Abstract
The main purpose of this paper is to provide information on how to create a convolutional neural network (CNN) for extracting features from EEG signals. Our task was to understand the primary aspects of creating and fine-tuning CNNs for various application scenarios. We considered the characteristics of EEG signals, coupled with an exploration of various signal processing and data preparation techniques. These techniques include noise reduction, filtering, encoding, decoding, and dimension reduction, among others. In addition, we conduct an in-depth analysis of well-known CNN architectures, categorizing them into four distinct groups: standard implementation, recurrent convolutional, decoder architecture, and combined architecture. This paper further offers a comprehensive evaluation of these architectures, covering accuracy metrics, hyperparameters, and an appendix that contains a table outlining the parameters of commonly used CNN architectures for feature extraction from EEG signals.
1. Introduction
The beginning of investigation into the chemical processes of the brain is attributed to D. Reymond (Du Bois Reymond), who in 1849 demonstrated that the brain, like nerves and muscles, possesses electrogenic properties [1]. However, it was not until 75 years later, in 1924, that the German psychiatrist Hans Berger made the first recording of an electroencephalogram (EEG) [2]. In the years that followed, this term became widely used in medicine, creating a new scientific field known as neuroscience. In 1935–1936, Pauline and Hallowell Davis recorded the first known event-related potential (ERP) in awake individuals, and their results were published a few years later in 1939 [3]. This significantly increased the popularity of using EEG for clinical purposes. ERPs are event-related voltage changes in EEG activity that are time-bound to sensory, motor, and cognitive events. Consequently, ERP analysis is used to identify and measure specific electrical signals associated with cognitive processes, and these signals can be used to control external devices such as prostheses or computers [4,5].
Electroencephalogram (EEG) signals are essential for various applications and serve as a valuable tool for understanding and interacting with the human brain [6]. Notable applications include sleep monitoring, which assists in sleep pattern analysis and diagnosing sleep disorders [7]. EEG signals contribute to emotion recognition by correlating patterns of brain activity with different emotional states [8]. In motor imagery tasks, the EEG facilitates the control of external devices, influencing rehabilitation and assistive technologies [9,10,11]. Neurofeedback therapy uses EEG to regulate brain activity, offering potential benefits for conditions such as anxiety [12]. In assessing cognitive load, EEG is used to measure mental workload during tasks important to human-computer interaction and design [13]. EEG is critical for diagnosing and monitoring epilepsy, assessing traumatic brain injury, and assessing the impact on brain function [14]. These applications highlight the versatility of EEG signals across a wide range of contexts related to the human brain. In addition, the EEG is one of the key components of brain-computer interfaces (BCI), providing direct communication between the brain and external devices [15]. Different activities, such as disease diagnosis or robot control, exhibit unique characteristics that necessitate a deep understanding of the underlying physical processes. For example, the control of sleep is influenced by various factors, with the frequency component playing a crucial role [16]. During sleep, humans go through six distinct stages: wakefulness, rapid eye movement sleep, and four non-REM sleep stages, each having its own characteristic frequency [17,18], as shown in Figure 1.
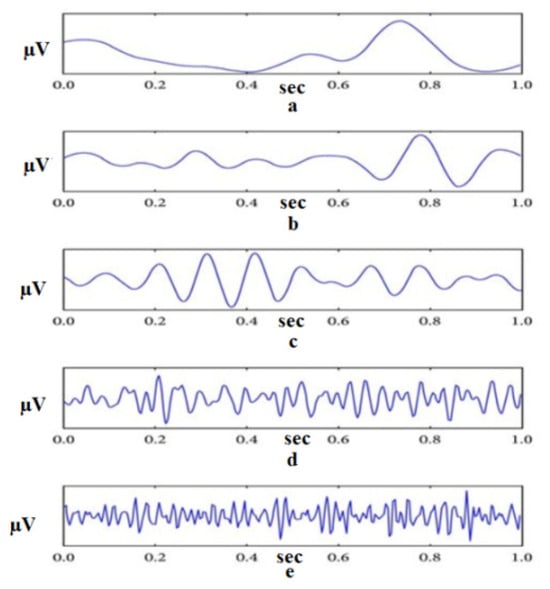
Figure 1.
Example of EEG decomposed into its frequency bands (From top to bottom: The delta, theta, alpha, beta, and gamma frequency bands) [19].
EEG signals exhibit non-linear characteristics, which pose challenges in their mathematical description, as indicated by previous studies [20,21]. Furthermore, a non-linear relationship exists between the object of interest and EEG data, which standard algorithms struggle to capture. While a linear relationship does exist between electrochemical synaptic activity (expressed as current density) and EEG data in accordance with Poisson’s equation and Maxwell’s equations [22,23,24], practical applications require a focus on underlying mental or cognitive processes rather than solely the electrochemical activity of the brain. For example, emotions do not linearly correspond to brain activity. This, combined with the linear mapping of brain activity to EEG data, implies that emotions do not have a linear relationship with EEG signals.
Recent developments in CNNs have shown their prowess in handling non-linear dependencies and effectively decomposing them into their characteristic frequency components [25]. As a result, many developed architectures can operate on raw EEG data without the need for any additional processing. However, the use of CNNs poses a limitation, given their “black box” nature [26,27], whereby an output is obtained without comprehending how it occurred. Nevertheless, despite CNNs finding numerous applications across various fields, we maintain the perspective that comprehending the data processing is crucial for fully grasping the significance of their architecture [28].
1.1. Research Problem Statement
In existing review articles on machine learning for EEG, authors typically avoid discussing general topics like “review of EEG algorithms” due to their broad nature. Instead, they often focus on how machine learning can be applied to address specific diseases [29]. However, we believe that this approach limits the potential of machine learning and does not show how a single machine learning algorithm can be adapted to similar tasks of extracting features from EEG signals. In this manuscript, our primary goal is therefore to illustrate how CNN architectures can be adapted to suit individual needs in response to changing tasks. To achieve this goal, we aim to provide readers with a comprehensive understanding of the signal processing within a CNN and examine all aspects of the CNN tuning process as it pertains to EEG signals. We have chosen to focus on CNNs as they are one of the most promising areas in the field of artificial intelligence [30,31] and are particularly well-suited for real-time processing, which is crucial when dealing with signals that exhibit non-linear and non-stationary properties. Instead of classifying CNNs based on the tasks of their application, we have categorized them into the following groups for analysis:
- -
- The standard implementation of CNNs
- -
- RCNN-based architectures
- -
- Decoder-based architectures
- -
- Cascade Architecture.
To gain a better understanding of specific CNN architectures, it is important to have a basic understanding of signal processing. By doing so, we hope to provide readers with the necessary background knowledge to fully comprehend and appreciate the significance of various CNN architectures.
1.2. Machine Learning for EEG. Why CNN?
The field of machine learning is incredibly diverse, encompassing a wide range of subfields, as evident from the numerous existing classification schemes and terminologies outlined [32]. In the context of EEG, traditional machine learning algorithms, such as decision trees, support vector machines, and random forests, are commonly employed to classify EEG data based on different features, including the amplitude, frequency, and coherence [33,34]. These algorithms are widely used and have proven to be effective in various applications related to EEG analysis.
CNNs belong to the class of artificial neural networks (ANNs) and are primarily used for visual image analysis [35]. They represent a regularized version of multilayer perceptrons. Multilayer perceptrons typically refer to fully connected networks, whereby every neuron in one layer is connected to every neuron in the next layer. Researchers such as Albawi [36] and Indolia [37] have aimed to provide an understanding of CNNs and have considered general information about their structure. In a more closely related context to our topic, CNNs have been applied in medical image understanding [38] and in understanding the behavior of 3D CNNs for the diagnosis of Alzheimer’s disease based on brain imaging [39]. In this context, ANNs, including CNNs, can analyze complex non-linear relationships between EEG inputs and output classification labels [40,41]. This analysis is used to classify EEG signals based on different cognitive states, such as wakefulness, sleep stages, or various types of epileptic seizures [42,43].
CNNs can collect temporal information, automatically extract features, scale large datasets, and have the flexibility to adapt to various EEG applications. At the same time, it is often believed that for many data analysis tasks that require detection and dominance of the frequency range in EEG signals, band-pass filters may be used. However, CNNs automatically learn and extract complex features from raw input data, whereas bandpass filters can only capture simple frequency patterns.
There are numerous review articles on the topic of machine learning for EEG that provide a comprehensive assessment of various architectures. In a well-known publication, Lotte et al. [16] offered a comprehensive overview of machine learning for EEG. Later, an updated analysis was conducted after 11 years, providing insights into the field in a more modern context [44]. The updated work presented a wide range of methods for processing EEG signals but provided fewer details on feature extraction and gave insufficient attention to deep learning architectures. As a result, the article described some architectures that are not commonly used in practice, potentially causing confusion. Furthermore, significant advances have occurred in the field of neural networks over the last 5 years, which are not fully reflected in the article.
Numerous detailed reviews have explored the use of machine learning in EEG signal analysis for disease detection and other applications. For instance, Maitin et al. [45] provided an insightful review of the use of machine learning for Parkinson’s disease detection, while Rodrigues et al. [46] presented a comprehensive overview of the utilization of machine learning for the detection of alcoholism and other diseases. In addition, Rasheed et al. [47] reviewed various machine learning architectures for epilepsy detection from EEG signals, while Lucas et al. [48] explored the use of machine learning for detecting pathologies in EEG signals. In the field of emotion recognition, Bazgir et al. [49], Xiao-Wei et al. [50], and Nedelcu et al. [51] conducted thorough reviews of various techniques for removing artifacts from EEG signals using machine learning. Aggarwal et al. [52], on the other hand, focused on brain-computer interfaces and the application of machine learning in signal processing, including feature extraction and real-time processing.
There are also many smaller-scale reviews, such as “Brain-Computer Interface and Emotions” [53], along with numerous other review papers investigating the use of machine learning for disease detection via EEG signals [54,55,56,57]. Additionally, papers by Roman et al. [58] and Shedeed et al. [59] have explored the application of machine learning for signal processing in EEG analysis.
While reviews of pre-existing architectures can be helpful in determining which architecture to use for a specific case, they may not provide a complete understanding of how a convolutional neural network (CNN) is constructed or how to modify its architecture to accommodate changes in EEG measurement conditions. Although there are many review papers on the topic of feature extraction from EEG signals using machine learning, the information presented in these papers is typically not consistent in format or evaluation criteria. Therefore, in this work, we aim to provide a comprehensive guide on how to prepare, process, and build CNN architectures for EEG signals, as well as how to tune hyperparameters, estimate models, run in real-time, and address other important considerations.
In recent years, there has been a trend toward the development of new types of brain computer interfaces with the goal of making the process of measuring EEG signals more comfortable [60,61,62]. One promising approach is the use of ear-based devices for measuring EEG signals [63]. While these devices offer greater convenience, the quality of the signals they pick up is typically lower due to increased noise levels. Another alternative that has gained popularity in recent times is dry electrodes, which are also known to produce noisier signals. Consequently, the need for the development of improved machine learning algorithms for extracting useful features from EEG signals becomes even more crucial.
2. Signal Processing
The process of detecting EEG potentials involves measuring the potentials of electrodes placed on the surface of the head relative to the potential of an electrode on the earlobe, expressed in microvolts. The potential of the reference electrode should remain constant over time, but the scalp’s electrically active conduction system can introduce variations in potential measurements. One of the simplest solutions to address this issue is to recalculate the EEG signal drifts relative to the total averaged reference [64]. However, due to the non-stationary nature of EEG and its high susceptibility to various types of noise, especially electrical noise, the task of denouncing raw EEG data to obtain useful information poses a significant challenge [65]. In fact, the noise problem remains one of the primary obstacles to extending the application of EEG beyond the laboratory setting [66].
Pre-processing the EEG is a crucial step in preparing it for further analysis. This involves a range of techniques aimed at reducing noise and removing artifacts to ensure a clean signal is ready for subsequent steps. The initial step in this process is the removal of noise originating from external electromagnetic fields [67]. Next, motion artifacts need to be addressed, as they can have a negative impact on the EEG signal. One of the most popular tools for removing artifacts and noise from non-linear EEG signals is Principal Component Analysis (PCA) [68]. PCA reduces dimensionality and redundancy by combining original variables in a way that maximizes variance, effectively removing artifacts from the data. Independent Component Analysis (ICA) is another popular technique used for EEG signal pre-processing [69]. When dealing with multi-channel signals, ICA separates the multi-component EEG signal into its independent parts, thereby removing noise and interference caused by blinks, eye movements, heart contractions, and muscle activity. This technique has proven to be particularly effective in addressing artifacts in multi-channel EEG recordings. Canonical Correlation Analysis (CCA) is yet another technique that enhances EEG signal quality. CCA identifies linear transformations to maximize the correlations between two datasets. It has been employed to improve brain-computer interface (BCI) performance in various scenarios, such as code-modulated visual evoked potentials, steady-state visual evoked potentials, and event-related potentials like P300 and error-related potentials.
2.1. Signal Processing with Machine Learning
Research on EEG artifact removal methods has spanned over 55 years, yet there is still no consensus on which algorithm is optimal for a particular application [70]. Nonetheless, EEG artifact removal techniques are crucial for fully utilizing EEG data and can be implemented through both automatic and manual online and offline methods. Two of the most popular methods for EEG artifact removal are the support vector machine (SVM) [71,72] and PCA [73,74]. While these methods are commonly used, numerous other techniques are available, and several review papers aim to explore different approaches for artifact removal [75,76,77,78]. These review papers provide a valuable resource for researchers seeking to evaluate the effectiveness of different artifact removal methods. Despite the lack of consensus on the optimal algorithm, ongoing research in EEG artifact removal methods continues to enhance EEG data quality and improve analysis accuracy. The use of various techniques, including SVM and PCA, can effectively mitigate artifacts in EEG signals, enabling comprehensive utilization of EEG data in a wide range of applications.
In recent years, several researchers have explored the use of CNNs for EEG noise reduction, demonstrating promising results. In a 2020 study, Sun et al. [79] introduced CNN-1D-ResCNN, one of the first applications of CNN for EEG noise reduction. Similarly, Yang et al. [80] employed CNN and an auto-encoder, incorporating weights into an objective function to remove artifacts without compromising the EEG field signal. These studies demonstrate the potential for CNNs to effectively reduce noise in EEG signals.
Another promising approach for EEG noise reduction involves the use of recurrent neural networks (RNNs) in conjunction with CNNs. Zhang et al. [81] presented an architecture that combines CNNs and a recurrent neural network (LSTM) to eliminate dangling artifacts, naming it EEGdenoiseNet. The incorporation of RNNs in this architecture enables the network to consider temporal dependencies in the EEG signal, enhancing the effectiveness of the noise reduction process. Furthermore, Mashhadi et al. [82] successfully transformed each EEG signal into an image for input into a model designed for image segmentation tasks. This model, based on a convolutional neural network architecture known as U-NET [83], allowed for the selection of weights and filters that removed artifacts from the EEG signal, underscoring the versatility of CNNs in processing EEG data. Overall, the use of CNNs, RNNs, and other deep learning techniques shows promise in reducing noise and artifacts in EEG signals. As research progresses, these methods are likely to become increasingly effective in improving the quality of EEG data for analysis.
Despite the promising results of using machine learning techniques for EEG artifact removal, there are still some limitations prior to their widespread application. One of the primary challenges stems from the non-linear nature of artifacts, which complicates the task of isolating artifacts while preserving the valuable information within EEG signals. Another significant constraint is the substantial computing power required for machine learning algorithms, rendering them less accessible and practical for many researchers. Consequently, researchers often opt to employ machine learning to extract features from EEG signals rather than direct artifact removal. This approach allows for a hybrid model, where artifacts are eliminated in the initial layers while subsequent layers extract features. Nonetheless, the application of machine learning for EEG artifact removal remains an active area of research, with ongoing advancements in computational capabilities and machine learning algorithms expected to enhance the effectiveness and accessibility of these methods. Therefore, it is anticipated that machine learning techniques will assume an increasingly significant role in EEG artifact removal in the future.
In addition to artifacts, EEG signals can also be affected by noise stemming from external sources, such as electromagnetic interference [84], suboptimal skin-electrode contact, or low electrode quality [85]. To address these issues, researchers have developed denoising techniques aimed at eliminating unwanted noise while preserving the EEG signal’s non-linear characteristics [86].
Denoising techniques typically involve signal processing methods, including filtering, averaging, and wavelet decomposition. It is imperative to apply these methods with care to ensure that the denoising process does not distort the inherent EEG signal or remove any critical information. Therefore, researchers must diligently assess the effectiveness of various denoising techniques and select the most suitable method for their specific application. In summary, denoising constitutes a crucial step in the EEG signal processing pipeline and is indispensable for obtaining precise and dependable results in EEG experiments.
2.2. Frequency and Spatial Components in EEG Signals
Spectral analysis is a widely used method for extracting valuable information from EEG signals. By analyzing the power spectral density (power spectrum) of the signal, spectral analysis can provide insight into the frequency composition or the distribution of signal power over frequency. This information aids researchers in understanding the underlying neural processes responsible for generating the EEG signal and in identifying patterns that may be associated with specific cognitive states or behaviors. The power spectrum can be computed through various techniques, including Fourier transform, wavelet transform, and autoregressive modeling. These methods enable researchers to analyze the power of different frequency bands in the EEG signal, such as the alpha, beta, theta, and delta bands. By examining changes in the power spectrum over time or across experimental conditions, researchers can gain valuable insights into how brain activity is modulated by various factors [87,88,89]. Additionally, the phase synchronization method is also rooted in the frequency component. This approach measures the tendency of two time series signals to maintain a constant phase separation over a period [90,91].
The coherence method is a widely used technique for examining the relationship between EEG signals originating from various regions of the brain [92,93]. It provides valuable physiological information about functional connectivity patterns, which can enhance the performance of EEG-based biometric systems [94]. Additionally, this approach can detect alterations in the information flow between cortical areas across different frequency bands [95].
Moreover, the connection between these signals can also be represented using the symmetric matrix method, which illustrates the covariance of each pair of variables. The values within the covariance matrix indicate the magnitude and direction of the distribution of multivariate data within a multidimensional space. By manipulating these values, it becomes possible to extract information about how the data are distributed across any two dimensions [96]. Examples of the implementation of a symmetric matrix are illustrated in Figure 2.
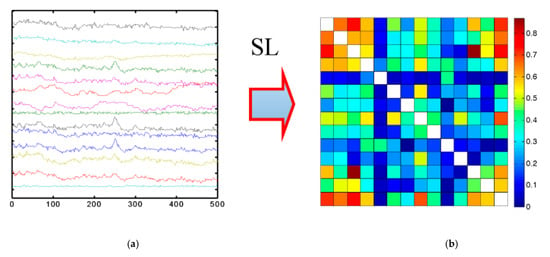
Figure 2.
Implementation of symmetric matrix for EEG signals: (a) Initial data, (b) Symmetric matrix [97].
The symmetric matrix can be complemented with additional methods, such as Riemannian geometry [98], to improve the performance of EEG data analysis for low-dimensional problems [99,100]. These methods enable a clearer tracing of the relationship between signals in the cerebral cortex using spatial filters. A spatial filter is an algorithm utilized for multi-channel electroencephalogram (EEG) analysis [101], often employed to extract features from EEG data based on variance. These features are then integrated into a deep neural network for classification [102].
One particularly effective spatial filter is the Common Spatial Patterns (CSPs), which has demonstrated success in extracting sensorimotor rhythms and can be employed in real-time in brain-computer interfaces. International competitions have further demonstrated the effectiveness of spatial filters in conjunction with machine learning models [103,104,105]. In addition, their application in classification problems, spatial filters can also enhance the signal-to-noise ratio in regression-based problems [106] or accentuate differences in power between various imaging conditions [107], as shown in Figure 3.
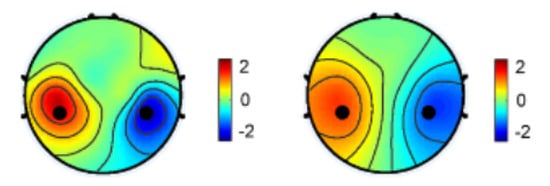
Figure 3.
Spatial distribution of EEG power difference for the left and right hands [107].
3. Feature Selection and Feature Extraction
EEG data are measured in microvolts, but their amplitudes can vary significantly, necessitating data scaling. The most commonly used tools for this purpose are normalization and standardization [108]. Normalization typically involves rescaling values to the range [0, 1], while standardization usually means rescaling the data to have a mean of 0 and a standard deviation of 1 (unit variance). In practice, normalization is a more frequently employed method of data preparation [109,110,111,112], though standardization is still used [113]. One notable variation on normalization is stratified normalization [114], introduced by Fdez [115], which is particularly useful for training deep neural networks to classify emotions across subjects using EEG signals. This method effectively removes between-participant variability while preserving emotional information in the data, compared to less popular methods like batch normalization [116].
Feature selection is a crucial step in enhancing the performance of machine learning algorithms by eliminating unnecessary, redundant, or noisy features from feature vectors. In EEG signal processing, statistical features such as mean, median, variance, standard deviation, and skewness are often used as the simplest features [116]. Frequency domain features (FDF) can also be computed using the discrete Fourier transform of raw EEG signals [117].
In addition to these standard methods, custom approaches have also been developed. For example, Duan et al. [29] introduced a new decision tree-based feature selection method for EEG signals, involving a feature space search and automatic selection of optimal features using a decision tree algorithm. Another approach is to utilize PCA for feature extraction from the EEG signal and subsequently employ a decision tree-based selection process to automatically select the optimal features. This method has been shown to effectively reduce the dimensionality of the EEG data while preserving important information. The EEG signal is a non-linear graph containing an extensive amount of information, with each aspect potentially representing an independent feature useful for specific functions. Therefore, feature selection is crucial in EEG signal processing to identify the most important features for a given analysis. Several survey projects have been conducted in this area [118]. It is worth noting that EEG devices can have up to 1024 electrodes, making processing all channels a computationally intensive task. To address this challenge, Alotaiby et al. [119] presented a survey of algorithms for channel selection for machine learning models. These algorithms aim to select the most informative channels while minimizing the computational burden.
Feature extraction is a critical step in EEG signal analysis, involving the conversion of raw data into numerical features that can be processed while retaining the information in the original dataset. Various popular methods are used for feature extraction in EEG signal processing, including Wavelet Transform (WT), Fast Fourier Transform (FFT), Time Frequency Distributions (TFD), Eigenvector Methods (EM), and Auto-Regressive Methods (AR), which are described in detail in, e.g., Refs. [120,121].
One well-known method for feature extraction is the Hilbert–Huang transform, a time-frequency method that decomposes EEG signals into empirical modes or intrinsic mode functions (IMF) to obtain instantaneous frequency data [122], as shown in Figure 4. Unlike the Fourier transform [123] used in harmonic analysis [124], instead of decomposing a signal into its constituent frequencies, the Hilbert–Huang transform aims to decompose data into its AM-FM intrinsic monocomponent modal functions, perceiving the locality of information. See Figure 4 for an illustration.
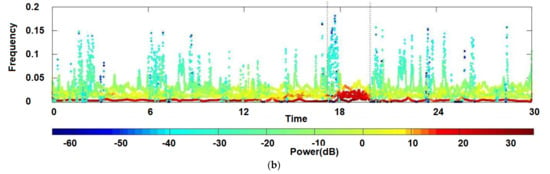
Figure 4.
Hilbert transform of a set of real-filtered EEGs: (a) The analyzed time series; (b) The signal power plotted in the time-frequency domain [125] Once the data has been processed and transformed into numerical features, it can serve as input for machine learning models. By choosing the right feature extraction method, researchers can derive informative and meaningful features from EEG signals, thereby enhancing the accuracy and efficiency of machine learning algorithms when applied to these signals.
4. Datasets and Transfer Learning in EEG
Acquiring suitable datasets for training and evaluating CNN models applied to EEG data presents unique challenges compared to popular tasks like machine vision, primarily due to the inherent characteristics of biodata.
EEG recordings, in particular, are known for their inherent noise, erratic behavior, susceptibility to artifacts, and significant variability from person to person and session to session. These issues often introduce biases, confounders, and limitations that can impact the accurate estimation of the CNN architecture’s performance.
One of the primary challenges in collecting EEG datasets for CNNs is ensuring the quality and reliability of the recorded data. Various forms of noise, including environmental noise, electrode artifacts, and muscle or eye movement artifacts, can distort EEG signals and compromise the accuracy of subsequent analysis. Addressing these issues necessitates stringent electrode placement protocols, precise equipment calibration, and diligent artifact detection and removal methods. Ideally, all data should be included in datasets, but in practice, this is not always the case. Moreover, the spatial resolution of EEG signals is limited, further complicating dataset acquisition. The scalp-based nature of the EEG recording makes it challenging to precisely localize nerve sources and differentiate activities in neighboring brain regions. Proper preprocessing and data completion are crucial for CNN analysis of EEG datasets. Effective filtering techniques, artifact removal algorithms, and appropriate data segmentation methods are needed to enhance the signal-to-noise ratio and provide meaningful and robust inputs for training CNN models.
The reliability, interpretability, and performance of CNN models when working with EEG data are critical factors. This chapter will focus on available datasets and cover several important aspects, some of which are less commonly addressed.
4.1. Analysis of Datasets
While numerous research papers present their own datasets for evaluating machine learning models applied to EEG signals [126,127], it is worth noting that these datasets are often narrowly defined and may lack the broad recognition associated with well-established benchmark datasets, like the Microsoft-COCO datasets [128] used in machine vision tasks. Approximately a decade ago, there were popular brain-computer interface (BCI) competitions in which researchers competed to develop machine learning architectures for feature extraction from EEG signals [129,130,131]. Some parts of the datasets used in these competitions are still available. Additionally, several review papers have attempted to generalize datasets for application problems [132,133]. Additionally, datasets and architecture evaluations can be accessed through platforms such as Kaggle competitions (https://www.kaggle.com/competitions?searchQuery=EEG, accessed on 26 April 2023). However, each EEG dataset is typically unique, varying in the number of channels, electrode locations, data acquisition frequency, and other factors, making it challenging to adapt them for use with different CNN models. To address this issue, transfer learning is employed, which involves using the trained weights obtained from one dataset with another related dataset. In a review paper, Wan et al. [134] and Zhang et al. [135] conducted a search of the literature from 2010 to 2020 on the use of transfer learning in EEG decoding for brain-computer interfaces.
Another approach is self-supervised learning (SSL) [136], which can generate a dataset from the data themselves, simplifying the dataset collection process. Creating an EEG dataset using self-monitoring reduces the need for time-consuming EEG annotation [137]. In principle, SSL algorithms aim to derive everything they need from the data itself [138]. However, self-monitoring systems require a substantial amount of data, and the architecture must be efficient in terms of runtime and memory requirements. To date, SSL in EEG has been widely used in tasks such as anomaly detection in electroencephalography [139] and especially in sleep phase detection tasks [140,141,142,143]. SSL is a promising method for data label detection and has the potential to be applied to a wider range of tasks for feature extraction from EEG signals [144,145,146]. However, the application of SSL to EEG signals is not as widespread, and it is often used to identify events with prominent frequency components.
4.2. Overfitting in EEG Data
Overfitting in machine learning is a phenomenon in which the constructed model performs well in explaining examples from the training set but exhibits relatively poor performance when tested on new examples not included in the training set. This issue is particularly prevalent in EEG data analysis for several reasons. Firstly, the use of a large number of channels (e.g., 1024) and a high frequency (1000 Hz) results in a vast amount of data, much of which may not be relevant for practical tasks. Consequently, machine learning models can inadvertently learn patterns from noise rather than the actual events of interest within the dataset.
One of the commonly employed methods to solve the problem of overfitting is cross-validation [147,148]. Nevertheless, King et al. [149] demonstrated that cross-validation is not always a comprehensive solution for overfitting issues when dealing with EEG data, and it cannot serve as a universal method. As expected, many authors concur that one of the fundamental principles for addressing overfitting problems is to utilize a larger amount of EEG data [150,151].
To address the challenge of overfitting in CNN architectures, one approach is to reduce the number of hidden layers in the network. Zhang et al. [152] introduced a highly accurate neural network that reduces overfitting by incrementally increasing feature sizes and downsampling time series to eliminate muscle artifacts. Regularization is another commonly employed technique to prevent overfitting in EEG, as demonstrated by Zhang et al. [153] and Raduntz et al. [154]. Ying et al. [155] conducted an extensive review of contemporary methods for addressing overfitting issues in EEG.
4.3. Dimension Reduction of EEG Data
Reducing the sampling frequency is a common approach to mitigating overfitting, as typical sampling frequencies for reading data are around 1000 Hz [156]. However, it is important to note that upsampling can negatively impact the performance of a convolutional neural network [157], and downsampling may result in a loss of information unless it is known that the relevant data lies within a specific frequency band accessible through the Nyquist frequency. A simple algorithm for dimensionality reduction in EEG signals has been presented by Pagnotta et al. [158], but it should always be considered that lower sampling rates can lead to a reduction in model quality and connectivity estimate accuracy. Balancing the trade-off between mitigating overfitting and preserving signal quality requires careful consideration.
One of the most widely employed methods for dimensionality reduction in EEG data are PCA, which has been extensively utilized in various studies [159,160,161]. PCA reduces the dimensionality of a data set with “m” features into a subspace with fewer “n” features while retaining most of the information (or variance) from the original data set.
In addition to PCA, there are other techniques for dimensionality reduction that can be employed to prevent overestimation, including lock-boxes [162], blind signal analyses [163], pre-registrations [164], and nested cross-validation [165]. Pooling in convolutional neural networks [166,167] can also be used to reduce dimensionality. For example, Nakagome et al. [168] demonstrated that downsampling neural network-based decoders can effectively reduce dimensionality in recurrent networks. While Tang provided an overview of methods for dimensionalizing EEG signals [169], it is important to note that there are numerous techniques available, and some of them may be beyond the realm of neuroscience [170].
4.4. Data Representation in Different Dimensions
EEG data can be represented in various formats, depending on the algorithmic requirements and the research questions being addressed. One common representation is the 1D format, which is utilized when convolution kernels move along a single dimension, such as when analyzing EEG data over time. Another representation is the 2D format, which allows convolution kernels to move along two dimensions, facilitating the depiction of EEG data as matrices. The 3D format involves a convolution kernel convolving with the input layer, generating an output tensor. Figure 5 illustrates how EEG data can be fed into CNNs using the standard implementations for 1D, 2D, and 3D formats, with the electrode locations on the matrix corresponding to their positions on the scalp.
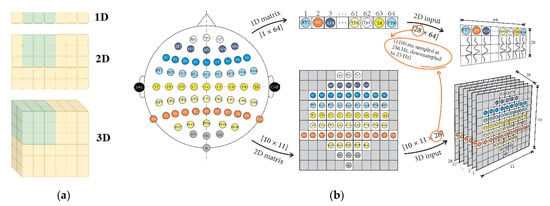
Figure 5.
Standard implementation of data transfer for 1D, 2D and 3D formats (a) Chollet et al. [171] location of electrodes on the matrix according to the placement on the scalp, (b) Sugi et al. [172].
Sugi et al. [172] introduced a CNN model that utilized 3D input for stimulus presentation intervals of 500, 400, and 300 ms, achieving remarkable P300 classification accuracy rates exceeding 80%. Similarly, Cho et al. [173] developed an emotion recognition method employing 3D convolutional neural networks (3D CNNs) to efficiently represent spatiotemporal features of EEG signals. Specifically, the authors reconstructed raw EEG signals as stacks of one-dimensional (1D) time series data into two-dimensional (2D) EEG frames based on their initial electrode positions. They then combined these frames with the time axis to obtain the 3D EEG stream, which they analyzed using 3D CNNs. Figure 6 illustrates these 3D reconstructions and their use in feature representation from spatiotemporal data.
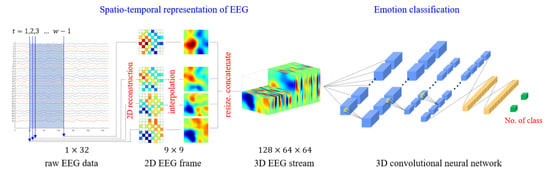
Figure 6.
A 3D CNN for emotion recognition tasks.
Recent research has explored various formats for presenting EEG data in CNNs for classification tasks, including 1D, 2D, and 3D formats. For instance, Oralhan et al. [174] presented EEG data in all three formats for classifying visual evoked potentials in a wireless brain-computer interface system, with the 3D CNN achieving the best results at an average classification accuracy of 93.75%. While the 2D format is more commonly used in the reviewed literature, some authors have suggested that 3D formats are better suited for studying spectral and spatial information in CNNs, as well as tasks that involve time and relationships, such as word recognition tasks [175,176]. However, it is crucial to recognize that the CNN architecture remains the most significant factor in determining their performance, and there is no evident correlation between data format and classification accuracy.
5. CNNs for EEG
The theory of CNN has been comprehensively covered in various works, such as the book authored by Francois Chollet, the developer of the Keras framework [177]. This book explains the theory of CNNs and the mechanisms for configuring them using the Python programming language [178]. Before employing CNN algorithms in EEG signal classification, one of the primary challenges lies in the quality of the recorded signal. EEG signals encompass numerous concurrent brain activities, rendering them complex and noisy to work with. Consequently, proper pre-filtering of raw EEG signals is essential to eliminate noise and artifacts. Additionally, the selection of extracted features should be based on their correlation with the desired outcome to enhance classification performance. CNNs consist of several crucial components, including layers of convolutional filters, activations, pooling, and fully connected layers. Pooling is employed to downsample the data effectively, reducing the number of parameters and mitigating overfitting. Convolution layers extract features from the data matrix, while filters increase the depth of the feature space and help learn more levels of abstract structures. The inclusion of fully connected layers aids in learning non-linear combinations of high-level features within the output of a convolutional layer. Each EEG channel represents a non-linear plot depicting EEG data measured in microvolts, illustrating variations in magnitude over time. Frequently, the overall input signal is initially transformed into a series of 2D time-frequency images. The time series data are represented as a 1D signal on one axis, while the signal’s frequency content is represented on the other axis. The work in [179] provides a clear guide on how to convert data into heat maps, as depicted in Figure 7.
Figure 7.
DWT waveforms for EEG data [179].
Convolutional filters are subsequently applied to each of these 2D images to extract local features, such as spikes or peaks, as shown in the heat plot in Figure 7, all of which are relevant to the task at hand. The output of the convolutional layer is then passed through one or more fully connected layers, which perform a non-linear feature transformation into the output categories.
For example, in machine vision tasks, recognizing a cat in an image involves a CNN learning to identify key visual features characteristic of cats, such as pointy ears, whiskers, and a furry body. Through a series of convolutions, activations, and merging operations, such as pooling, the CNN progressively transforms raw pixel values into a set of high-level features that capture these distinctive characteristics. After passing the image through the CNN layers, the fully connected output layer computes a set of class scores based on these features, with the highest score corresponding to the “cat” class. However, unlike searching for a cat in an image, working with EEG data are far more intricate and challenging to explain, given the data’s multidimensionality and interdependence. It is exceedingly difficult to visually identify the features requiring extraction from the graph. Consequently, the most effective approach involves the utilization of a CNN, which employs a series of trainable filters and layers. This CNN methodology enables the extraction of high-level features from the input image, facilitating the classification of these features into distinct categories among several possibilities.
5.1. Hyperparameters
Hyperparameters are an important component of CNN models, as they provide control over the training process and significantly impact the model’s performance and accuracy [180,181,182]. These hyperparameters encompass various factors, such as the number and size of kernels in each convolutional layer, the step size, and the size of kernels in the pooling layer [183]. While there has been extensive research on the influence of hyperparameters on CNN performance [184], the authors often provide only general guidance [185]. Determining the optimal hyperparameters for a specific model remains a challenging task, and the existing literature sometimes lacks practical insights on this matter. To address this challenge, various libraries are available for hyperparameter tuning, such as hose in Python [186], including the Scikit-learn library’s GridSearchCV and RandomizedSearchCV methods [187,188]. GridSearchCV explores all possible hyperparameter combinations to identify the best model, while RandomizedSearchCV tests random combinations of hyperparameters, making it a more efficient choice for larger datasets. Although some authors still employ RandomizedSearchCV [189], most researchers opt for GridSearchCV [190,191]. These methods prove invaluable in optimizing CNN model performance by identifying the ideal hyperparameters for a specific architecture while simultaneously reducing the risk of overfitting.
5.2. Kernel Size
Another important hyperparameter for CNN tuning is the kernel size, represented by the weight matrix used to filter the input data. Typically, small filters are employed to detect high-frequency objects, while larger kernels are utilized to identify low-frequency objects. A larger kernel size implies a less detailed examination of the data but may result in a more generalized representation of the input data. The EEG is characterized by non-linearity and non-stationarity, making it challenging to analyze comprehensively. Therefore, the selection of the kernel filter size is crucial and should be treated as if it were stationary within a specific time interval. This brings up the question of which size to choose for the kernel. Google researchers have made strides in addressing this issue by introducing a novel layer architecture called Inception [192,193]. The fundamental concept behind the Inception module is to perform multiple operations in parallel, such as combination and convolution, using filters of various sizes (3 × 3, 5 × 5, …). Figure 8 shows a convolution operation with 16 filters of sizes 1 × 1, 3 × 3. The resulting output tensor includes dimensions of 32 × 32 × 16, 32 × 32 × 32, and 32 × 32 × 64, where the last number corresponds to the number of resulting feature maps, equal to the number of filters collapsed on the image.
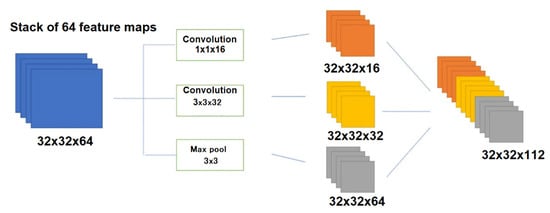
Figure 8.
Illustration of the Inception module implemented with input data of 32 × 32 × 64 and output data of 32 × 32 × 16, 32 × 32 × 32, and 32 × 32 × 64.
However, this type of model usually requires substantial computational resources, including a higher number of parameters and a longer training period [194,195].
6. Popular CNN Architectures for EEG
The EEG signal carries valuable information at specific frequency ranges: alpha (8–13 Hz), beta (14–40 Hz), theta (4–8 Hz), delta (0.5–3 Hz), gamma (above 40 Hz), and more. Each of these frequencies has its own unique characteristics and applications [196]. Since the frequency ranges and their characteristics have been extensively studied [197,198], many researchers have decomposed EEG signals into their frequency components and then fed the data into a CNN. It is important to note that one of the advantages of a CNN is its ability to discover dependencies that may elude human observation. Therefore, it is advisable to examine the raw data and exercise caution to prevent overfitting. In light of this, Zhang et al. [199] employed the short-time Fourier transform (STFT) method for frequency decomposition of the EEG signal to detect motor activity. The resulting data were then input into a 7-layer CNN designed for classification tasks with various core layers of 3 × 3, 2 × 2, and 3 × 3. In this CNN, the last two layers were fully connected layers, comprising 100 and 2 neurons, respectively, with the SoftMax classifier, as shown in Figure 9.
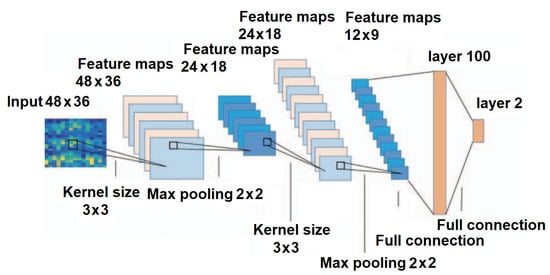
Figure 9.
Structural diagram of the CNN architecture for motion detection [199].
The use of CNNs in analyzing EEG signals has demonstrated promising results, particularly in the detection of frequency patterns [4,33]. Some researchers continue to rely on conventional methods to decompose EEG signals before feeding the data to CNNs. While these methods may offer faster results, their accuracy needs to be thoroughly validated. Lawhern et al. [200] introduced a specialized architecture known as EEGNet, designed specifically for brain-computer interfaces. In Figure 10, it can be observed how a CNN can decompose EEG signals. The CNN starts with a time convolution to learn frequency filters, followed by a depth convolution that learns dependent frequency mass filters. The final convolution is a basic convolution that generates a temporal summary for each feature map within the sequence. This is followed by a point convolution that learns the regularity of the feature map set, with more details about the model provided. These architectures are capable of learning hierarchical features from sequential data, making them well-suited for capturing patterns in the time domain of EEG signals. The main advantage of the EEGNet is the compactness of the CNN architecture. And the model can be easily adapted for different scenarios and integrated into other architectures of ML [201].
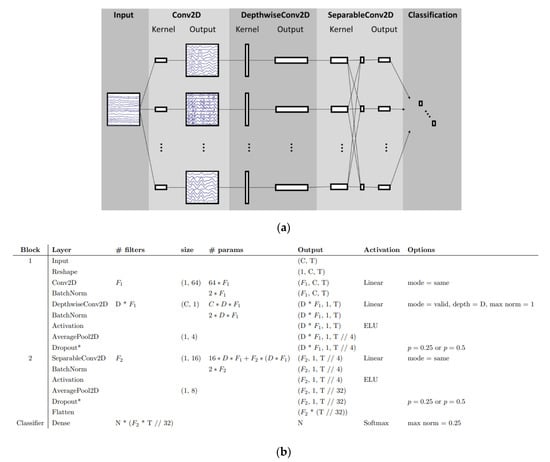
Figure 10.
EEGNet. (a) Block diagram; (b) Model forms as proposed in [200].
Wang et al. [202] introduced an 8-layer CNN designed for emotion recognition. The model’s input size was determined as width × height, where the width corresponded to 32 (representing the number of electrode channels), and the height was set at 3 × 128 = 384 (calculated as the product of the window size, 3 s, and the sampling frequency, 128 Hz). The batch size used for the model was set to 128, indicating the amount of data used in each batch. In the proposed model, Conv2 represents a multidimensional (2D) convolutional layer, Pooling2D stands for maximum 2D pooling, BatchNorm2d denotes 2D batch normalization, and Liner signifies a fully connected layer. Each convolutional layer is followed by an activation layer, Leaky ReLu. As a result, the architecture comprises eight convolutional layers, four batch normalizations, four dropout layers with a probability of 0.25, three maximum pooling layers, and two fully connected layers. The 5 × 1 convolution kernel folds the data in the temporal direction, while another 1 × 3 convolution kernel handles data folding in the spatial direction. The first three convolution blocks are connected to the maximum pooling layers at the end, and the architecture culminates in a fully connected layer utilized for emotion recognition classification. This comprehensive, lightweight model has a high degree of generalization and versatility, with an emphasis on real-time wearable applications. The visual representation of this architecture is shown in Figure 11.
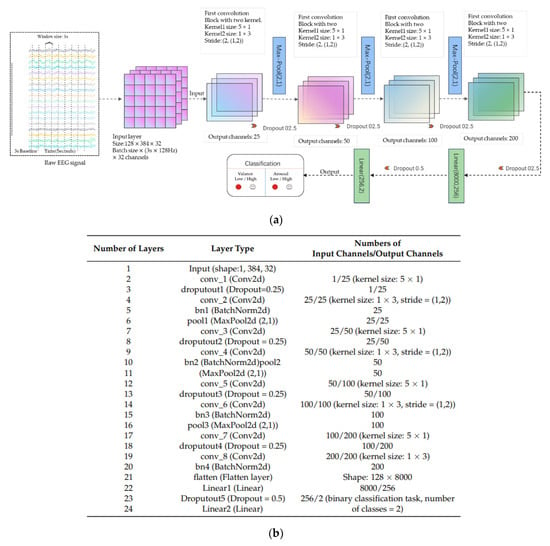
Figure 11.
CNN architecture for emotion recognition: (a) Block diagram; (b) Model forms proposed in [202].
Lun et al. [203] presented a 5-layer CNN structure designed for classifying physiological activity. The authors employed a 4-layer maximum pooling and a fully connected (FC) layer for classification. To mitigate the risk of overfitting, they incorporated dropout and batch normalization techniques. This architecture predominantly relies on 1D convolution, which is well-suited for extracting essential local features between neighboring element values of a feature vector. This model allows the decoding of raw EEG signals, providing reliability and adaptability, which simplifies the design of BCI systems for application applications. This is shown in Figure 12.
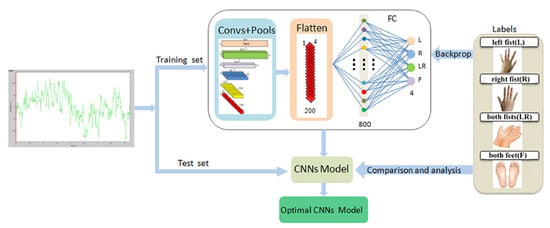
Figure 12.
CNN structure for the task of classifying physiological activity [203].
The primary challenge in EEG classification for MI tasks is its specificity. This means that each individual may exhibit unique characteristics that influence the system’s ability to correctly classify MI movement. To address this issue, multi-scale, multi-branch, or parallel architectures can be employed, enhancing the model’s generality.
The use of standard CNN architectures for EEG signal processing has demonstrated promising results in various applications, including motion image classification and seizure detection. Nevertheless, selecting optimal architecture and hyperparameters remains a task that is inherently tailored to the specific EEG signal and classification objective.
6.1. Architectures with Encoders and Decoders
In the context of EEG signal classification, it is common to utilize encoders and decoders to transform data into scales more suitable for CNNs [204]. The primary objective of these encoders is to reduce the dimensionality of the original feature space. Unlike PCA techniques, decoders are typically integrated into the CNN architecture and not used as a separate component of the data preparation process. Several studies have demonstrated that autoencoders outperform PCA in preparing data for CNNs [205,206]. By leveraging the CNN training procedure, autoencoders can effectively capture the salient features of objects, facilitating the recovery of the original sample objects.
Decoders are frequently employed in Generative Adversarial Networks (GANs) for various applications, including image generation [207,208]. More recently, GANs have been extended to time-series data [209], yielding promising results. Successful applications of the GAN architecture include generating synthetic data for use in LSTM networks [210], removing noise from data [211], and detecting sleep stages.
Supervised and unsupervised CNNs serve different purposes in the analysis of EEG (electroencephalography) data. Supervised CNNs are employed in EEG classification tasks where output labels are known. The networks require labeled training data to learn the relationship between EEG inputs and corresponding output labels. Supervised CNNs find applications in various domains, including emotion recognition, seizure detection, and sleep staging within EEG analysis. In contrast, unsupervised CNNs are utilized to explore EEG data, extract patterns, and identify underlying structures for research purposes. These networks are designed to learn the internal structure of EEG data without the need for labeled data. Unsupervised CNNs can be suitable for tasks such as clustering, dimensionality reduction, and anomaly detection. Autoencoders represent a specific type of unsupervised CNN used for studying EEG characteristics. The network is trained to recover input data at the output layer, with a bottleneck in the middle that learns a compressed representation of the input data. The learned features can subsequently be used for classification tasks. Therefore, supervised CNNs are employed for EEG classification tasks, while unsupervised CNNs are utilized for data exploration and feature learning.
6.2. Recurrent Neural Networks
Recurrent neural networks (RNNs) are well-suited for analyzing EEG data due to their ability to capture temporal dependencies, which are crucial for understanding brain dynamics [212]. The cyclic connections in RNNs allow the network to maintain an internal state or memory that can be updated with new data. This memory enables the network to process sequential data, such as EEG signals, while preserving temporal relationships between the data points. However, RNNs face challenges with vanishing gradients when dealing with long-length data. This issue has been addressed with the Long Short-Term Memory (LSTM) architecture [212]. Ma et al. [213] developed an architecture for predicting decision-making behavior from EEG signals using the t-SNE method. This method employs a stepwise iterative approach to represent the original data in a low-dimensional manner while preserving information about its local neighborhood. The architecture includes the t-SNE algorithm for feature extraction from EEG signals in the first stage and a recurrent neural network with a LSTM layer for predicting decision behavior in the second stage, as shown in Figure 13.
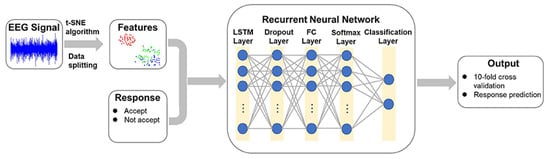
Figure 13.
RNN based algorithm for predicting decision behavior [213].
LSTMs, as well as CNNs, are applicable for sleep stage detection. Mousavi et al. [214] implemented a CNN with two sections for extracting temporal and frequency information. The input signals consist of a sequence of 30 s EEG epochs, and the output data represents the corresponding stages or classes. The encoder processes the input sequence, while the decoder computes the category of each individual channel of the 30 s EEG input sequence. The encoder consists of long short-term memory (LSTM) blocks that capture complex and long-term short-term contextual dependencies between inputs and targets, as shown in Figure 14. This algorithm proposed new approaches to calculating losses, which helped reduce the impact of the class imbalance problem and improve sleep stage detection performance. These LSTM blocks address non-linear dependencies across the entire time series when predicting the target.
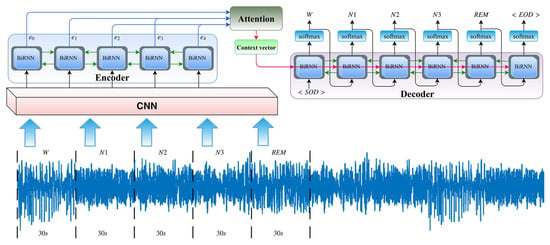
Figure 14.
CNN architecture for automatic estimation of sleep stages [214].
Fu et al. [215] introduced an architecture that employs a bidirectional recurrent neural network (BiRNN), consisting of an encoding and decoding module, for sleep phase detection. This approach combines time- and frequency-domain feature extraction using a CNN to expand the feature extraction domain while preserving the original EEG feature information. Time series information is extracted using the BiRNN encoding-decoding module, and automatic sleep stage discrimination of the EEG signal is performed using the SoftMax function. The block diagram of the network is shown in Figure 15.
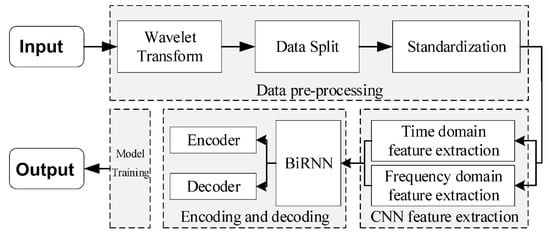
Figure 15.
Block diagram based on BiRNN for sleep stage detection [215].
In recent years, BiRNN has gained popularity for predicting speech from EEG signals. Schuster et al. [216] introduced the BiRNN architecture in 1997 as an extension of a RNN to a bidirectional recurrent neural network (BRNN), as shown in Figure 16. The authors showed that in the task of extracting a feature from EEG signals, the BRNN structure leads to better results than other ANN structures, while the training time for BRNN is approximately the same as for other RNNs.
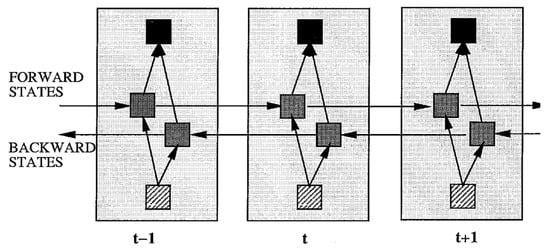
Figure 16.
Structuring the BIRNN schema shown unfolded in time for three time steps [216].
Unlike the standard RNN, the BRNN divides neurons into two directions: one for the positive time direction (forward state) and the other for the negative time direction (backward state).
6.3. Cascaded Architecture
Many papers exist in which the authors have presented architectures that combine multiple architectures to solve a task, often referred to as a “cascaded architecture”. The advantage of this approach is that it allows for the use of, for example, parts of an architecture designed to extract frequency patterns with another part of the CNN designed for extracting spatial filters. For instance, Altuwaijri et al. [217] employed the EEGNet architecture in the first stage to work on the frequency components of the signal, as described above for image signal classification. They included a block for altering the data dimension, referred to as Squeeze–Excitation, and only in the final part of the classification task did they use a few layers of custom CNN architecture.
Li et al. [218] proposed a solution to the motion classification problem by introducing the Temporal-Spectral Fusion of Squeeze and Excitation Functions (TS-SEFFNet). In this combined architecture, a deep temporal convolution block (DT-Conv block) was used to extract multivariate temporal representations from raw EEG data alongside a parallel multispectral convolution block (MS-Conv block). The use of multilayer wavelet convolutions enabled the extraction of information regarding the spectral component of the signal. A feature fusion block (SE-Feature-Fusion block) was employed to merge deep temporal and multispectral data into complex merged feature maps. Experimental results confirm that this architecture can effectively decode EEG, which can be considered a powerful tool for MI-EEG-based BCI. Finally, for motion classification, a classification block was utilized, as illustrated in Figure 17.
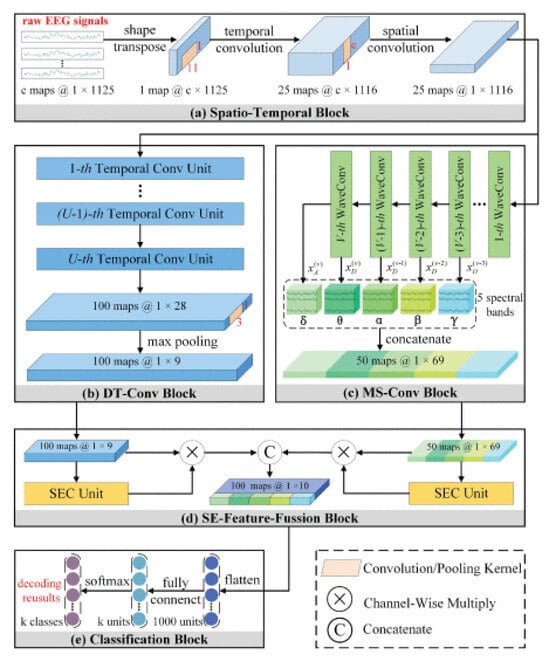
Figure 17.
Structural diagram of a combined architecture for motor imaging classification problems [218].
Several papers exist in which authors have presented complex combined deep architectures for the purpose of extracting features from EEG signals. These models are particularly intriguing because they are composed of distinct blocks, each of which can be adapted for use in a customized architecture. For example, Kostas et al. [219] employed a self-directed learning model for speech recognition from EEG signals. The model utilizes a multilevel convolutional feature encoder consisting of multiple blocks. Each block includes time convolution, followed by level normalization and a GELU activation function. The raw signal entering the encoder is normalized to have a mean of zero and a unit variance. The total encoder step determines the number of time steps provided to the converter. The output of the feature encoder is then directed into the context network, which follows the transformer architecture. This context network incorporates a convolutional layer for encoding absolute position information. Finally, the authors append the convolution output, followed by GELU, to the input and then apply layer normalization.
7. Details of CNNs in the Context of EEG Signals
The primary building blocks of CNNs include convolutional layers, subsampling (pooling) layers, activation layers, and fully connected layers. As part of EEG signal processing, we will consider hyperparameters, activation functions, and loss functions.
The loss function, in the theory of statistical decision-making, characterizes the losses incurred due to incorrect decisions based on observed data. Machine learning inherently revolves around optimization, and as in any optimization problem, we need to determine how far our predictions deviate to make the necessary adjustments. Loss functions take predictions and compare them to actual values or data labels, providing an error metric. Loss functions are fundamental components of any architecture and have been well-studied [220]. Thiyagarajan et al. [221] utilized a triplet-based loss function for clustering EEG data in their CNN, while Zhang et al. [222] applied the central loss function to improve the deep learning performance for EEG signal classification. Zhao et al. implemented focal loss for EEG-based seizure detection using a linear graph convolution network with focal loss [223]. Luo et al. [224] for EEG signal reconstruction using GAN with Wasserstein distance used temporal-spatial-frequency loss. This TSF-MSE-based loss function reconstructs signals by calculating MSE based on time series characteristics, general spatial structure characteristics, and power spectral density characteristics. Several researchers have introduced their custom loss functions, which, however, tend to be specialized and challenging to adapt to other architectures [225,226,227]. Brophy et al. [228] used a custom loss function to improve the denoising of electrode motion artifacts in ECG using convolutional neural networks. The choice of the optimal loss function for an architecture designed to work with EEG signals remains an ongoing challenge. Commonly used approaches include Mean Normalized Error (MNE) for extracting frequency patterns and Softmax loss for extracting spatial patterns.
Optimisers play a crucial role in CNN training, aiding in achieving increasingly accurate predictions. Optimizers determine the optimal set of model parameters, such as weights and biases, so that the model performs best for a given problem. The gradient descent algorithm [229,230] is a widely used optimization technique. A review of the papers showed that Adam is a commonly employed optimizer for both classification and prediction in the EEG field.
One of the stages in the development of a CNN is the choice of the activation function of the neurons. The type of activation function largely determines the functionality of the architecture and the method of training the model. The classic backpropagation algorithm [231] works well for CNNs with a few layers but encounters challenges as network depth increases, notably due to the problem of gradient attenuation [232]. The attenuation of gradients refers to the diminishing or vanishing of gradient values as they are propagated backward through the layers during the training process. In the context of the classic backpropagation algorithm, the attenuation of gradients can impede the effective updating of the network’s weights, especially in deep architectures. As the error propagates from the output layer to the input layer, the current result is multiplied by the derivative of the activation function at each layer. Different activation functions are employed in fully connected layers (FC), with the Rectified Linear Unit (ReLU) activation function being a common choice for neural network layers, especially for tasks in the frequency domain and spatial problem-solving, such as those addressed by Softmax. Dubey et al. [233] wrote about activation functions in the deep learning field; Mehta et al. [234] considered activation functions in the context of CNN; and Hao et al. [235] considered function activations more locally for EEG signal classification. In machine learning problems, metrics are used to evaluate the model quality and compare different algorithms. Understanding these metrics is crucial, as their values are used to evaluate the developed architecture. Choosing the right metrics is essential to avoid misinterpretation of the work of the CNN architecture. Carvalho et al. [236], R. Padilla et al. [237], and Saeedeh Ziyabari et al. [238] considered metrics in the context of machine learning, CNNs, and EEG signal processing.
8. Progress in Hardware
Laboratory equipment for non-real-time EEG data analysis faces no issues with computing power [239]. However, in recent years, there has been a growing interest in brain-computer interface devices that operate in real-time and are not highly powerful. Simultaneously, affordable consumer-grade EEG devices based on microcontrollers can measure EEG signals with the same quality as laboratory equipment [240,241]. This necessitated the implementation of machine learning algorithms directly on the microcontroller. The concept of Edge AI enables the utilization of machine learning algorithms directly on chips like Kneron, Kendryte, K210, and RISC-V [242,243]. For example, Fang et al. [244] employed Edge AI in a system-on-chip design for an EEG-based real-time emotion recognition task. TensorFlow introduced a platform for machine learning on embedded devices, known as TinyML [245], which gained popularity for microcontrollers [246,247,248]. STMicroelectronics has introduced a framework, X-CUBE-AI, for implementing machine learning algorithms on STM32 series microcontrollers [249,250,251].
Wang et al. [252] presented an EEGNet-based motor visualization brain-computer interface for low-power edge computing. To implement the EEGNet model on the ARM Cortex-M family of microcontrollers, Wang et al. [252] downsized the input feature map by reducing temporal and spatial dimensions and narrowing the time window, which relaxed the memory requirements.
Mezzina et al. [253] developed an Embedded convolutional NN (E-CNN) using two 1D convolutional layers, an intermediate batch normalization step to counter data covariate shift, and two dropout sections to mitigate overfitting phenomena. The batch size for the stochastic gradient descent was set to 128, and the optimal number of epochs to prevent overfitting was set to 50. The model was tested on STM32 microcontrollers with quantization crosses.
As the market for embedded devices continues to grow, along with the growing scope of EEG data, the significance of machine learning architectures for real-time EEG signal processing and feature extraction in embedded systems is becoming increasingly relevant.
9. Conclusions
In this paper, we have addressed the design of CNNs for custom tasks in the field of feature extraction from EEG signals. Our analysis encompasses several popular algorithms and explores the data preparation and hyperparameter tuning processes. One limitation in evaluating the models we have reviewed is that different datasets were used by the authors for their assessments. In neuroscience, comparing the effectiveness of models involved in different tasks is not as straightforward as in domains like computer vision, which employ widely recognized datasets such as COCO [254]. For this reason, we have provided a table in Appendix A detailing the parameters for different architectures. This table will help those who are starting to develop a custom architecture and will allow, at the initial stage, the selection of hyperparameters and the determination of the structure of the created architecture.
Recurrent neural networks, particularly LTSM, have found extensive application in event prediction from EEG signals. Encoders and decoders, serving as alternatives to PCA methods, have become effective in reducing dimensionality. These blocks are already part of the CNN architecture, and their placement within the network hierarchy depends on the architecture type.
Popular architectures can be categorized into two application areas: (a) identification of frequency patterns (e.g., sleep, emotions) and (b) spatial analysis, usually used for prediction tasks (e.g., motor imagery, speech). Standard implementations of CNNs are well suited for extracting frequency components, as evidenced by numerous papers with minor architectural and hyperparameter variations. Models based on recurrent neural networks, in particular LTSM, have been widely applied to the task of event prediction from EEG signals. Encoders and decoders, serving as alternatives to PCA methods, have become effective in reducing dimensionality. These blocks are already part of the CNN architecture, and their placement within the network hierarchy depends on the architecture type. The block architecture has shown that different model blocks can be easily implemented into new models, and the EEGNet model has become particularly popular in this direction, often being used as the first block for the task of decomposing the EEG signal into frequency components. While some papers use data preparation methods like PCA and ICA to enhance classification accuracy, many others work with raw data. CNNs exhibit an advantage in classification and prediction tasks, but one drawback when working with EEG signals is their limited generalizability, unlike the machine vision domain. In the future, we expect to see more approaches employing LSTM and more complex cascade models, with a new framework developed that will allow resource-intensive CNNs to run on hardware without large computing power.
In upcoming research, we plan to extend this article by implementing different CNN architectures (e.g., Similarity Learning Network, Multi-task learning) on the same dataset to facilitate a more direct comparison of their effectiveness. We will also consider well-established CNN architectures implemented outside the field of neuroscience. CNNs are more advanced in machine vision tasks and tuned for object detection tasks, yet the popular Yolov model [255], implemented for machine vision tasks, has also been used to extract features from EEG [256]. Therefore, it is logical for neuroscientists to look for new architects beyond the EEG domain. The accuracy of feature extraction from EEG data depends not only on the CNN architecture but also on various external factors, such as the number of EEG channels. More electrodes can provide enhanced spatial resolution, aiding in localizing neural activity and producing ERP. High-quality electrodes are effective in reducing electrical noise, and optimizing impedance matching between the electrodes and the scalp further improves the signal-to-noise ratio. Additionally, advanced data analysis techniques like machine learning and multivariate analysis can reveal subtle patterns in EEG data that are difficult to detect using traditional analysis methods. Combining EEG with other neuroimaging techniques, such as fMRI or MEG, can provide additional information about the neural processes underlying ERP.
Author Contributions
Writing—original draft, I.R.; Writing—review & editing, I.R., M.-S.D., A.N. and D.M.; Validation, A.N.; Funding acquisition, Supervision, D.M. All authors have read and agreed to the published version of the manuscript.
Funding
I.R., A.N. and D.M.’s work was supported in part by a USSOCOM grant EESB P85655.
Acknowledgments
We would like to thank Robert Oostenveld from Radboud University for his comments in the process of preparing the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Applying CNN to Feature Extraction from EEG Signals.
Table A1.
Applying CNN to Feature Extraction from EEG Signals.
| № | Tasks | Dataset | CNN | Learning Type | Steps | Structure | Optimization | Activation Function | Function Loss | Evaluation Metrics | Framework | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Classification task | Sleep stage annotations | Physionet Sleep-EDF dataset | SleepEEGNet | Supervised | Decomposition of data into frequency components and subsequent classification | 2DCNN and BiRNN | RMSProp optimizer | ReLU | lMFE | k-fold cross validation. overall accuracy, precision, recall (sensitivity), specificity, and F1-score. | Python 3.7–3.10, TensorFlow 2.8 | [256] |
| 2 | Emotion recognition | DEAP dataset 39 | EEG-Based Emotion Recognition Using a 2D CNN | Supervised | Decomposition of data into frequency components and subsequent classification | 2D CNN | Particle Swarm Optimization | LeakyReLU Outpit—Softmax | Cross Entropy | 85% | Python 3.7–3.10 | [214] | |
| 3 | Motor Imagery Signals Classification | BCI Competition IV 2a (BCI-IV2a), High Gamma (HGD) | MBEEGSE | Supervised | MBEEGSE architecture. Divided into three branches, each with EEGNet and SE block | EEGNet and Squeeze-and-Excitation (SE) Block | Adam optimizer | Softmax | Cross Entropy | 70% | Keras 3.0.4, Python 3.6, 3.7, 3.8, 3.9 | [202] | |
| 4 | Motor Imagery EEG Decoding | BCI Competition 2008 IV 2a Dataset High Gamma Dataset: (HGD) | TS-SEFFNet | Supervised | First, the deep temporal convolution block (DT-Conv block). Second, multispectral convolution block (MS-Conv block) is then run in parallel using multilevel wavelet convolutions. Finally, block (SE-Feature-Fusion block) displays the depth-time and multispectral features into complex pooled feature maps that extract the feature responses across channels. | DT-Conv block, MS-Conv block, SE-Feature-Fusion block | The Optimization Steps of the Proposed TS- SEFFNet Method | Softmax | Custom loss function | 93.25% | Torch 1.4, Python 3.8 | [217] | |
| 5 | Sleep stage annotations | Physionet Challenge dataset | Self-supervised learning (SSL) | Unsupervised | The first step is a sampling process by which examples are extracted from the time series S (EEG recording). The following describes a learning process where sample examples are used to train the feature extractor end-to-end. | Relative positioning (RP). Temporal shuffing (TS), Contrastive predective coding (CPC) | Adam optimizer | Rectified Linear Unit (ReLU) | Cross-entropy loss function | 72.3% | Torch 1.4, Python 3.9 | [218] | |
| 6 | Prediction task | EEG Imaginary Speech Recognition | Kara One database | - | Supervised | a CNN containing two convolutional layers with 64 and 128 filters connected to a dense layer with 64 neurons for input signal spectrum of a 0.25 s window | 2D CNN | Adam optimizer | Linear | Categorical cross-entropy | 37% | - | [257] |
| 7 | EEG-Speech Recognition | Custom dataset (not available) | - | Supervised | ResNet18/50/101 with 2 layers of managed recurrent units—Gated Recurrent Unit (GRU). And after that ResNet18 operation are fed to the input of a recurrent neural network containing 1024 hidden GRUs. | CNN and RNN | Adam optimizer | Softmax | - | 85% | - | [258] | |
| 8 | EEG speech recognition | Custom dataset (not available) | - | Supervised | The architecture used includes the already trained VGG Net CNN design and the target CNN design, while the already trained VGG Net CNN design extracts global features for general image classification work, and the target CNN design aims at efficient and accurate categorization of EEG signals using already trained Model VGG Net CNN. | Deep Residual–encoder–based VGG net CNN | - | Softmax | Softmax cross-entropy | 95% | - | [259] | |
| 9 | Seizure prediction | CHB-MIT and Kaggle | - | Supervised | a hybrid network that can combine the additional benefits of CNN and Transformer. The CNN is used to extract local information that contains two 3 × 3 convolutions with stride 1 and another 3 × 3 convolution with stride 2 to reduce the size of the input features. Each convolutional layer is followed by a GELU activation and a batch normalization (BN) layer. The model has two stages for extracting multiscale features from the EEG spectrum. Each stage consists of a set of Transformer blocks applied to extract long-term dependencies. | CNN and transformer | Adam | Softmax | Cross-entropy | 95% | Torch 1.4, Python 3.8 | [260] | |
| 10 | Predicting Human Intention-Behavior | BCI competition IV Dataset 2b | - | Supervised | The multi-scale CNN model has seven layers, which are one input layer, two convolutional layers, one max-pooling layer, one multi-scale layer, one full connection layer and one softmax output layer. The input layer in the multi-scale CNN model is fed with a time-frequency image with the size of 40 × 32 × 3 after EEG signals are preprocessed by STFT | Multi-Scale CNN Model | - | Linear | Cross-entropy | 73.9% | Python 3.8, Keras 3 | [261] | |
| 11 | Artifact Removal | EEG Artifact Detection and Correction | Costum dataset, not available | - | Unsupervised | modification of a feed-forward neural network that uses weight sharing and exhibits translation invariance. Learning in the CNNs operates on the same principle as a traditional feed-forward neural network where an error from output layer is back-propagated through the network and weights of the network are proportionally updated to the gradient of error. | CNN | Adam | - | Cross-entropy | - | Python 3.9, Keras 3 | [262] |
| 12 | Remove Muscle Artifacts from EEG | EEGdenoiseNet | - | Supervised | CNN for myogenic artifact reduction contains seven similar blocks. In each of the first six blocks, two 1D-convolution layers with small 1*3 kernels, 1 stride, and a ReLU activation function are followed by a 1D-Average pooling layer with pool size equal to two. In the seventh block, two 1D-convolution layers are followed by a flatten layer.The network gradually reduce the EEG signal sampling rate by the 1D-Average pooling layer. | CNN | RMSprop | ReLU | mean squared error (MSE) | - | Python 3.10, Tensorflow 2.8 | [263] | |
| 13 | Denoise EEG signal from artifacts | EEGdenoiseNet | MultiResUNet3+ | Supervised | Net3+ consists of full-blown pass-through connections that aggregate connections between encoders and decoders and internal connections between decoder subnets. Instead of directly combining the encoder and decoder functions, the encoder functions go through several convolutional levels with residual connections | CNN, encoders | Adam | Rely | mean squared error (MSE) | - | - | [152] | |
References
- Blanc, Y.; Dimanico, U. History of the Study of Skeletal Muscle Function with Emphasis on Kinesiological Electromyography. Open Rehabil. J. 2010, 3, 84–93. [Google Scholar] [CrossRef]
- Britton, J.W.; Frey, L.C.; Hopp, J.L.; Korb, P.; Koubeissi, M.Z.; Lievens, W.E.; Pestana-Knight, E.M.; St Louis, E.K. Electroencephalography (EEG): An Introductory Text and Atlas of Normal and Abnormal Findings in Adults, Children, and Infants. Am. Epilepsy Soc. 2016, 4. [Google Scholar]
- Nidal, K.; Malik, A. EEG/ERP Analysis: Methods and Applications; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Winkler, I.; Debener, S.; Muller, K.-R.; Tangermann, M. On the influence of high-pass filtering on ICA-based artifact reduction in EEG-ERP. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 4101–4105. [Google Scholar]
- Wang, Y.; Wang, Z.; Clifford, W.; Markham, C.; Ward, T.E.; Deegan, C. Validation of low-cost wireless EEG system for measuring event-related potentials. In Proceedings of the 2018 29th Irish Signals and Systems Conference (ISSC), Belfast, UK, 21–22 June 2018; pp. 1–6. [Google Scholar]
- Thompson, T.; Steffert, T.; Ros, T.; Leach, J.; Gruzelier, J. EEG applications for sport and performance. Methods 2008, 45, 279–288. [Google Scholar] [CrossRef]
- Armitage, R.; Hoffmann, R.F. Sleep EEG, depression and gender. Sleep Med. Rev. 2001, 5, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Masood, N.; Farooq, H. Investigating EEG Patterns for Dual-Stimuli Induced Human Fear Emotional State. Sensors 2019, 19, 522. [Google Scholar] [CrossRef] [PubMed]
- Al-Quraishi, M.S.; Elamvazuthi, I.; Daud, S.A.; Parasuraman, S.; Borboni, A. EEG-Based Control for Upper and Lower Limb Exoskeletons and Prostheses: A Systematic Review. Sensors 2018, 18, 3342. [Google Scholar] [CrossRef] [PubMed]
- Vasiljevic, G.A.M.; de Miranda, L.C. Brain–Computer Interface Games Based on Consumer-Grade EEG Devices: A Systematic Literature Review. Int. J. Hum. Comput. Interact. 2019, 36, 105–142. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, B.; Kong, W.; Xi, X.; Zhou, Y.; Gao, F. Tensor-based dynamic brain functional network for motor imagery classification. Biomed. Signal Process. Control 2021, 69, 102940. [Google Scholar] [CrossRef]
- Sanchez-Reyes, L.-M.; Rodriguez-Resendiz, J.; Avecilla-Ramirez, G.N.; Garcia-Gomar, M.-L.; Robles-Ocampo, J.-B. Impact of EEG Parameters Detecting Dementia Diseases: A Systematic Review. IEEE Access 2021, 9, 78060–78074. [Google Scholar] [CrossRef]
- Tao, D.; Tan, H.; Wang, H.; Zhang, X.; Qu, X.; Zhang, T. A Systematic Review of Physiological Measures of Mental Workload. Int. J. Environ. Res. Public Health. 2019, 16, 2716. [Google Scholar] [CrossRef]
- Stocker, R.A. Intensive Care in Traumatic Brain Injury Including Multi-Modal Monitoring and Neuroprotection. Med. Sci. 2019, 7, 37. [Google Scholar] [CrossRef]
- Ang, K.K.; Guan, C. EEG-Based Strategies to Detect Motor Imagery for Control and Rehabilitation. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 25, 392–401. [Google Scholar] [CrossRef]
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain–computer interfaces. J. Neural Eng. 2007, 4, R1–R13. [Google Scholar] [CrossRef]
- Hori, T.; Wolpert, M.D. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects’, the Rechtschaffen & Kales (1968) standard. Psychiatry Clin. Neurosci. 2001, 55, 305–310. [Google Scholar] [CrossRef]
- Carley, D.W.; Farabi, S.S. Physiology of Sleep. Diabetes Spectr. 2016, 29, 5–9. [Google Scholar] [CrossRef]
- Miah, O. Prediction of Motor Imagery Tasks from Multi-Channel EEG Data for Brain-Computer Interface Applications. bioRxiv 2020. [Google Scholar] [CrossRef]
- Pereda, E.; Gamundi, A.; Rial, R.; González, J. Non-linear behaviour of human EEG: Fractal exponent versus correlation dimension in awake and sleep stages. Neurosci. Lett. 1998, 250, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Elger, C.E.; Widman, G.; Andrzejak, R.; Arnhold, J.; David, P.; Lehnertz, K. Nonlinear EEG Analysis and Its Potential Role in Epileptology. Epilepsia 2000, 41, S34–S38. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Ding, L. Electrophysiological Mapping and Neuroimaging. In Neural Engineering; He, B., Ed.; Springer: Boston, MA, USA, 2013. [Google Scholar] [CrossRef]
- Riera, J.J.; Ogawa, T.; Goto, T.; Sumiyoshi, A.; Nonaka, H.; Evans, A.; Miyakawa, H.; Kawashima, R.; Halnes, G.; Mäki-Marttunen, T.; et al. Pitfalls in the dipolar model for the neocortical EEG sources. J. Neurophysiol. 2012, 108, 956–975. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Oh, S.; Shyamkumar, P.; Ramasamy, M.; Harbaugh, R.E.; Varadan, V.K. Nano- Bio- Textile Sensors with Mobile Wireless Platform for Wearable Health Monitoring of Neurological and Cardiovascular Disorders. J. Electrochem. Soc. 2013, 161, B3116–B3150. [Google Scholar] [CrossRef]
- Sridhar, C.; Lih, O.S.; Jahmunah, V.; Koh, J.E.W.; Ciaccio, E.J.; San, T.R.; Arunkumar, N.; Kadry, S.; Acharya, U.R. Accurate detection of myocardial infarction using non linear features with ECG signals. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 3227–3244. [Google Scholar] [CrossRef]
- Correia-Silva, J.R.; Berriel, R.F.; Badue, C.; De Souza, A.F.; Oliveira-Santos, T. Copycat CNN: Are random non-Labeled data enough to steal knowledge from black-box models? Pattern Recognit. 2021, 113, 107830. [Google Scholar] [CrossRef]
- Wang, B.; Ma, R.; Kuang, J.; Zhang, Y. How Decisions Are Made in Brains: Unpack “Black Box” of CNN With Ms. Pac-Man Video Game. IEEE Access 2020, 8, 142446–142458. [Google Scholar] [CrossRef]
- Ferrone, E.; Araneo, R.; Notargiacomo, A.; Pea, M.; Rinaldi, A. ZnO Nanostructures and Electrospun ZnO-Polymeric Hybrid Nanomaterials in Biomedical, Health, and Sustainability Applications. Nanomaterials 2019, 9, 1449. [Google Scholar] [CrossRef]
- Duan, L.; Ge, H.; Ma, W.; Miao, J. EEG feature selection method based on decision tree. Bio-Med. Mater. Eng. 2015, 26, S1019–S1025. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A Survey of Convolutional Neural Networks: Analysis, Applications, and Prospects. IEEE Trans. Neural Networks Learn. Syst. 2021, 33, 6999–7019. [Google Scholar] [CrossRef]
- Tirumala, S.S.; Ali, S.; Ramesh, C.P. Evolving deep neural networks: A new prospect. In Proceedings of the 2016 12th International Conference on Natural Computation and 13th Fuzzy Systems and Knowledge Discovery (ICNC-FSKD), Changsha, China, 13–15 August 2016; pp. 69–74. [Google Scholar]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Guler, I.; Ubeyli, E.D. Multiclass Support Vector Machines for EEG-Signals Classification. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 117–126. [Google Scholar] [CrossRef]
- Panda, R.; Khobragade, P.S.; Jambhule, P.D.; Jengthe, S.N.; Pal, P.; Gandhi, T.K. Classification of EEG signal using wavelet transform and support vector machine for epileptic seizure diction. In Proceedings of the 2010 International Conference on Systems in Medicine and Biology, Kharagpur, India, 16–18 December 2010; pp. 405–408. [Google Scholar] [CrossRef]
- Valueva, M.; Nagornov, N.; Lyakhov, P.; Valuev, G.; Chervyakov, N. Application of the residue number system to reduce hardware costs of the convolutional neural network. Math. Comput. Simul. 2020, 177, 232–243. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Indolia, S.; Goswami, A.K.; Mishra, S.; Asopa, P. Conceptual Understanding of Convolutional Neural Network- A Deep Learning Approach. Procedia Comput. Sci. 2018, 132, 679–688. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Islam, J.; Zhang, Y. Understanding 3D CNN Behavior for Alzheimer’s Disease Diagnosis from Brain PET Scan. arXiv 2019, arXiv:1912.04563. [Google Scholar] [CrossRef]
- Li, Y.-J.; Fan, F.-Y. Classification of Schizophrenia and Depression by EEG with ANNs. In Proceedings of the 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, Shanghai, China, 17–18 January 2006; pp. 2679–2682. [Google Scholar] [CrossRef]
- Srinivasan, V.; Eswaran, C.; Sriraam, N. Approximate Entropy-Based Epileptic EEG Detection Using Artificial Neural Networks. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-H.; Yu, B.-W.; Lee, D.-H.; Lee, S.-W. Classification of Drowsiness Levels Based on a Deep Spatio-Temporal Convolutional Bidirectional LSTM Network Using Electroencephalography Signals. Brain Sci. 2019, 9, 348. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Tiwari, J.; Acharya, U.R. Automatic Sleep-Stage Scoring in Healthy and Sleep Disorder Patients Using Optimal Wavelet Filter Bank Technique with EEG Signals. Int. J. Environ. Res. Public Heal. 2021, 18, 3087. [Google Scholar] [CrossRef] [PubMed]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain–computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef]
- Maitin, A.M.; Muñoz, J.P.R.; García-Tejedor, A.J. Survey of Machine Learning Techniques in the Analysis of EEG Signals for Parkinson’s Disease: A Systematic Review. Appl. Sci. 2022, 12, 6967. [Google Scholar] [CrossRef]
- Rodrigues, J.d.C.; Filho, P.P.R.; Peixoto, E.; Arun, K.N.; de Albuquerque, V.H.C. Classification of EEG signals to detect alcoholism using machine learning techniques. Pattern Recognit. Lett. 2019, 125, 140–149. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Qadir, J.; Sivathamboo, S.; Kwan, P.; Kuhlmann, L.; O’Brien, T.; Razi, A. Machine Learning for Predicting Epileptic Seizures Using EEG Signals: A Review. IEEE Rev. Biomed. Eng. 2021, 14, 139–155. [Google Scholar] [CrossRef]
- Gemein, L.A.; Schirrmeister, R.T.; Chrabąszcz, P.; Wilson, D.; Boedecker, J.; Schulze-Bonhage, A.; Hutter, F.; Ball, T. Machine-learning-based diagnostics of EEG pathology. NeuroImage 2020, 220, 117021. [Google Scholar] [CrossRef]
- Bazgir, O.; Mohammadi, Z.; Habibi, S.A.H. Emotion Recognition with Machine Learning Using EEG Signals. In Proceedings of the 2018 25th National and 3rd International Iranian Conference on Biomedical Engineering (ICBME), Qom, Iran, 29–30 November 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Wang, X.-W.; Nie, D.; Lu, B.-L. Emotional state classification from EEG data using machine learning approach. Neurocomputing 2013, 129, 94–106. [Google Scholar] [CrossRef]
- Nedelcu, E.; Portase, R.; Tolas, R.; Muresan, R.; Dinsoreanu, M.; Potolea, R. Artifact detection in EEG using machine learning. In Proceedings of the 13th IEEE International Conference on Intelligent Computer Communication and Processing (ICCP), Cluj-Napoca, Romania, 7–9 September 2017; pp. 77–83. [Google Scholar]
- Aggarwal, S.; Chugh, N. Review of Machine Learning Techniques for EEG Based Brain Computer Interface. Arch. Comput. Methods Eng. 2022, 29, 3001–3020. [Google Scholar] [CrossRef]
- Müller, K.-R.; Tangermann, M.; Dornhege, G.; Krauledat, M.; Curio, G.; Blankertz, B. Machine learning for real-time single-trial EEG-analysis: From brain–computer interfacing to mental state monitoring. J. Neurosci. Methods 2008, 167, 82–90. [Google Scholar] [CrossRef]
- Hosseini, M.-P.; Hosseini, A.; Ahi, K. A Review on Machine Learning for EEG Signal Processing in Bioengineering. IEEE Rev. Biomed. Eng. 2021, 14, 204–218. [Google Scholar] [CrossRef]
- Wu, W.; Nagarajan, S.; Chen, Z. Bayesian Machine Learning: EEG\/MEG signal processing measurements. IEEE Signal Process. Mag. 2015, 33, 14–36. [Google Scholar] [CrossRef]
- Fernández-Varela, I.; Hernández-Pereira, E.; Álvarez-Estévez, D.; Moret-Bonillo, V. Combining machine learning models for the automatic detection of EEG arousals. Neurocomputing 2017, 268, 100–108. [Google Scholar] [CrossRef]
- Saeidi, M.; Karwowski, W.; Farahani, F.V.; Fiok, K.; Taiar, R.; Hancock, P.A.; Al-Juaid, A. Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sci. 2021, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Roman-Gonzalez, A. EEG Signal Processing for BCI Applications. Human-Computer Systems Interaction: Backgrounds and Applications 2. In Advances in Intelligent and Soft Computing; Hippe, Z.S., Kulikowski, J.L., Mroczek, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 98, pp. 571–591. [Google Scholar] [CrossRef]
- Shedeed, H.A.; Issa, M.F.; El-Sayed, S.M. Brain EEG signal processing for controlling a robotic arm. In Proceedings of the 2013 8th International Conference on Computer Engineering & Systems (ICCES), Cairo, Egypt, 26–28 November 2013; pp. 152–157. [Google Scholar] [CrossRef]
- Kawala-Sterniuk, A.; Browarska, N.; Al-Bakri, A.; Pelc, M.; Zygarlicki, J.; Sidikova, M.; Martinek, R.; Gorzelanczyk, E.J. Summary of over Fifty Years with Brain-Computer Interfaces—A Review. Brain Sci. 2021, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Rakhmatulin, I.; Parfenov, A.; Traylor, Z.; Nam, C.S.; Lebedev, M. Low-cost brain computer interface for everyday use. Exp. Brain Res. 2021, 239, 3573–3583. [Google Scholar] [CrossRef] [PubMed]
- Baek, H. Ergonomic Issues in Brain-Computer Interface Technologies: Current Status, Challenges, and Future Direction. Comput. Intell. Neurosci. 2019, 2020, 5427154. [Google Scholar] [CrossRef] [PubMed]
- Occhipinti, E.; Davies, H.J.; Hammour, G.; Mandic, D.P. Hearables: Artefact removal in Ear-EEG for continuous 24/7 monitoring. In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–23 July 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Rakhmatulin, I. The electronic board to replace the reference voltage on the earlobe for EEG measurement. Measurement 2021, 173, 108673. [Google Scholar] [CrossRef]
- Pijn, J.P.; Van Neerven, J.; Noest, A.; da Silva, F.H.L. Chaos or noise in EEG signals; dependence on state and brain site. Electroencephalogr. Clin. Neurophysiol. 1991, 79, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.X. Where Does EEG Come from and What Does It Mean? Trends Neurosci. 2017, 40, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.B.; Marino, A.A.; Chesson, A.L. Frequency-specific responses in the human brain caused by electromagnetic fields. J. Neurol. Sci. 1994, 123, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Kottaimalai, R.; Rajasekaran, M.P.; Selvam, V.; Kannapiran, B. EEG signal classification using Principal Component Analysis with Neural Network in Brain Computer Interface applications. In Proceedings of the 2013 International Conference on Emerging Trends in Computing, Communication and Nanotechnology (ICE-CCN), Tirunelveli, India, 25–26 March 2013; pp. 227–231. [Google Scholar] [CrossRef]
- Rivet, B.; Souloumiac, A.; Attina, V.; Gibert, G. xDAWN Algorithm to Enhance Evoked Potentials: Application to Brain–Computer Interface. IEEE Trans. Biomed. Eng. 2009, 56, 2035–2043. [Google Scholar] [CrossRef]
- Mumtaz, W.; Rasheed, S.; Irfan, A. Review of challenges associated with the EEG artifact removal methods. Biomed. Signal Process. Control. 2021, 68, 102741. [Google Scholar] [CrossRef]
- Yasoda, K.; Ponmagal, R.S.; Bhuvaneshwari, K.S.; Venkatachalam, K. Automatic detection and classification of EEG artifacts using fuzzy kernel SVM and wavelet ICA (WICA). Soft Comput. 2020, 24, 16011–16019. [Google Scholar] [CrossRef]
- Shao, S.-Y.; Shen, K.-Q.; Ong, C.J.; Wilder-Smith, E.P.V.; Li, X.-P. Automatic EEG Artifact Removal: A Weighted Support Vector Machine Approach with Error Correction. IEEE Trans. Biomed. Eng. 2009, 56, 336–344. [Google Scholar] [CrossRef]
- Kaczorowska, M.; Plechawska-Wojcik, M.; Tokovarov, M.; Dmytruk, R. Comparison of the ICA and PCA methods in correction of EEG signal artefacts. In Proceedings of the 2017 10th International Symposium on Advanced Topics in Electrical Engineering (ATEE), Bucharest, Romania, 23–25 March 2017; pp. 262–267. [Google Scholar] [CrossRef]
- Hamaneh, M.B.; Chitravas, N.; Kaiboriboon, K.; Lhatoo, S.D.; Loparo, K.A. Automated Removal of EKG Artifact from EEG Data Using Independent Component Analysis and Continuous Wavelet Transformation. IEEE Trans. Biomed. Eng. 2014, 61, 1634–1641. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Hsu, S.-H.; Pion-Tonachini, L.; Jung, T.-P. Evaluation of Artifact Subspace Reconstruction for Automatic EEG Artifact Removal. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1242–1245. [Google Scholar] [CrossRef]
- Jiang, X.; Bian, G.-B.; Tian, Z. Removal of Artifacts from EEG Signals: A Review. Sensors 2019, 19, 987. [Google Scholar] [CrossRef]
- Kang, G.; Jin, S.-H.; Kim, D.K.; Kang, S.W. EEG artifacts removal using machine learning algorithms and independent component analysis. Clin. Neurophysiol. 2018, 129, e24. [Google Scholar] [CrossRef]
- Stalin, S.; Roy, V.; Shukla, P.K.; Zaguia, A.; Khan, M.M.; Shukla, P.K.; Jain, A. A Machine Learning-Based Big EEG Data Artifact Detection and Wavelet-Based Removal: An Empirical Approach. Math. Probl. Eng. 2021, 2021, 2942808. [Google Scholar] [CrossRef]
- Sun, W.; Su, Y.; Wu, X.; Wu, X. A novel end-to-end 1D-ResCNN model to remove artifact from EEG signals. Neurocomputing 2020, 404, 108–121. [Google Scholar] [CrossRef]
- Yang, B.; Duan, K.; Fan, C.; Hu, C.; Wang, J. Automatic ocular artifacts removal in EEG using deep learning. Biomed. Signal Process. Control 2018, 43, 148–158. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, M.; Wei, C.; Mantini, D.; Li, Z.; Liu, Q. EEGdenoiseNet: A benchmark dataset for deep learning solutions of EEG denoising. J. Neural Eng. 2021, 18, 056057. [Google Scholar] [CrossRef]
- Mashhadi, N.; Khuzani, A.Z.; Heidari, M.; Khaledyan, D. Deep learning denoising for EOG artifacts removal from EEG signals. In Proceedings of the 2020 IEEE Global Humanitarian Technology Conference (GHTC), Seattle, WA, USA, 29 October–1 November 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science; Navab, N., Hornegger, J., Wells, W., Frangi, A., Eds.; Springer: Cham, Switzerland, 2015; Volume 9351. [Google Scholar] [CrossRef]
- Jaiswal, A.; Nenonen, J.; Parkkonen, L. On electromagnetic head digitization in MEG and EEG. Sci. Rep. 2023, 13, 3801. [Google Scholar] [CrossRef]
- Goyal, K.; Borkholder, D.A.; Day, S.W. Dependence of Skin-Electrode Contact Impedance on Material and Skin Hydration. Sensors 2022, 22, 8510. [Google Scholar] [CrossRef]
- Grobbelaar, M.; Phadikar, S.; Ghaderpour, E.; Struck, A.F.; Sinha, N.; Ghosh, R.; Ahmed, M.Z.I. A Survey on Denoising Techniques of Electroencephalogram Signals Using Wavelet Transform. Signals 2022, 3, 577–586. [Google Scholar] [CrossRef]
- Brouwer, A.-M.; Hogervorst, M.A.; van Erp, J.B.F.; Heffelaar, T.; Zimmerman, P.H.; Oostenveld, R. Estimating workload using EEG spectral power and ERPs in the n-back task. J. Neural Eng. 2012, 9, 045008. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, M.I.; Ghilardi, M.F.; Kelly, S.P.; Blangero, A. Machine learning for EEG-based biomarkers in Parkinson’s disease. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2661–2665. [Google Scholar] [CrossRef]
- Orban, M.; Elsamanty, M.; Guo, K.; Zhang, S.; Yang, H. A Review of Brain Activity and EEG-Based Brain–Computer Interfaces for Rehabilitation Application. Bioengineering 2022, 9, 768. [Google Scholar] [CrossRef]
- Sun, J.; Hong, X.; Tong, S. Phase Synchronization Analysis of EEG Signals: An Evaluation Based on Surrogate Tests. IEEE Trans. Biomed. Eng. 2012, 59, 2254–2263. [Google Scholar] [CrossRef]
- Roach, B.J.; Mathalon, D.H. Event-Related EEG Time-Frequency Analysis: An Overview of Measures and An Analysis of Early Gamma Band Phase Locking in Schizophrenia. Schizophr. Bull. 2008, 34, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.C. An introduction to the coherence function and its use in EEG signal analysis. J. Med Eng. Technol. 1981, 5, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ren, D.; Li, K.; Feng, Y.; Ma, D.; Yan, X.; Wang, G. Epileptic Seizure Detection in Long-Term EEG Recordings by Using Wavelet-Based Directed Transfer Function. IEEE Trans. Biomed. Eng. 2018, 65, 2591–2599. [Google Scholar] [CrossRef]
- La Rocca, D.; Campisi, P.; Vegso, B.; Cserti, P.; Kozmann, G.; Babiloni, F.; Fallani, F.D.V. Human Brain Distinctiveness Based on EEG Spectral Coherence Connectivity. IEEE Trans. Biomed. Eng. 2014, 61, 2406–2412. [Google Scholar] [CrossRef]
- Babiloni, F.; Cincotti, F.; Babiloni, C.; Carducci, F.; Mattia, D.; Astolfi, L.; Basilisco, A.; Rossini, P.; Ding, L.; Ni, Y.; et al. Estimation of the cortical functional connectivity with the multimodal integration of high-resolution EEG and fMRI data by directed transfer function. NeuroImage 2005, 24, 118–131. [Google Scholar] [CrossRef]
- Olias, J.; Martin-Clemente, R.; Sarmiento-Vega, M.A.; Cruces, S. EEG Signal Processing in MI-BCI Applications with Improved Covariance Matrix Estimators. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 895–904. [Google Scholar] [CrossRef]
- Wang, F.; Wang, H.; Fu, R. Real-Time ECG-Based Detection of Fatigue Driving Using Sample Entropy. Entropy 2018, 20, 196. [Google Scholar] [CrossRef]
- Guan, S.; Zhao, K.; Yang, S. Motor Imagery EEG Classification Based on Decision Tree Framework and Riemannian Geometry. Brain-Inspired Intell. Syst. Dly. Assist. 2019, 2019, 5627156. [Google Scholar] [CrossRef]
- Yger, F.; Lotte, F.; Sugiyama, M. Averaging covariance matrices for EEG signal classification based on the CSP: An empirical study. In Proceedings of the 2015 23rd European Signal Processing Conference (EUSIPCO), Nice, France, 31 August–4 September 2015; pp. 2721–2725. [Google Scholar] [CrossRef]
- Van Vliet, M.; Salmelin, R. Post-hoc modification of linear models: Combining machine learning with domain information to make solid inferences from noisy data. NeuroImage 2019, 204, 116221. [Google Scholar] [CrossRef]
- Srinivasan, R.; Nunez, P.; Silberstein, R. Spatial filtering and neocortical dynamics: Estimates of EEG coherence. IEEE Trans. Biomed. Eng. 1998, 45, 814–826. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, A.; Mamun, K.; Tsunoda, T. A Deep Learning Approach for Motor Imagery EEG Signal Classification. In Proceedings of the 2016 3rd Asia-Pacific World Congress on Computer Science and Engineering (APWC on CSE), Nadi, Fiji, 5–6 December 2016; pp. 34–39. [Google Scholar] [CrossRef]
- Blankertz, B.; Muller, K.-R.; Krusienski, D.; Schalk, G.; Wolpaw, J.; Schlogl, A.; Pfurtscheller, G.; Millan, J.; Schroder, M.; Birbaumer, N. The BCI competition III: Validating alternative approaches to actual BCI problems. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 153–159. [Google Scholar] [CrossRef]
- Tangermann, M.; Müller, K.-R.; Aertsen, A.; Birbaumer, N.; Braun, C.; Brunner, C.; Leeb, R.; Mehring, C.; Miller, K.J.; Müller-Putz, G.R.; et al. Review of the BCI Competition IV. Front. Neurosci. 2012, 6, 55. [Google Scholar] [CrossRef]
- Delorme, A.; Makeig, S. EEGLAB: An Open Source Toolbox for Analysis of Single-Trial EEG Dynamics Including Independent Component Analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef]
- Wu, D.; King, J.-T.; Chuang, C.-H.; Lin, C.-T.; Jung, T.-P. Spatial Filtering for EEG-Based Regression Problems in Brain–Computer Interface (BCI). arXiv 2017, arXiv:1702.02914. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.-T.; Jung, T.-P. Translation of EEG Spatial Filters from Resting to Motor Imagery Using Independent Component Analysis. PLoS ONE 2012, 7, e37665. [Google Scholar] [CrossRef]
- Ghasemzadeh, P.; Kalbkhani, H.; Shayesteh, M.G. Sleep stages classification from EEG signal based on Stockwell transform. IET Signal Process. 2019, 13, 242–252. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Baloglu, U.B.; Acharya, U.R. A deep convolutional neural network model for automated identification of abnormal EEG signals. Neural Comput. Appl. 2018, 32, 15857–15868. [Google Scholar] [CrossRef]
- Williamson, J.R.; Bliss, D.W.; Browne, D.W.; Narayanan, J.T. Seizure prediction using EEG spatiotemporal correlation structure. Epilepsy Behav. 2012, 25, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yao, L.; Zhang, D.; Wang, X.; Sheng, Q.Z.; Gu, T. Multi-Person Brain Activity Recognition via Comprehensive EEG Signal Analysis. In Proceedings of the 14th EAI International Conference on Mobile and Ubiquitous Systems: Computing, Networking and Services, Melbourne, Australia, 7–10 November 2017; pp. 28–37. [Google Scholar] [CrossRef]
- Amin, H.U.; Mumtaz, W.; Subhani, A.R.; Saad, M.N.M.; Malik, A.S. Classification of EEG Signals Based on Pattern Recognition Approach. METHODS article. Front. Comput. Neurosci. 2017, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Farzan, F.; Atluri, S.; Frehlich, M.; Dhami, P.; Kleffner, K.; Price, R.; Lam, R.W.; Frey, B.N.; Milev, R.; Ravindran, A.; et al. Standardization of electroencephalography for multi-site, multi-platform and multi-investigator studies: Insights from the canadian biomarker integration network in depression. Sci. Rep. 2017, 7, 7473. [Google Scholar] [CrossRef]
- Wang, J.; Wang, M. Review of the emotional feature extraction and classification using EEG signals. Cogn. Robot. 2021, 1, 29–40. [Google Scholar] [CrossRef]
- Fdez, J.; Guttenberg, N.; Witkowski, O.; Pasquali, A. Cross-Subject EEG-Based Emotion Recognition Through Neural Networks with Stratified Normalization. Front. Neurosci. Sec. Brain Imaging Methods 2021, 15, 626277. [Google Scholar] [CrossRef]
- Liu, M.; Wu, W.; Gu, Z.; Yu, Z.; Qi, F.; Li, Y. Deep learning based on Batch Normalization for P300 signal detection. Neurocomputing 2017, 275, 288–297. [Google Scholar] [CrossRef]
- Shih, M.-T.; Doctor, F.; Fan, S.-Z.; Jen, K.-K.; Shieh, J.-S. Instantaneous 3D EEG Signal Analysis Based on Empirical Mode Decomposition and the Hilbert–Huang Transform Applied to Depth of Anaesthesia. Entropy 2015, 17, 928–949. [Google Scholar] [CrossRef]
- Rakhmatulin, I. Review of EEG Feature Selection by Neural Networks. Int. J. Sci. Bus. 2020, 4, 101–112. [Google Scholar] [CrossRef]
- Alotaiby, T.; El-Samie, F.E.A.; A Alshebeili, S.; Ahmad, I. A review of channel selection algorithms for EEG signal processing. EURASIP J. Adv. Signal Process. 2015, 2015, 66. [Google Scholar] [CrossRef]
- Molla, M.K.I.; Ahamed, S.; Almassri, A.M.M.; Wagatsuma, H. Classification of Motor Imagery Using Trial Extension in Spatial Domain with Rhythmic Components of EEG. Mathematics 2023, 11, 3801. [Google Scholar] [CrossRef]
- Riyadi, M.A.; Setiawan, I.; Amir, A. EEG Multiclass Signal Classification Based on Subtractive Clustering-ANFIS and Wavelet Packet Decomposition. In Proceedings of the 2021 International Conference on Electrical and Information Technology (IEIT), Malang, Indonesia, 14–15 September 2021; pp. 81–86. [Google Scholar] [CrossRef]
- Boonyakitanont, P.; Lek-Uthai, A.; Chomtho, K.; Songsiri, J. A review of feature extraction and performance evaluation in epileptic seizure detection using EEG. Biomed. Signal Process. Control 2019, 57, 101702. [Google Scholar] [CrossRef]
- Svetlakov, M.; Kovalev, I.; Konev, A.; Kostyuchenko, E.; Mitsel, A. Representation Learning for EEG-Based Biometrics Using Hilbert–Huang Transform. Computers 2022, 11, 47. [Google Scholar] [CrossRef]
- Van Hal, B.; Rhodes, S.; Dunne, B.; Bossemeyer, R. Low-cost EEG-based sleep detection. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Chicago, IL, USA, 26–30 August 2014; pp. 4571–4574. [Google Scholar] [CrossRef]
- Azarbad, M. A Time-Frequency approach for EEG signal segmentation. J. Artif. Intell. Data Min. 2014, 2, 63–71. [Google Scholar] [CrossRef]
- Birvinskas, D.; Jusas, V.; Martisius, V.; Damasevicius, E. EEG Dataset Reduction and Feature Extraction Using Discrete Cosine Transform. In Proceedings of the Sixth UKSim/AMSS European Symposium on Computer Modeling and Simulation, Malta, Malta, 14–16 November 2012; pp. 199–204. [Google Scholar] [CrossRef]
- Lan, Z.; Liu, Y.; Sourina, O.; Wang, L.; Scherer, R.; Müller-Putz, G. SAFE: An EEG dataset for stable affective feature selection. Adv. Eng. Inform. 2020, 44, 101047. [Google Scholar] [CrossRef]
- Iancu, B.; Soloviev, V.; Zelioli, L.; Lilius, J. ABOships—An Inshore and Offshore Maritime Vessel Detection Dataset with Precise Annotations. Remote Sens. 2021, 13, 988. [Google Scholar] [CrossRef]
- Blankertz, B.; Curio, G.; Müller, K. Classifying Single Trial EEG: Towards Brain Computer Interfacing. In Proceedings of the Advances in Neural Information Processing Systems 14 (NIPS 01), Vancouver, BC, Canada, 3–8 December 2001; The MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Wolpaw, J.R.; Birbaumer, N.; McFarland, D.J.; Pfurtscheller, G.; Vaughan, T.M. Brain–computer interfaces for communication and control. Clin. Neurophysiol. 2002, 113, 767–791. [Google Scholar] [CrossRef] [PubMed]
- Dornhege, G.; Blankertz, B.; Curio, G.; Muller, K.-R. Boosting bit rates in noninvasive EEG single-trial classifications by feature combination and multiclass paradigms. IEEE Trans. Biomed. Eng. 2004, 51, 993–1002. [Google Scholar] [CrossRef]
- Cimtay, Y.; Ekmekcioglu, E. Investigating the Use of Pretrained Convolutional Neural Network on Cross-Subject and Cross-Dataset EEG Emotion Recognition. Sensors 2020, 20, 2034. [Google Scholar] [CrossRef]
- Bouallegue, G.; Djemal, R.; Belwafi, K. Artificial EEG signal generated by a network of neurons with one and two dendrites. Results Phys. 2020, 20, 103699. [Google Scholar] [CrossRef]
- Wan, Z.; Yang, R.; Huang, M.; Zeng, N.; Liu, X. A review on transfer learning in EEG signal analysis. Neurocomputing 2020, 421, 1–14. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, G.; Zheng, X.; Li, H.; Zhang, S.; Yu, Y.; Liang, R. Application of Transfer Learning in EEG Decoding Based on Brain-Computer Interfaces: A Review. Sensors 2020, 20, 6321. [Google Scholar] [CrossRef]
- Montero Quispe, K.G.; Utyiama, D.M.S.; dos Santos, E.M.; Oliveira, H.A.B.F.; Souto, E.J.P. Applying Self-Supervised Representation Learning for Emotion Recognition Using Physiological Signals. Sensors 2022, 22, 9102. [Google Scholar] [CrossRef]
- Chato, L.; Regentova, E. Survey of Transfer Learning Approaches in the Machine Learning of Digital Health Sensing Data. J. Pers. Med. 2023, 13, 1703. [Google Scholar] [CrossRef]
- Malekzadeh, A.; Zare, A.; Yaghoobi, M.; Kobravi, H.-R.; Alizadehsani, R. Epileptic Seizures Detection in EEG Signals Using Fusion Handcrafted and Deep Learning Features. Sensors 2021, 21, 7710. [Google Scholar] [CrossRef]
- Xu, J.; Zheng, Y.; Mao, Y.; Wang, R.; Zheng, W.-S. Anomaly Detection on Electroencephalography with Self-supervised Learning. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Republic of Korea, 16–19 December 2020; pp. 363–368. [Google Scholar] [CrossRef]
- Jiang, X.; Zhao, J.; Du, B.; Yuan, Z. Self-supervised Contrastive Learning for EEG-based Sleep Staging. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Yang, C.; Xiao, D.; Westover, M.B.; Sun, J. Self-supervised EEG Representation Learning for Automatic Sleep Staging. arXiv 2021, arXiv:2110.15278. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, J.; Ye, J.; Zhang, H.; Bu, Y.; Zhang, Y.; Wu, H. Self-Supervised Learning for Sleep Stage Classification with Predictive and Discriminative Contrastive Coding. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1290–1294. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, Z.; Mo, R.; Chen, Z.; Zheng, W.S.; Wang, R. Task-Oriented Self-supervised Learning for Anomaly Detection in Electroencephalography. In Proceedings of the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI 2022, Singapore, 18–22 September 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13438, pp. 193–203. [Google Scholar] [CrossRef]
- Rafiei, M.H.; Gauthier, L.V.; Adeli, H.; Takabi, D. Self-Supervised Learning for Electroencephalography. IEEE Trans. Neural Networks Learn. Syst. 2022, 2022, 3190448. [Google Scholar] [CrossRef]
- Banville, H.; Albuquerque, I.; Hyvärinen, A.; Moffat, G.; Engemann, D.-A.; Gramfort, A. Self-Supervised Representation Learning from Electroencephalography Signals. In Proceedings of the 2019 IEEE 29th International Workshop on Machine Learning for Signal Processing (MLSP), Pittsburgh, PA, USA, 13–16 October 2019; IEEE: Piscataway, NJ, USA, 2019; Volume 10, pp. 1–6. [Google Scholar] [CrossRef]
- Islam, T.; Washington, P. Individualized Stress Mobile Sensing Using Self-Supervised Pre-Training. Appl. Sci. 2023, 13, 12035. [Google Scholar] [CrossRef]
- Mattiev, J.; Sajovic, J.; Drevenšek, G.; Rogelj, P. Assessment of Model Accuracy in Eyes Open and Closed EEG Data: Effect of Data Pre-Processing and Validation Methods. Bioengineering 2023, 10, 42. [Google Scholar] [CrossRef]
- Kingphai, K.; Moshfeghi, Y. On Time Series Cross-Validation for Deep Learning Classification Model of Mental Workload Levels Based on EEG Signals. In Proceedings of the Machine Learning, Optimization, and Data Science, Certosa di Pontignano, Italy, 19–22 September 2022; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2023; Volume 13811, pp. 402–416. [Google Scholar] [CrossRef]
- King, R.D.; Orhobor, O.I.; Taylor, C.C. Cross-validation is safe to use. Nat. Mach. Intell. 2021, 3, 276. [Google Scholar] [CrossRef]
- Saqib, M.; Zhu, Y.; Wang, M.; Beaulieu-Jones, B. Regularization of Deep Neural Networks for EEG Seizure Detection to Mitigate Overfitting. In Proceedings of the 2020 IEEE 44th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 13–17 July 2020; pp. 664–673. [Google Scholar] [CrossRef]
- Lashgari, E.; Liang, D.; Maoz, U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods 2020, 346, 108885. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, C.; Zhao, M.; Liu, Q.; Wu, H. A Novel Convolutional Neural Network Model to Remove Muscle Artifacts from EEG. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1265–1269. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, G.; Jin, J.; Zhao, Q.; Wang, X.; Cichocki, A. Sparse Bayesian Classification of EEG for Brain–Computer Interface. IEEE Trans. Neural Networks Learn. Syst. 2015, 27, 2256–2267. [Google Scholar] [CrossRef]
- Radüntz, T.; Scouten, J.; Hochmuth, O.; Meffert, B. Automated EEG artifact elimination by applying machine learning algorithms to ICA-based features. J. Neural Eng. 2017, 14, 046004. [Google Scholar] [CrossRef] [PubMed]
- Ying, X. An Overview of Overfitting and its Solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Park, C.; Looney, D.; Kidmose, P.; Ungstrup, M.; Mandic, D.P. Time-Frequency Analysis of EEG Asymmetry Using Bivariate Empirical Mode Decomposition. IEEE Trans. Neural Syst. Rehabilitation Eng. 2011, 19, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liu, D.; Yang, H.; Li, H.; Wu, F. Convolutional Neural Network-Based Block Up-Sampling for HEVC. IEEE Trans. Circuits Syst. Video Technol. 2019, 29, 3701–3715. [Google Scholar] [CrossRef]
- Pagnotta, M.F.; Plomp, G. Time-varying MVAR algorithms for directed connectivity analysis: Critical comparison in simulations and benchmark EEG data. PLoS ONE 2018, 13, e0198846. [Google Scholar] [CrossRef] [PubMed]
- Haufe, S.; Dähne, S.; Nikulin, V.V. Dimensionality reduction for the analysis of brain oscillations. NeuroImage 2014, 101, 583–597. [Google Scholar] [CrossRef]
- Artoni, F.; Delorme, A.; Makeig, S. Applying dimension reduction to EEG data by Principal Component Analysis reduces the quality of its subsequent Independent Component decomposition. NeuroImage 2018, 175, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, C.; Koenig, T.; Jelic, V.; Prichep, L.; John, R.E.; Wahlund, L.-O.; Dodge, Y.; Dierks, T. Application and comparison of classification algorithms for recognition of Alzheimer’s disease in electrical brain activity (EEG). J. Neurosci. Methods 2007, 161, 342–350. [Google Scholar] [CrossRef]
- Christie, S.A.; Hubbard, A.E.; Callcut, R.A.M.; Hameed, M.; Dissak-Delon, F.N.; Mekolo, D.; Saidou, A.; Mefire, A.C.; Nsongoo, P.; Dicker, R.A.; et al. Machine learning without borders? An adaptable tool to optimize mortality prediction in diverse clinical settings. J. Trauma Inj. Infect. Crit. Care 2018, 85, 921–927. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Hollcraft, N.; Alessio, A.M. Blind analysis of CT image noise using residual denoised images. In Proceedings of the 2015 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), San Diego, CA, USA, 31 October–7 November 2015; pp. 1–4. [Google Scholar] [CrossRef]
- Bakker, M.; Veldkamp, C.L.S.; Akker, O.R.v.D.; van Assen, M.A.L.M.; Crompvoets, E.; Ong, H.H.; Wicherts, J.M. Recommendations in pre-registrations and internal review board proposals promote formal power analyses but do not increase sample size. PLoS ONE 2020, 15, e0236079. [Google Scholar] [CrossRef]
- Hang, S.T.; Aono, M. Bi-linearly weighted fractional max pooling. Multimed. Tools Appl. 2017, 76, 22095–22117. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Krichen, M. Convolutional Neural Networks: A Survey. Computers 2023, 12, 151. [Google Scholar] [CrossRef]
- Nakagome, S.; Luu, T.P.; He, Y.; Ravindran, A.S.; Contreras-Vidal, J.L. An empirical comparison of neural networks and machine learning algorithms for EEG gait decoding. Sci. Rep. 2020, 10, 4372. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, D.; Li, X. Dimensionality Reduction Methods for Brain Imaging Data Analysis. ACM Comput. Surv. 2022, 54, 87. [Google Scholar] [CrossRef]
- Sorzano, C.; Vargas, J.; Monata, P. A survey of dimensionality reduction techniques. arXiv 2014, arXiv:1403.2877. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python; Manning Publications: New York, NY, USA, 2017. [Google Scholar]
- Sugi, M.; Nambu, I.; Wada, Y. Recent Developments in Deep Learning for Engineering Applications. Comput. Intell. Neurosci. 2017, 2018, 8163949. [Google Scholar] [CrossRef]
- Cho, J.; Hwang, H. Spatio-Temporal Representation of an Electoencephalogram for Emotion Recognition Using a Three-Dimensional Convolutional Neural Network. Sensors 2020, 20, 3491. [Google Scholar] [CrossRef]
- Oralhan, Z.; Oralhan, B.; Khayyat, M.M.; Abdel-Khalek, S.; Mansour, R.F. 3D Input Convolutional Neural Network for SSVEP Classification in Design of Brain Computer Interface for Patient User. Comput. Math. Methods Med. 2022, 2022, 8452002. [Google Scholar] [CrossRef]
- Gao, J.; Yang, C.; Liu, F.; Qi, J. Emotion Prediction of EEG Signals based on 1D Convolutional Neural Network. J. Phys. Conf. Ser. 2021, 2024, 012044. [Google Scholar] [CrossRef]
- Kwak, Y.; Kong, K.; Song, W.-J.; Min, B.-K.; Kim, S.-E. Multilevel Feature Fusion With 3D Convolutional Neural Network for EEG-Based Workload Estimation. IEEE Access 2020, 8, 16009–16021. [Google Scholar] [CrossRef]
- Chollet, F. Keras. 2015. Available online: https://github.com/keras-team/keras (accessed on 4 December 2023).
- Sha’abani, M.; Fuad, N.; Jamal, N.; Ismail, M.F. kNN and SVM Classification for EEG: A Review. In InECCE2019, Proceedings of the 5th International Conference on Electrical, Control & Computer Engineering, Kuantan, Pahang, Malaysia, 29 July 2019; Lecture Notes in Electrical Engineering; Springer: Cham, Switzerland, 2020; Volume 632. [Google Scholar] [CrossRef]
- Constable, P.A.; Marmolejo-Ramos, F.; Gauthier, M.; Lee, I.O.; Skuse, D.H.; Thompson, D.A. Discrete Wavelet Transform Analysis of the Electroretinogram in Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder. Front. Neurosci. 2022, 16, 890461. [Google Scholar] [CrossRef]
- Hilde, P.; Andreas, C.; Vanschoren, J. Importance of Tuning Hyperparameters of Machine Learning Algorithms. arXiv 2020, arXiv:2007.07588. [Google Scholar] [CrossRef]
- Fatyanosa, T.N.; Aritsugi, M. Effects of the Number of Hyperparameters on the Performance of GA-CNN. In Proceedings of the 2020 IEEE/ACM International Conference on Big Data Computing, Applications and Technologies (BDCAT), Leicester, UK, 7–10 December 2020; pp. 144–153. [Google Scholar] [CrossRef]
- Talaat, F.M.; Gamel, S.A. RL based hyper-parameters optimization algorithm (ROA) for convolutional neural network. J. Ambient. Intell. Humaniz. Comput. 2022, 14, 13349–13359. [Google Scholar] [CrossRef]
- Eom, J.; Kim, H.; Lee, S.H.; Kim, S. DNN-Assisted Cooperative Localization in Vehicular Networks. Energies 2019, 12, 2758. [Google Scholar] [CrossRef]
- Bernard, S.; Heutte, L.; Adam, S. Influence of Hyperparameters on Random Forest Accuracy. Lect. Notes Comput. Sci. 2009, 5519, 171–180. [Google Scholar] [CrossRef]
- Cooney, C.; Korik, A.; Folli, R.; Coyle, D. Evaluation of Hyperparameter Optimization in Machine and Deep Learning Methods for Decoding Imagined Speech EEG. Sensors 2020, 20, 4629. [Google Scholar] [CrossRef]
- Bergstra, J.; Komer, B.; Eliasmith, C.; Yamins, D.; Cox, D.D. Hyperopt: A Python library for model selection and hyperparameter optimization. Comput. Sci. Discov. 2015, 8, 014008. [Google Scholar] [CrossRef]
- García, E.M.; Alberti, M.G.; Arcos Álvarez, A.A. Measurement-While-Drilling Based Estimation of Dynamic Penetrometer Values Using Decision Trees and Random Forests. Appl. Sci. 2022, 12, 4565. [Google Scholar] [CrossRef]
- Kaleri, N.; Akkaldevi, S.; Krishna Teja, K. Heart Failure Survival Prediction using Various Machine Learning Approaches. In Data Engineering and Intelligent Computing; Lecture Notes in Networks and Systems; Bhateja, V., Khin Wee, L., Lin, J.C.W., Satapathy, S.C., Rajesh, T.M., Eds.; Springer: Cham, Switzerland, 2022; Volume 446, pp. 73–81. [Google Scholar] [CrossRef]
- Asadi-Pooya, A.A.; Kashkooli, M.; Asadi-Pooya, A.; Malekpour, M.; Jafari, A. Machine learning applications to differentiate comorbid functional seizures and epilepsy from pure functional seizures. J. Psychosom. Res. 2022, 153, 110703. [Google Scholar] [CrossRef]
- Avinash, M.; Nithya, M.; Aravind, S. Automated Machine Learning-Algorithm Selection with Fine-Tuned Parameters. In Proceedings of the 2022 6th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 25–27 May 2022; pp. 1175–1180. [Google Scholar] [CrossRef]
- Schiratti, J.-B.; Le Douget, J.-E.; Le Van Quyen, M.; Essid, S.; Gramfort, A. An Ensemble Learning Approach to Detect Epileptic Seizures from Long Intracranial EEG Recordings. In Proceedings of the ICASSP 2018—2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 856–860. [Google Scholar] [CrossRef]
- Zheng, G.; Han, G.; Soomro, N.Q. An inception module CNN classifiers fusion method on pulmonary nodule diagnosis by signs. Tsinghua Sci. Technol. 2020, 25, 368–383. [Google Scholar] [CrossRef]
- Mauro, D.; Appice, N.; Basile, N. Activity Prediction of Business Process Instances with Inception CNN Models. 2019—Advances in Artificial Intelligence. In Proceedings of the International Conference of the Italian Association for Artificial Intelligence—AI*IA 2019, Rende, Italy, 19–22 Novembre 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11946, pp. 348–361. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A.; Liu, W.; et al. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Liao, J.J.; Luo, J.J.; Yang, T.; So, R.Q.Y.; Chua, M.C.H. Effects of local and global spatial patterns in EEG motor-imagery classification using convolutional neural network. Brain Comput. Interfaces 2020, 7, 47–56. [Google Scholar] [CrossRef]
- Ayvazyan, H.L.; Antonyan, S.V.; Makaryan, A.H.; Hovhannisyan, B.A.; Sivolenko, E.R.; Tsaturyan, G.A. Registration of Brain Radio Signals and Their Bispectral Analysis. J. Contemp. Phys. Armen. Acad. Sci. 2022, 57, 87–90. [Google Scholar] [CrossRef]
- Mahmud, T.; Khan, I.A.; Ibn Mahmud, T.; Fattah, S.A.; Zhu, W.-P.; Ahmad, M.O. Sleep Apnea Detection from Variational Mode Decomposed EEG Signal Using a Hybrid CNN-BiLSTM. IEEE Access 2021, 9, 102355–102367. [Google Scholar] [CrossRef]
- Krishnan, K.K.; Soman, K.P. CNN based classification of motor imaginary using variational mode decomposed EEG-spectrum image. Biomed. Eng. Lett. 2021, 11, 235–247. [Google Scholar] [CrossRef]
- Zhang, J.; Yan, C.; Gong, X. Deep convolutional neural network for decoding motor imagery based brain computer interface. In Proceedings of the 2017 IEEE International Conference on Signal Processing, Communications and Computing (ICSPCC), Xiamen, China, 22–25 October 2017; pp. 1–5. [Google Scholar]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolutional Neural Network for EEG-Based Brain–Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Raza, H.; Chowdhury, A.; Bhattacharyya, S.; Samothrakis, S. Single-Trial EEG Classification with EEGNet and Neural Structured Learning for Improving BCI Performance. In Proceedings of the International Joint Conference on Neural Networks (IJCNN 2020), Glasgow, UK, 19–24 July 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Xia, P.; Wang, P.; Chen, X.; Du, L.; Fang, Z.; Du, M. EEG-Based Emotion Recognition Using a 2D CNN with Different Kernels. Bioengineering 2022, 9, 231. [Google Scholar] [CrossRef]
- Lun, X.; Yu, Z.; Chen, T.; Wang, F.; Hou, Y. A Simplified CNN Classification Method for MI-EEG via the Electrode Pairs Signals. Front. Hum. Neurosci. 2020, 14, 338. [Google Scholar] [CrossRef]
- Xu, L.; Xu, M.; Jung, T.-P.; Ming, D. Review of brain encoding and decoding mechanisms for EEG-based brain–computer interface. Cogn. Neurodynamics 2021, 15, 569–584. [Google Scholar] [CrossRef]
- Liang, Z. EEGFuseNet: Hybrid Unsupervised Deep Feature Characterization and Fusion for High-Dimensional EEG with An Application to Emotion Recognition. arXiv 2021, arXiv:2102.03777v2. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Luo, Y.; Lu, B.-L. EEG Data Augmentation for Emotion Recognition Using a Conditional Wasserstein GAN. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 2535–2538. [Google Scholar] [CrossRef]
- Hwang, S.; Hong, K.; Son, G.; Byun, H. EZSL-GAN: EEG-based Zero-Shot Learning approach using a Generative Adversarial Network. In Proceedings of the 2019 7th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Republic of Korea, 18–20 February 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Hartmann, K.; Schirrmeister, R.; Ball, T. EEG-GAN: Generative adversarial networks for electroencephalograhic (EEG) brain signals. arXiv 2018, arXiv:1806.01875. [Google Scholar]
- Jiao, Y.; Deng, Y.; Luo, Y.; Lu, B.-L. Driver sleepiness detection from EEG and EOG signals using GAN and LSTM networks. Neurocomputing 2020, 408, 100–111. [Google Scholar] [CrossRef]
- Sumiya, Y.; Horie, K.; Shiokawa, H.; Kitagawa, H. Noise Reduction GAN for Mice Electroencephalogram Signals. In ICBSP ’19, Proceedings of the 2019 4th International Conference on Biomedical Imaging, Signal Processing, Nagoya, Japan, 17–19 October 2019; pp. 94–101. [Google Scholar] [CrossRef]
- Sherstinsky, A. Fundamentals of Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM) Network. Phys. D Nonlinear Phenom. 2020, 404, 132306. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, M.; Hu, L.; Zhang, L.; Hua, Z. A Novel Recurrent Neural Network to Classify EEG Signals for Customers’ Decision-Making Behavior Prediction in Brand Extension Scenario. Front. Hum. Neurosci. 2021, 15, 610890. [Google Scholar] [CrossRef]
- Mousavi, S.; Afghah, F.; Acharya, U.R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, C.; Zhang, L.; Wang, S.; Zhang, Y. Deep Learning Model of Sleep EEG Signal by Using Bidirectional Recurrent Neural Network Encoding and Decoding. Electronics 2022, 11, 2644. [Google Scholar] [CrossRef]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Altuwaijri, G.A.; Muhammad, G.; Altaheri, H.; Alsulaiman, M. A Multi-Branch Convolutional Neural Network with Squeeze-and-Excitation Attention Blocks for EEG-Based Motor Imagery Signals Classification. Diagnostics 2022, 12, 995. [Google Scholar] [CrossRef]
- Li, Y.; Guo, L.; Liu, Y.; Liu, J.; Meng, F. A Temporal-Spectral-Based Squeeze-and- Excitation Feature Fusion Network for Motor Imagery EEG Decoding. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1534–1545. [Google Scholar] [CrossRef]
- Kostas, D.; Aroca-Ouellette, S.; Rudzicz, F. BENDR: Using transformers and a contrastive self-supervised learning task to learn from massive amounts of EEG data. arXiv 2021, arXiv:2101.12037. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, P.; Wang, Z.; Ye, X. A New Loss Function for CNN Classifier Based on Predefined Evenly-Distributed Class Centroids. IEEE Access 2019, 8, 10888–10895. [Google Scholar] [CrossRef]
- Thiyagarajan, R.; Curro, C.; Keene, S. A learned embedding space for EEG signal clustering. In Proceedings of the 2017 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 2 December 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Q. Using the center loss function to improve deep learning performance for EEG signal classification. In Proceedings of the 2018 Tenth International Conference on Advanced Computational Intelligence (ICACI), Xiamen, China, 29–31 March 2018; pp. 578–582. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, C.; Zhang, G.; Wang, Y.; Chen, X.; Jia, W.; Yuan, Q.; Xu, F.; Zheng, Y. EEG-Based Seizure detection using linear graph convolution network with focal loss. Comput. Methods Programs Biomed. 2021, 208, 106277. [Google Scholar] [CrossRef]
- Luo, T.-J.; Fan, Y.; Chen, L.; Guo, G.; Zhou, C. EEG Signal Reconstruction Using a Generative Adversarial Network with Wasserstein Distance and Temporal-Spatial-Frequency Loss. Front. Neurosci. 2020, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Zhang, K.; Li, Z.; Qiao, Y. A Discriminative Feature Learning Approach for Deep Face Recognition. In Computer Vision—ECCV 2016. ECCV 2016. Lecture Notes in Computer Science, 9911; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Jean, D. Improving the Prediction of Asset Returns with Machine Learning by Using a Custom Loss Function. 2022. Available online: https://ssrn.com/abstract=3973086 (accessed on 4 December 2023). [CrossRef]
- Vicar, T.; Hejc, J.; Novotna, P.; Ronzhina, M.; Janousek, O. ECG Abnormalities Recognition Using Convolutional Network with Global Skip Connections and Custom Loss Function. In Proceedings of the 2020 Computing in Cardiology Conference, Rimini, Italy, 13–16 September 2020. [Google Scholar] [CrossRef]
- Brophy, E.; Hennelly, B.; De Vos, M.; Boylan, G.; Ward, T. Improved Electrode Motion Artefact Denoising in ECG Using Convolutional Neural Networks and a Custom Loss Function. IEEE Access 2022, 10, 54891–54898. [Google Scholar] [CrossRef]
- Gupta, H.; Jin, K.H.; Nguyen, H.Q.; McCann, M.T.; Unser, M. CNN-Based Projected Gradient Descent for Consistent CT Image Reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yang, G. Modified Convolutional Neural Network Based on Dropout and the Stochastic Gradient Descent Optimizer. Algorithms 2018, 11, 28. [Google Scholar] [CrossRef]
- Li, F.; He, F.; Wang, F.; Zhang, D.; Xia, Y.; Li, X. A Novel Simplified Convolutional Neural Network Classification Algorithm of Motor Imagery EEG Signals Based on Deep Learning. Appl. Sci. 2020, 10, 1605. [Google Scholar] [CrossRef]
- Ferreira, J.; Aarts, R.; PCluitmans, P. Optimized moving-average filtering for gradient artefact correction during simultaneous EEG-fMRI. In Proceedings of the 5th ISSNIP-IEEE Biosignals and Biorobotics Conference: Biosignals and Robotics for Better and Safer Living (BRC), Salvador, Brazil, 26–28 May 2014; pp. 1–6. [Google Scholar] [CrossRef]
- Dubey, S.R.; Singh, S.K.; Chaudhuri, B.B. Activation functions in deep learning: A comprehensive survey and benchmark. Neurocomputing 2022, 503, 92–108. [Google Scholar] [CrossRef]
- Mehta, D.B.; Barot, P.A.; Langhnoja, S.G. Effect of Different Activation Functions on EEG Signal Classification based on Neural Networks. In Proceedings of the 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 11–13 March 2020; pp. 132–135. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Lou, Y.; Song, Z. The Role of Activation Function in CNN. In Proceedings of the 2020 2nd International Conference on Information Technology and Computer Application (ITCA), Guangzhou, China, 18–20 December 2020; pp. 429–432. [Google Scholar] [CrossRef]
- Carvalho, D.V.; Pereira, E.M.; Cardoso, J.S. Machine Learning Interpretability: A Survey on Methods and Metrics. Electronics 2019, 8, 832. [Google Scholar] [CrossRef]
- Padilla, R.; Netto, S.L.; da Silva, E.A.B. A Survey on Performance Metrics for Object-Detection Algorithms. In Proceedings of the 2020 International Conference on Systems, Signals and Image Processing (IWSSIP), Niteroi, Brazil, 1–3 July 2020; pp. 237–242. [Google Scholar] [CrossRef]
- Ziyabari, S. Objective evaluation metrics for automatic classification of EEG events. arXiv 2017, arXiv:1712.10107. [Google Scholar]
- Dadebayev, D.; Goh, W.W.; Tan, E.X. EEG-based emotion recognition: Review of commercial EEG devices and machine learning techniques. J. King Saud Univ. Comput. Inf. Sci. 2021, 34, 4385–4401. [Google Scholar] [CrossRef]
- Rezeika, A.; Benda, M.; Stawicki, P.; Gembler, F.; Saboor, A.; Volosyak, I. Brain–Computer Interface Spellers: A Review. Brain Sci. 2018, 8, 57. [Google Scholar] [CrossRef]
- Żyliński, M.; Nassibi, A.; Rakhmatulin, I.; Malik, A.; Papavassiliou, C.; Mandic, D.P. Deployment of Artificial Intelligence Models on Edge Devices: A Tutorial Brief. J. Latex Cl. Files 2023, 18. [Google Scholar] [CrossRef]
- Kalapothas, S.; Galetakis, M.; Flamis, G.; Plessas, F.; Kitsos, P. A Survey on RISC-V-Based Machine Learning Ecosystem. Information 2023, 14, 64. [Google Scholar] [CrossRef]
- Ingolfsson, T.M.; Wang, X.; Hersche, M.; Burrello, A.; Cavigelli, L.; Benini, L. ECG-TCN: Wearable Cardiac Arrhythmia Detection with a Temporal Convolutional Network. In Proceedings of the 2021 IEEE 3rd International Conference on Artificial Intelligence Circuits and Systems (AICAS), Washington, DC, USA, 6–9 June 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Fang, W.-C.; Wang, K.-Y.; Fahier, N.; Ho, Y.-L.; Huang, Y.-D. Development and Validation of an EEG-Based Real-Time Emotion Recognition System Using Edge AI Computing Platform with Convolutional Neural Network System-on-Chip Design. IEEE J. Emerg. Sel. Top. Circuits Syst. 2019, 9, 645–657. [Google Scholar] [CrossRef]
- Liu, S.-C.; Gao, C.; Kim, K.; Delbruck, T. Energy-efficient activity-driven computing architectures for edge intelligence. In Proceedings of the 2022 IEEE International Electron Devices Meeting (IEDM), San Francisco, CA, USA, 3–7 December 2022; pp. 21.2.1–21.2.4. [Google Scholar] [CrossRef]
- Pham, H.-T.; Nguyen, M.-A.; Sun, C.-C. AIoT Solution Survey and Comparison in Machine Learning on Low-cost Microcontroller. In Proceedings of the 2019 International Symposium on Intelligent Signal Processing and Communication Systems (ISPACS), Taipei, Taiwan, 3–6 December 2019; pp. 1–2. [Google Scholar] [CrossRef]
- Saha, S.S.; Sandha, S.S.; Srivastava, M. Machine Learning for Microcontroller-Class Hardware: A Review. IEEE Sensors J. 2022, 22, 21362–21390. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.P. A review on TinyML: State-of-the-art and prospects. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 1595–1623. [Google Scholar] [CrossRef]
- Rakhmatuiln, I.; Zhanikeev, M.; Parfenov, A. Raspberry PI Shield—For measure EEG (PIEEG). In Proceedings of the 2021 5th International Conference on Electrical, Electronics, Communication, Computer Technologies and Optimization Techniques (ICEECCOT), Mysuru, India, 10–11 December 2021; pp. 410–413. [Google Scholar] [CrossRef]
- Falbo, V.; Apicella, T.; Aurioso, D.; Danese, L.; Bellotti, F.; De Gloria, A. Analyzing Machine Learning on Mainstream Microcontrollers. In Applications in Electronics Pervading Industry, Environment and Society, ApplePies; Lecture Notes in Electrical Engineering; Saponara, S., De Gloria, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; p. 627. [Google Scholar] [CrossRef]
- Harika, N.; Kumar, T.K. Real Time Smart Music Player Using Facial Expression. In Proceedings of the 2022 IEEE International Conference on Signal Processing and Communications (SPCOM), Bangalore, India, 11–15 July 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Wang, X.; Hersche, M.; Tomekce, B.; Kaya, B.; Magno, M.; Benini, L. An Accurate EEGNet-based Motor-Imagery Brain–Computer Interface for Low-Power Edge Computing. In Proceedings of the 2020 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Bari, Italy, 1 June–1 July 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Mezzina, G.; De Venuto, D. Low-Complexity Unidimensional CNN based Brain Speller for Embedded Platforms. In Proceedings of the 2021 IEEE Nordic Circuits and Systems Conference (NorCAS), Oslo, Norway, 26–27 October 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Zitnick, C.L.; Dollár, P. Microsoft COCO: Common Objects in Context. In Proceedings of the 13th European Conference on Computer Vision—ECCV 2014, Zurich, Swityerland, 6–12 September 2014; Lecture Notes in Computer Science; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Cham, Switzerland, 2014; Volume 8693. [Google Scholar] [CrossRef]
- Rakhmatulin, I.; Duchowski, A.T. Deep Neural Networks for Low-Cost Eye Tracking. Procedia Computer Science 2020, 176, 685–694. [Google Scholar] [CrossRef]
- Ye, J.; Pandarinath, C. Representation learning for neural population activity with Neural Data Transformers. arXiv 2021, arXiv:2108.01210v1. [Google Scholar] [CrossRef]
- Banville, H.; Chehab, O.; Hyvärinen, A.; Engemann, D.-A.; Gramfort, A. Uncovering the structure of clinical EEG signals with self-supervised learning. J. Neural Eng. 2021, 18, 046020. [Google Scholar] [CrossRef]
- Rusnac, A.-L.; Grigore, O. CNN Architectures and Feature Extraction Methods for EEG Imaginary Speech Recognition. Sensors 2022, 22, 4679. [Google Scholar] [CrossRef] [PubMed]
- Vorontsova, D.; Menshikov, I.; Zubov, A.; Orlov, K.; Rikunov, P.; Zvereva, E.; Flitman, L.; Lanikin, A.; Sokolova, A.; Markov, S.; et al. Silent EEG-Speech Recognition Using Convolutional and Recurrent Neural Network with 85% Accuracy of 9 Words Classification. Sensors 2021, 21, 6744. [Google Scholar] [CrossRef] [PubMed]
- Chinta, B.; Moorthi, M. EEG-dependent automatic speech recognition using deep residual encoder based VGG net CNN. Comput. Speech Lang. 2023, 79, 101477. [Google Scholar] [CrossRef]
- Li, C.; Huang, X.; Song, R.; Qian, R.; Liu, X.; Chen, X. EEG-based seizure prediction via Transformer guided CNN. Measurement 2022, 203, 111948. [Google Scholar] [CrossRef]
- Huang, C.; Xiao, Y.; Xu, G. Predicting Human Intention-Behavior Through EEG Signal Analysis Using Multi-Scale CNN. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 1722–1729. [Google Scholar] [CrossRef]
- Saba-Sadiya, S.; Chantland, E.; Alhanai, T.; Liu, T.; Ghassemi, M.M. Unsupervised EEG Artifact Detection and Correction. Front. Digit Health 2021, 2, 608920. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).