIII-1-W1-2023, 457–463. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Junttila, S.; Luoma, V.; Calders, K.; Kankare, V.; Saarinen, N.; Kukko, A.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Capturing Seasonal Radial Growth of Boreal Trees with Terrestrial Laser Scanning. For. Ecol. Manag. 2023, 529, 120733. [Google Scholar] [CrossRef]
- Yrttimaa, T.; Luoma, V.; Saarinen, N.; Kankare, V.; Junttila, S.; Holopainen, M.; Hyyppä, J.; Vastaranta, M. Exploring Tree Growth Allometry Using Two-Date Terrestrial Laser Scanning. For. Ecol. Manag. 2022, 518, 120303. [Google Scholar] [CrossRef]
- CloudCompare. 2022. Available online: https://www.danielgm.net/cc/release/ (accessed on 21 December 2023).
- Python. 2023. Available online: https://www.python.org (accessed on 23 November 2023).








| Resolution (°) | 0.5 | 1 | 2 | 3 | 5 | 7.5 | 10 | 15 | 20 |
|---|---|---|---|---|---|---|---|---|---|
| ME (m) | −0.007 | −0.022 | −0.027 | −0.020 | −0.009 | 0.002 | 0.005 | 0.008 | 0.007 |
| MAE (m) | 0.026 | 0.033 | 0.034 | 0.029 | 0.022 | 0.021 | 0.022 | 0.023 | 0.022 |
| σ (m) | 0.033 | 0.034 | 0.031 | 0.027 | 0.027 | 0.029 | 0.027 | 0.029 | 0.028 |
| Resolution (°) | 0.5 | 1 | 2 | 3 | 5 | 7.5 | 10 | 15 | 20 |
|---|---|---|---|---|---|---|---|---|---|
| (m) | 0.172 | 0.179 | 0.205 | 0.222 | 0.234 | 0.226 | 0.216 | 0.206 | 0.227 |
| σ (m) | 0.130 | 0.145 | 0.163 | 0.161 | 0.148 | 0.132 | 0.121 | 0.115 | 0.124 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira-Obaya, D.; Cabo, C.; Ordóñez, C.; Rodríguez-Pérez, J.R. A Handheld Laser-Scanning-Based Methodology for Monitoring Tree Growth in Chestnut Orchards. Sensors 2024, 24, 1717. https://doi.org/10.3390/s24061717
Pereira-Obaya D, Cabo C, Ordóñez C, Rodríguez-Pérez JR. A Handheld Laser-Scanning-Based Methodology for Monitoring Tree Growth in Chestnut Orchards. Sensors. 2024; 24(6):1717. https://doi.org/10.3390/s24061717
Chicago/Turabian StylePereira-Obaya, Dimas, Carlos Cabo, Celestino Ordóñez, and José Ramón Rodríguez-Pérez. 2024. "A Handheld Laser-Scanning-Based Methodology for Monitoring Tree Growth in Chestnut Orchards" Sensors 24, no. 6: 1717. https://doi.org/10.3390/s24061717
APA StylePereira-Obaya, D., Cabo, C., Ordóñez, C., & Rodríguez-Pérez, J. R. (2024). A Handheld Laser-Scanning-Based Methodology for Monitoring Tree Growth in Chestnut Orchards. Sensors, 24(6), 1717. https://doi.org/10.3390/s24061717








