Negative Charge-Carrying Glycans Attached to Exosomes as Novel Liquid Biopsy Marker
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture Cultivation
2.3. Exosome Collection and Isolation
2.4. Nanoparticle-Tracking Analysis (NTA)
2.5. Dynamic Light-Scattering (DLS)
2.6. Microscopic Analysis (SEM and AFM)
2.7. Surface Plasmon Resonance (SPR)
2.8. Electrochemical Measurements
3. Results
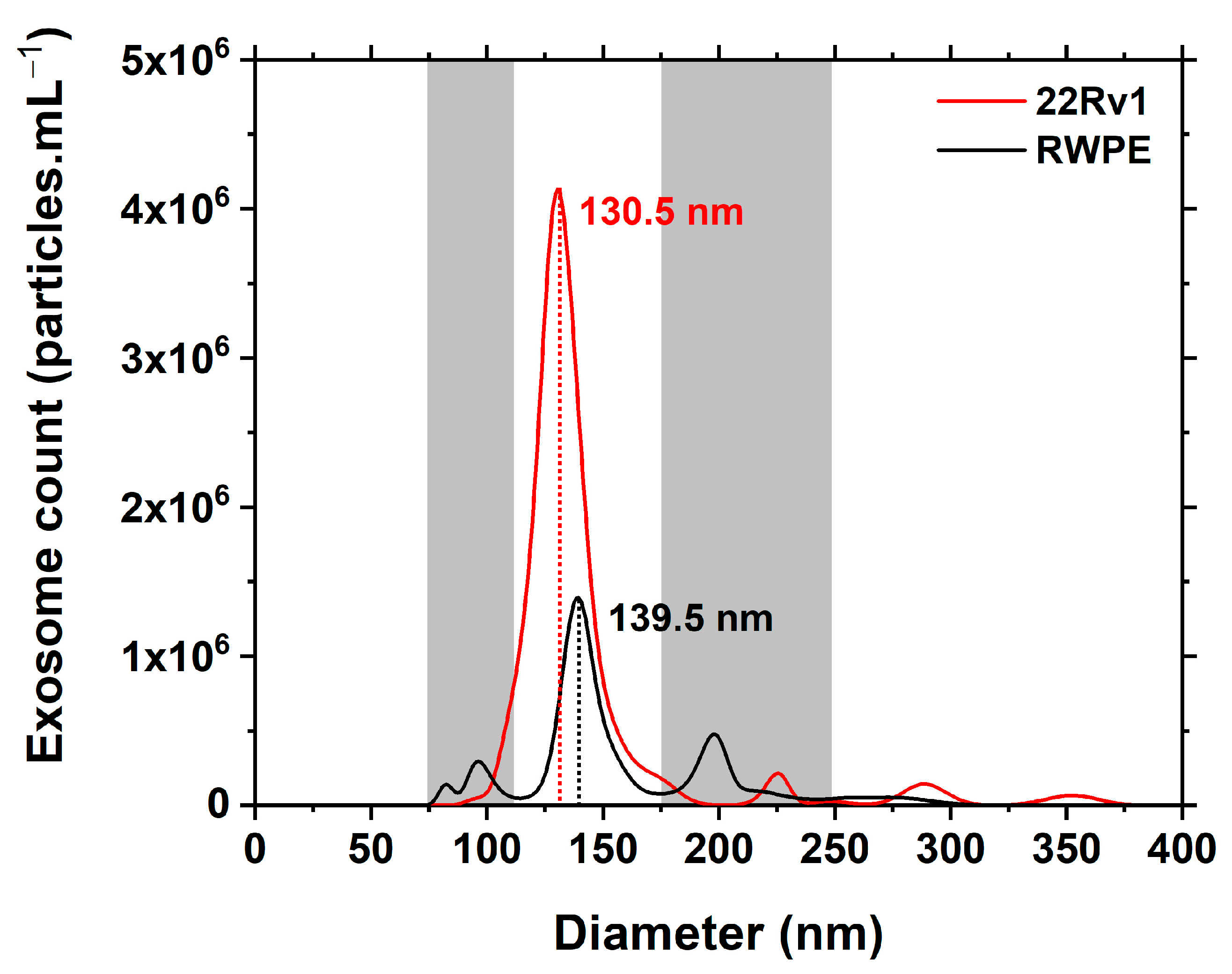
3.1. NTA Characterisation
3.2. DLS and ζ-Potential Characterisation
3.3. Microscopic Characterisation of Isolated Exosomes
3.4. SPR Glycoprofiling
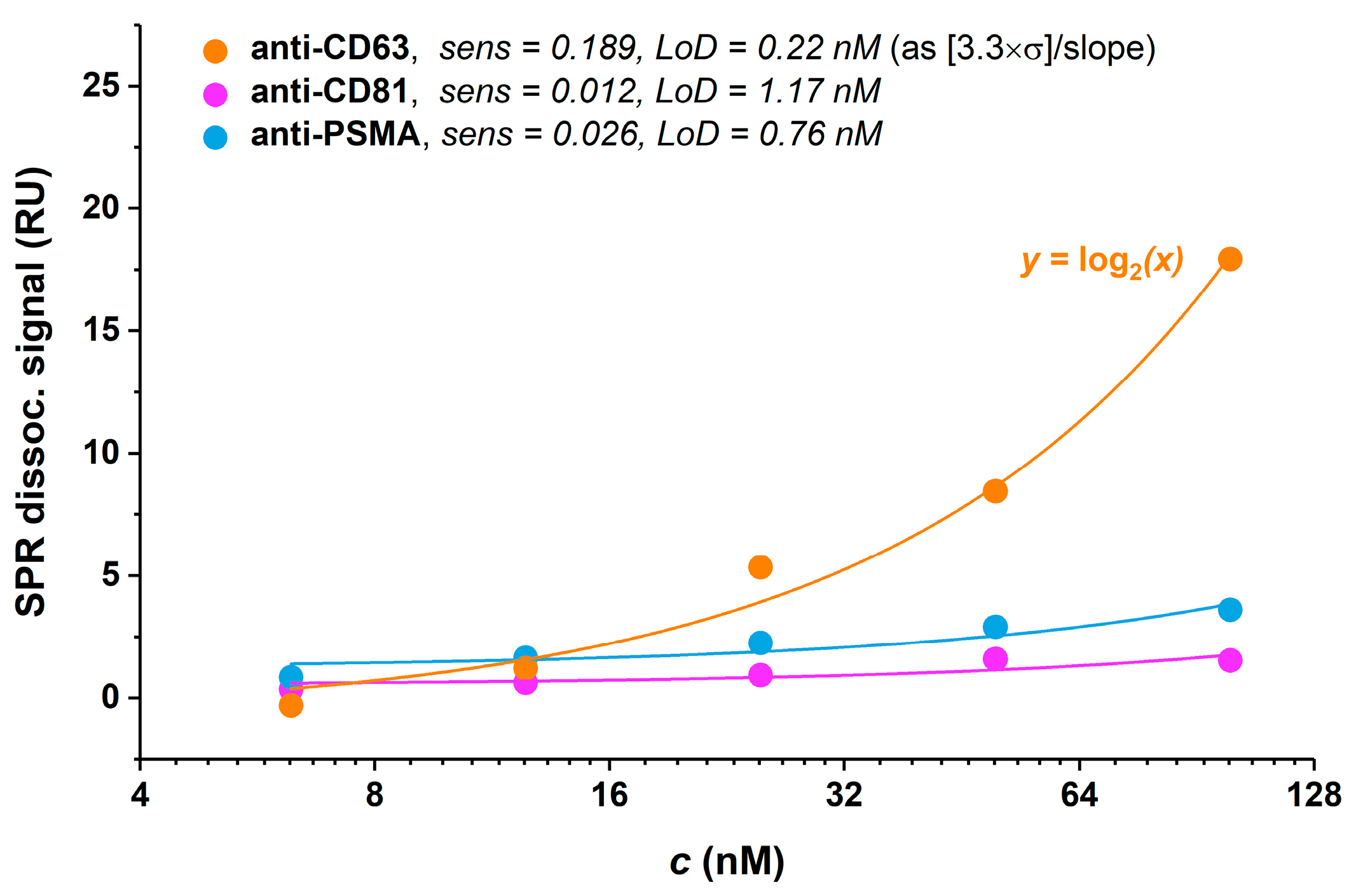
3.4.1. Antibody Selection
3.4.2. Lectin-Based In-Situ Glycoprofiling
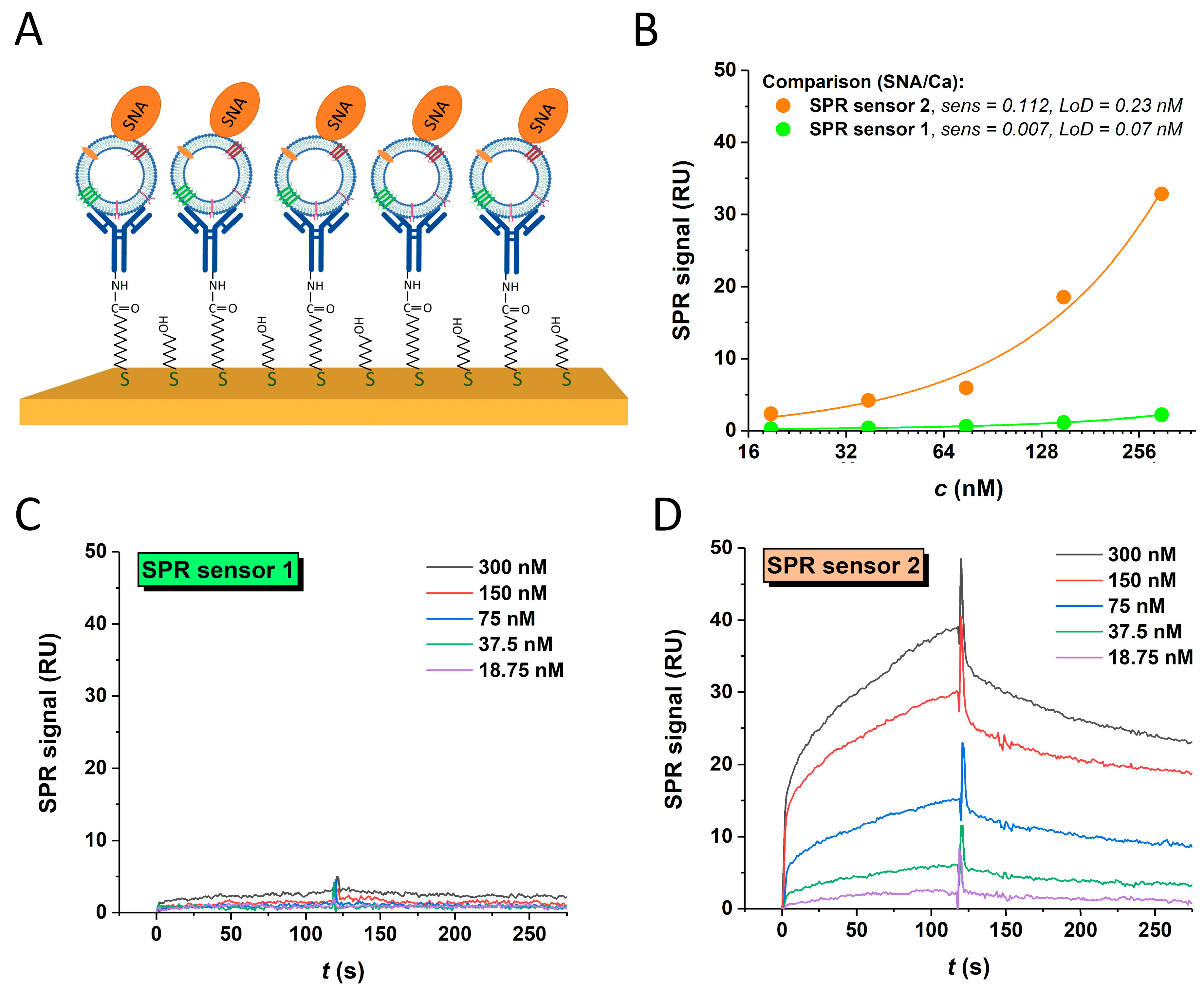
3.4.3. SPR Sandwich Configuration
3.5. Electrochemical Measurements
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bertok, T.; Jane, E.; Chrenekova, N.; Hroncekova, S.; Bertokova, A.; Hires, M.; Vikartovska, A.; Kubanikova, P.; Sokol, R.; Fillo, J.; et al. Analysis of serum glycome by lectin microarrays for prostate cancer patients—A search for aberrant glycoforms. Glycoconj. J. 2020, 37, 703–711. [Google Scholar] [CrossRef]
- Narimatsu, Y.; Joshi, H.J.; Nason, R.; Van Coillie, J.; Karlsson, R.; Sun, L.; Ye, Z.; Chen, Y.-H.; Schjoldager, K.T.; Steentoft, C.; et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol. Cell 2019, 75, 394–407.e395. [Google Scholar] [CrossRef] [PubMed]
- Molejon, M.I.; Weiz, G.; Breccia, J.D.; Vaccaro, M.I. Glycoconjugation: An approach to cancer therapeutics. World J. Clin. Oncol. 2020, 11, 110–120. [Google Scholar] [CrossRef]
- Gao, Y.; Luan, X.; Melamed, J.; Brockhausen, I. Role of Glycans on Key Cell Surface Receptors That Regulate Cell Proliferation and Cell Death. Cells 2021, 10, 1252. [Google Scholar] [CrossRef]
- Paul, A.; Segal, D.; Zacco, E. Glycans to improve efficacy and solubility of protein aggregation inhibitors. Neural Regen. Res. 2021, 16, 2215–2216. [Google Scholar] [CrossRef]
- Lee, H.S.; Qi, Y.; Im, W. Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Sci. Rep. 2015, 5, 8926. [Google Scholar] [CrossRef]
- Sarkar, A.; Wintrode, P.L. Effects of glycosylation on the stability and flexibility of a metastable protein: The human serpin α(1)-antitrypsin. Int. J. Mass Spectrom. 2011, 302, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Bertok, T.; Bertokova, A.; Hroncekova, S.; Chocholova, E.; Svecova, N.; Lorencova, L.; Kasak, P.; Tkac, J. Novel Prostate Cancer Biomarkers: Aetiology, Clinical Performance and Sensing Applications. Chemosensors 2021, 9, 205. [Google Scholar] [CrossRef]
- Bertok, T.; Bertokova, A.; Jane, E.; Hires, M.; Aguedo, J.; Potocarova, M.; Lukac, L.; Vikartovska, A.; Kasak, P.; Borsig, L.; et al. Identification of Whole-Serum Glycobiomarkers for Colorectal Carcinoma Using Reverse-Phase Lectin Microarray. Front. Oncol. 2021, 11, 735338. [Google Scholar] [CrossRef]
- Bertok, T.; Pinkova Gajdosova, V.; Bertokova, A.; Svecova, N.; Kasak, P.; Tkac, J. Breast cancer glycan biomarkers: Their link to tumour cell metabolism and their perspectives in clinical practice. Expert Rev. Proteom. 2021, 18, 881–910. [Google Scholar] [CrossRef]
- Vrablova, V.; Kosutova, N.; Blsakova, A.; Bertokova, A.; Kasak, P.; Bertok, T.; Tkac, J. Glycosylation in extracellular vesicles: Isolation, characterization, composition, analysis and clinical applications. Biotechnol. Adv. 2023, 67, 108196. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sun, H.T.; Wang, S.; Huang, S.L.; Zheng, Y.; Wang, C.Q.; Hu, B.Y.; Qin, W.; Zou, T.T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Lorencova, L.; Bertok, T.; Bertokova, A.; Gajdosova, V.; Hroncekova, S.; Vikartovska, A.; Kasak, P.; Tkac, J. Exosomes as a Source of Cancer Biomarkers: Advances in Electrochemical Biosensing of Exosomes. ChemElectroChem 2020, 7, 1956–1973. [Google Scholar] [CrossRef]
- Bertokova, A.; Svecova, N.; Kozics, K.; Gabelova, A.; Vikartovska, A.; Jane, E.; Hires, M.; Bertok, T.; Tkac, J. Exosomes from prostate cancer cell lines: Isolation optimisation and characterisation. Biomed. Pharmacother. 2022, 151, 113093. [Google Scholar] [CrossRef] [PubMed]
- Gil, B.; Keshavarz, M.; Wales, D.; Darzi, A.; Yeatman, E. Orthogonal Surface-Enhanced Raman Scattering/Field-Effect Transistor Detection of Breast and Colorectal Cancer-Derived Exosomes using Graphene as a Tag-Free Diagnostic Template. Adv. NanoBiomed Res. 2023, 3, 2300055. [Google Scholar] [CrossRef]
- Michela, B. Liquid Biopsy: A Family of Possible Diagnostic Tools. Diagnostics 2021, 11, 1391. [Google Scholar] [CrossRef]
- Mitchell, A.; Pickering, C.; Xu, G.; Rice, R.; Castellanos, A.; Bhadra, R.; Brcic, L.; Lindenmann, J.; Smolle, F.; Lindpaintner, K.; et al. Glycoproteomics as a powerful liquid biopsy-based screening tool for non-small cell lung cancer. J. Clin. Oncol. 2022, 40, e21148. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Nordström, T.; Vickers, A.; Assel, M.; Lilja, H.; Grönberg, H.; Eklund, M. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur. Urol. 2015, 68, 139–146. [Google Scholar] [CrossRef]
- Narain, T.A.; Sooriakumaran, P. Beyond Prostate Specific Antigen: New Prostate Cancer Screening Options. World J. Men’s Health 2022, 40, 66–73. [Google Scholar] [CrossRef]
- Penzkofer, T.; Tempany-Afdhal, C.M. Prostate cancer detection and diagnosis: The role of MR and its comparison with other diagnostic modalities—A radiologist’s perspective. NMR Biomed 2014, 27, 3–15. [Google Scholar] [CrossRef]
- Salciccia, S.; Capriotti, A.L.; Laganà, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef]
- Görgens, A.; Corso, G.; Hagey, D.W.; Jawad Wiklander, R.; Gustafsson, M.O.; Felldin, U.; Lee, Y.; Bostancioglu, R.B.; Sork, H.; Liang, X.; et al. Identification of storage conditions stabilizing extracellular vesicles preparations. J. Extracell. Vesicles 2022, 11, e12238. [Google Scholar] [CrossRef]
- Han, L.; Zhao, Z.; He, C.; Li, J.; Li, X.; Lu, M. Removing the stumbling block of exosome applications in clinical and translational medicine: Expand production and improve accuracy. Stem Cell Res. Ther. 2023, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; Di Raimo, R.; Mizzoni, D.; Fais, S. Immunocapture-based ELISA to characterize and quantify exosomes in both cell culture supernatants and body fluids. Methods Enzym. 2020, 645, 155–180. [Google Scholar] [CrossRef]
- Logozzi, M.; Angelini, D.F.; Giuliani, A.; Mizzoni, D.; Di Raimo, R.; Maggi, M.; Gentilucci, A.; Marzio, V.; Salciccia, S.; Borsellino, G.; et al. Increased Plasmatic Levels of PSA-Expressing Exosomes Distinguish Prostate Cancer Patients from Benign Prostatic Hyperplasia: A Prospective Study. Cancers 2019, 11, 1449. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R.; van Leeuwen, T.G. Refractive Index Determination of Nanoparticles in Suspension Using Nanoparticle Tracking Analysis. Nano Lett. 2014, 14, 6195–6201. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, D.; Kolářová, K.; Čech, J.; Haušild, P.; Kuliček, J.; Ukraintsev, E.; Stehlík, Š.; Dao, R.; Neuman, J.; Rezek, B. Correlative Atomic Force Microscopy and Scanning Electron Microscopy of Bacteria-Diamond-Metal Nanocomposites. Ultramicroscopy 2023, 258, 113909. [Google Scholar] [CrossRef] [PubMed]
- Neuman, J.; Novacek, Z.; Pavera, M.; Novotna, V. Correlative Probe and Electron Microscopy CPEM™–The Novel Technology for 3D Material Surface Analysis. Microsc. Microanal. 2019, 25, 430–431. [Google Scholar] [CrossRef]
- Švecová, E.; Ostatná, V.; Fojt, L.; Hermannová, M.; Velebný, V.; Ondreáš, F. Adsorption/desorption behavior of hyaluronic acid fragments at charged hydrophobic surface. Carbohydr. Polym. 2022, 277, 118831. [Google Scholar] [CrossRef]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunn-Moore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Li, D.; Gu, J.; Qian, D.; Qin, X.; Chen, Y. Exosome carrying PSGR promotes stemness and epithelial-mesenchymal transition of low aggressive prostate cancer cells. Life Sci. 2021, 264, 118638. [Google Scholar] [CrossRef]
- Ukraintsev, E.; Hematian, H.; Neuman, J.; Rezek, B. AFM-in-SEM analyses of thiorphan as-sembly on ZnO polar and nonpolar surfaces. In Proceedings of the Nanocon 2023 Conference, Brno, Czech Republic, 18–20 October 2023. accepted. [Google Scholar]
- Kamat, V.; Boutot, C.; Rafique, A.; Granados, C.; Wang, J.; Badithe, A.; Torres, M.; Chatterjee, I.; Olsen, O.; Olson, W.; et al. High affinity human Fc specific monoclonal antibodies for capture kinetic analyses of antibody-antigen interactions. Anal. Biochem. 2022, 640, 114455. [Google Scholar] [CrossRef]
- Sha, S.; Wang, Y.; Liu, M.; Liu, G.; Fan, N.; Li, Z.; Dong, W. Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis. Cells 2022, 11, 2453. [Google Scholar] [CrossRef]
- Hagiwara, K.; Tobisawa, Y.; Kaya, T.; Kaneko, T.; Hatakeyama, S.; Mori, K.; Hashimoto, Y.; Koie, T.; Suda, Y.; Ohyama, C.; et al. Wisteria floribunda Agglutinin and Its Reactive-Glycan-Carrying Prostate-Specific Antigen as a Novel Diagnostic and Prognostic Marker of Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Al Saoud, R.; Hamrouni, A.; Idris, A.; Mousa, W.K.; Abu Izneid, T. Recent advances in the development of sialyltransferase inhibitors to control cancer metastasis: A comprehensive review. Biomed. Pharmacother. 2023, 165, 115091. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, E.; Macauley, M.S. Hypersialylation in Cancer: Modulation of Inflammation and Therapeutic Opportunities. Cancers 2018, 10, 207. [Google Scholar] [CrossRef]
- Palecek, E.; Tkác, J.; Bartosík, M.; Bertók, T.; Ostatná, V.; Palecek, J. Electrochemistry of Nonconjugated Proteins and Glycoproteins. Toward Sensors for Biomedicine and Glycomics. Chem. Rev. 2015, 115, 2045–2108. [Google Scholar] [CrossRef]
- Jastrząb, P.; Narejko, K.; Car, H.; Wielgat, P. Cell Membrane Sialome: Sialic Acids as Therapeutic Targets and Regulators of Drug Resistance in Human Cancer Management. Cancers 2023, 15, 5103. [Google Scholar] [CrossRef]
- Li, K.; Chen, Y.; Li, A.; Tan, C.; Liu, X. Exosomes play roles in sequential processes of tumor metastasis. Int. J. Cancer 2019, 144, 1486–1495. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, G.; Guan, F. Biological Functions and Analytical Strategies of Sialic Acids in Tumor. Cells 2020, 9, 273. [Google Scholar] [CrossRef]
- Büll, C.; den Brok, M.H.; Adema, G.J. Sweet escape: Sialic acids in tumor immune evasion. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2014, 1846, 238–246. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Riechelmann, H.; Skvortsova, I.I. The role of exosomes in cancer metastasis. Semin. Cancer Biol. 2017, 44, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Hong, C.S.; Ludwig, S.; Azambuja, J.H.; Sharma, P.; Theodoraki, M.N.; Whiteside, T.L. Isolation and Analysis of Tumor-Derived Exosomes. Curr. Protoc. Immunol. 2019, 127, e91. [Google Scholar] [CrossRef] [PubMed]
- Suthar, J.; Taub, M.; Carney, R.P.; Williams, G.R.; Guldin, S. Recent developments in biosensing methods for extracellular vesicle protein characterization. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1839. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, A.L.; Johansson, K.; Mossberg, M.; Kahn, R.; Karpman, D. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr. Nephrol. 2019, 34, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Giuliani, A.; Maggi, M.; Sciarra, A.; Fais, S. Plasmatic Exosome Number and Size Distinguish Prostate Cancer Patients from Healthy Individuals: A Prospective Clinical Study. Front. Oncol. 2021, 11, 727317. [Google Scholar] [CrossRef]
- Hood, J.L.; Scott, M.J.; Wickline, S.A. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014, 448, 41–49. [Google Scholar] [CrossRef]
- Brunetti, J.; Depau, L.; Falciani, C.; Gentile, M.; Mandarini, E.; Riolo, G.; Lupetti, P.; Pini, A.; Bracci, L. Insights into the role of sulfated glycans in cancer cell adhesion and migration through use of branched peptide probe. Sci. Rep. 2016, 6, 27174. [Google Scholar] [CrossRef]
- Pomin, V.H.; Mulloy, B. Current structural biology of the heparin interactome. Curr. Opin. Struct. Biol. 2015, 34, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, J.J.; Ballauff, M.; Zimmermann, R.; Freudenberg, U.; Werner, C. Thermodynamic Analysis of the Interaction of Heparin with Lysozyme. Biomacromolecules 2020, 21, 4615–4625. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.; Boelaars, K.; Brown, K.; Eveline Li, R.J.; Kruijssen, L.; Bruijns, S.C.M.; van Ee, T.; Schetters, S.T.T.; Crommentuijn, M.H.W.; van der Horst, J.C.; et al. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021, 12, 1270. [Google Scholar] [CrossRef]
- Guo, X.; Elkashef, S.M.; Loadman, P.M.; Patterson, L.H.; Falconer, R.A. Recent advances in the analysis of polysialic acid from complex biological systems. Carbohydr. Polym. 2019, 224, 115145. [Google Scholar] [CrossRef]
- Kawakami, K.; Fujita, Y.; Kato, T.; Horie, K.; Koie, T.; Umezawa, K.; Tsumoto, H.; Miura, Y.; Katagiri, Y.; Miyazaki, T.; et al. Diagnostic potential of serum extracellular vesicles expressing prostate-specific membrane antigen in urologic malignancies. Sci. Rep. 2021, 11, 15000. [Google Scholar] [CrossRef] [PubMed]
- Øverbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, A.; Kajau, H.; Margolis, E.; Tutrone, R.; Grimm, T.; Trottmann, M.; Stief, C.; Stoll, G.; Fischer, C.A.; Flinspach, C.; et al. Validation of a CE-IVD, urine exosomal RNA expression assay for risk assessment of prostate cancer prior to biopsy. Sci. Rep. 2022, 12, 4777. [Google Scholar] [CrossRef]
- Ho, W.-L.; Hsu, W.-M.; Huang, M.-C.; Kadomatsu, K.; Nakagawara, A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J. Hematol. Oncol. 2016, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, Y.; Wang, Y.; Hu, X.; Koh, K.; Chen, H. Tunable Au@SiO2/Au Film Metasurface as Surface Plasmon Resonance Enhancer for Direct and Ultrasensitive Detection of Exosomes. Anal. Chem. 2023, 95, 9663–9671. [Google Scholar] [CrossRef] [PubMed]
- Haizan, I.; Park, D.H.; Choi, M.Y.; Lee, H.; Choi, J.-H. Nanomaterials-Based Exosomes for the Diagnostics and Drug Deliveries of Central Nervous System Diseases. BioChip J. 2023, 17, 293–307. [Google Scholar] [CrossRef]



| Lectin/Agglutinin (Abbreviation) | Specificity | SU/GBS | Carcinoma Exosomes | Control Exosomes |
|---|---|---|---|---|
| Aleuria aurantia (AAL) | fucose, arabinose | 2 subunits/5 fucose BS | − | − |
| Galanthus nivalis (GNL) | mannose | 4 subunits/12 mannose BS | − | − |
| Maackia amurensis (MAAII) | α-2,3 linked Sia | 2 subunits | + | + |
| Phaseolus vulgaris (PHA-E) | galactose, bisecting GlcNac, biantennary N-glycans | 4 E subunits | − | + |
| Phaseolus vulgaris (PHA-L) | branched N-glycans | 4 L type subunits | − | + |
| Pisum sativum (PSA) | mannose, glucose | 4 subunits | − | − |
| Sambucus nigra (SNA) | α-2,6 linked Sia | 2 subunits | + | + |
| Wisteria floribunda (WFL) | N-Acetylgalactosamine | 4 subunits | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosutova, N.; Lorencova, L.; Hires, M.; Jane, E.; Orovcik, L.; Kollar, J.; Kozics, K.; Gabelova, A.; Ukraintsev, E.; Rezek, B.; et al. Negative Charge-Carrying Glycans Attached to Exosomes as Novel Liquid Biopsy Marker. Sensors 2024, 24, 1128. https://doi.org/10.3390/s24041128
Kosutova N, Lorencova L, Hires M, Jane E, Orovcik L, Kollar J, Kozics K, Gabelova A, Ukraintsev E, Rezek B, et al. Negative Charge-Carrying Glycans Attached to Exosomes as Novel Liquid Biopsy Marker. Sensors. 2024; 24(4):1128. https://doi.org/10.3390/s24041128
Chicago/Turabian StyleKosutova, Natalia, Lenka Lorencova, Michal Hires, Eduard Jane, Lubomir Orovcik, Jozef Kollar, Katarina Kozics, Alena Gabelova, Egor Ukraintsev, Bohuslav Rezek, and et al. 2024. "Negative Charge-Carrying Glycans Attached to Exosomes as Novel Liquid Biopsy Marker" Sensors 24, no. 4: 1128. https://doi.org/10.3390/s24041128
APA StyleKosutova, N., Lorencova, L., Hires, M., Jane, E., Orovcik, L., Kollar, J., Kozics, K., Gabelova, A., Ukraintsev, E., Rezek, B., Kasak, P., Cernocka, H., Ostatna, V., Blahutova, J., Vikartovska, A., Bertok, T., & Tkac, J. (2024). Negative Charge-Carrying Glycans Attached to Exosomes as Novel Liquid Biopsy Marker. Sensors, 24(4), 1128. https://doi.org/10.3390/s24041128







