Domain Adversarial Convolutional Neural Network Improves the Accuracy and Generalizability of Wearable Sleep Assessment Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Data Preprocessing
2.3. Model Architecture
2.4. Model Performance
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AHI | Apnea–hypopnea index |
| CI | Confidence interval |
| CNN | Convolutional neural network |
| DACNN | Domain adversarial convolutional neural network |
| EEG | Electroencephalogram |
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| MAE | Mean average error |
| MESA | Multi-Ethnic Study of Atherosclerosis |
| PPG | Photoplethysmography |
| PSG | Polysomnography |
| RMSE | Root mean square error |
| ROC | Receiver operating characteristic |
| SDB | Sleep-disordered breathing |
| SE | Sleep efficiency |
| WASO | Wake after sleep onset |
References
- Nieto, F.J.; Petersen, D. (Eds.) Foundations of Sleep Health, 1st ed.; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar]
- Pillai, J.A.; Leverenz, J.B. Sleep and Neurodegeneration: A Critical Appraisal. Chest 2017, 151, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Worley, S.L. The Extraordinary Importance of Sleep: The Detrimental Effects of Inadequate Sleep on Health and Public Safety Drive an Explosion of Sleep Research. Pharm. Ther. 2018, 43, 758–763. [Google Scholar]
- Piwek, L.; Ellis, D.A.; Andrews, S.; Joinson, A. The Rise of Consumer Health Wearables: Promises and Barriers. PLoS Med. 2016, 13, e1001953. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.B.; Kripke, D.F.; Messin, S.; Mullaney, D.J.; Wyborney, G. An Activity-Based Sleep Monitor System for Ambulatory Use. Sleep 1982, 5, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Djanian, S.; Bruun, A.; Nielsen, T.D. Sleep Classification Using Consumer Sleep Technologies and AI: A Review of the Current Landscape. Sleep Med. 2022, 100, 390–403. [Google Scholar] [CrossRef]
- Neishabouri, A.; Nguyen, J.; Samuelsson, J.; Guthrie, T.; Biggs, M.; Wyatt, J.; Cross, D.; Karas, M.; Migueles, J.H.; Khan, S.; et al. Quantification of Acceleration as Activity Counts in ActiGraph Wearable. Sci. Rep. 2022, 12, 11958. [Google Scholar] [CrossRef]
- Cole, R.J.; Kripke, D.F.; Gruen, W.; Mullaney, D.J.; Gillin, J.C. Automatic Sleep/Wake Identification From Wrist Activity. Sleep 1992, 15, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Palotti, J.; Mall, R.; Aupetit, M.; Rueschman, M.; Singh, M.; Sathyanarayana, A.; Taheri, S.; Fernandez-Luque, L. Benchmark on a Large Cohort for Sleep-Wake Classification with Machine Learning Techniques. NPJ Digit. Med. 2019, 2, 50. [Google Scholar] [CrossRef]
- Sadeh, A.; Sharkey, M.; Carskadon, M.A. Activity-Based Sleep-Wake Identification: An Empirical Test of Methodological Issues. Sleep 1994, 17, 201–207. [Google Scholar] [CrossRef]
- Zhai, B.; Perez-Pozuelo, I.; Clifton, E.A.D.; Palotti, J.; Guan, Y. Making Sense of Sleep: Multimodal Sleep Stage Classification in a Large, Diverse Population Using Movement and Cardiac Sensing. Proc. ACM Interact. Mob. Wearable Ubiquitous Technol. 2020, 4, 67:1–67:33. [Google Scholar] [CrossRef]
- Sundararajan, K.; Georgievska, S.; Te Lindert, B.H.W.; Gehrman, P.R.; Ramautar, J.; Mazzotti, D.R.; Sabia, S.; Weedon, M.N.; van Someren, E.J.W.; Ridder, L.; et al. Sleep Classification from Wrist-Worn Accelerometer Data Using Random Forests. Sci. Rep. 2021, 11, 24. [Google Scholar] [CrossRef]
- van Hees, V.T.; Sabia, S.; Anderson, K.N.; Denton, S.J.; Oliver, J.; Catt, M.; Abell, J.G.; Kivimäki, M.; Trenell, M.I.; Singh-Manoux, A. A Novel, Open Access Method to Assess Sleep Duration Using a Wrist-Worn Accelerometer. PLoS ONE 2015, 10, e0142533. [Google Scholar] [CrossRef] [PubMed]
- de Zambotti, M.; Cellini, N.; Menghini, L.; Sarlo, M.; Baker, F.C. Sensors Capabilities, Performance, and Use of Consumer Sleep Technology. Sleep Med. Clin. 2020, 15, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Patterson, M.R.; Nunes, A.A.S.; Gerstel, D.; Pilkar, R.; Guthrie, T.; Neishabouri, A.; Guo, C.C. 40 Years of Actigraphy in Sleep Medicine and Current State of the Art Algorithms. NPJ Digit. Med. 2023, 6, 51. [Google Scholar] [CrossRef]
- Chen, X.; Wang, R.; Zee, P.; Lutsey, P.L.; Javaheri, S.; Alcántara, C.; Jackson, C.L.; Williams, M.A.; Redline, S. Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep 2015, 38, 877–888. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The National Sleep Research Resource: Towards a Sleep Data Commons. J. Am. Med. Inf. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Gal, Y.; Ghahramani, Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. arXiv 2016, arXiv:1506.02142. [Google Scholar] [CrossRef]
- Wu, H.; Gu, X. Max-Pooling Dropout for Regularization of Convolutional Neural Networks. arXiv 2015, arXiv:1512.01400. [Google Scholar] [CrossRef]
- Prechelt, L. Early Stopping—But When? In Neural Networks: Tricks of the Trade; Goos, G., Hartmanis, J., Van Leeuwen, J., Orr, G.B., Müller, K.R., Eds.; Springer: Berlin/Heidelberg, Germany, 1998; Volume 1524, pp. 55–69. [Google Scholar] [CrossRef]
- Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; Marchand, M. Domain-Adversarial Neural Networks. arXiv 2015, arXiv:1412.4446. [Google Scholar] [CrossRef]
- Sicilia, A.; Zhao, X.; Hwang, S.J. Domain Adversarial Neural Networks for Domain Generalization: When It Works and How to Improve. Mach. Learn. 2023, 112, 2685–2721. [Google Scholar] [CrossRef]
- Lee, P.; Tse, C.Y. Calibration of Wrist-Worn ActiWatch 2 and ActiGraph wGT3X for Assessment of Physical Activity in Young Adults. Gait Posture 2019, 68, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Redline, S.; Budhiraja, R.; Kapur, V.; Marcus, C.L.; Mateika, J.H.; Mehra, R.; Parthasarthy, S.; Somers, V.K.; Strohl, K.P.; Sulit, L.G.; et al. The Scoring of Respiratory Events in Sleep: Reliability and Validity. J. Clin. Sleep Med. 2007, 3, 169–200. [Google Scholar] [CrossRef]
- van Hees, V.T.; Sabia, S.; Jones, S.E.; Wood, A.R.; Anderson, K.N.; Kivimäki, M.; Frayling, T.M.; Pack, A.I.; Bucan, M.; Trenell, M.I.; et al. Estimating Sleep Parameters Using an Accelerometer without Sleep Diary. Sci. Rep. 2018, 8, 12975. [Google Scholar] [CrossRef] [PubMed]
- Lockley, S.W.; Skene, D.J.; Arendt, J. Comparison between Subjective and Actigraphic Measurement of Sleep and Sleep Rhythms. J. Sleep Res. 1999, 8, 175–183. [Google Scholar] [CrossRef]
- Migueles, J.H.; Rowlands, A.V.; Huber, F.; Sabia, S.; van Hees, V.T. GGIR: A Research Community–Driven Open Source R Package for Generating Physical Activity and Sleep Outcomes From Multi-Day Raw Accelerometer Data. J. Meas. Phys. Behav. 2019, 2, 188–196. [Google Scholar] [CrossRef]
- van Hees, V.T.; Fang, Z.; Langford, J.; Assah, F.; Mohammad, A.; da Silva, I.C.M.; Trenell, M.I.; White, T.; Wareham, N.J.; Brage, S. Autocalibration of Accelerometer Data for Free-Living Physical Activity Assessment Using Local Gravity and Temperature: An Evaluation on Four Continents. J. Appl. Physiol. 2014, 117, 738–744. [Google Scholar] [CrossRef]
- Fekedulegn, D.; Andrew, M.E.; Shi, M.; Violanti, J.M.; Knox, S.; Innes, K.E. Actigraphy-Based Assessment of Sleep Parameters. Ann. Work. Expo. Health 2020, 64, 350–367. [Google Scholar] [CrossRef] [PubMed]
- Jean-Louis, G.; Kripke, D.F.; Mason, W.J.; Elliott, J.A.; Youngstedt, S.D. Sleep Estimation from Wrist Movement Quantified by Different Actigraphic Modalities. J. Neurosci. Methods 2001, 105, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab, M.; Busso, C. Domain Adversarial for Acoustic Emotion Recognition. IEEE/ACM Trans. Audio Speech Lang. Process. 2018, 26, 2423–2435. [Google Scholar] [CrossRef]
- Ghosh, A. Improved Vertex Finding in the MINERvA Passive Target Region with Convolutional Neural Networks and Deep Adversarial Neural Network. In Proceedings of the 19th International Workshop on Neutrinos from Accelerators NUFACT2017—PoS(NuFact2017), Uppsala, Sweden, 25–30 September 2017. [Google Scholar] [CrossRef]
- Jing, F.; Zhang, S.W.; Zhang, S. Prediction of Enhancer–Promoter Interactions Using the Cross-Cell Type Information and Domain Adversarial Neural Network. BMC Bioinform. 2020, 21, 507. [Google Scholar] [CrossRef]
- Lafarge, M.W.; Pluim, J.P.W.; Eppenhof, K.A.J.; Moeskops, P.; Veta, M. Domain-Adversarial Neural Networks to Address the Appearance Variability of Histopathology Images. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Québec City, QC, Canada, 14 September 2017; Cardoso, M.J., Arbel, T., Carneiro, G., Syeda-Mahmood, T., Tavares, J.M.R., Moradi, M., Bradley, A., Greenspan, H., Papa, J.P., Madabhushi, A., et al., Eds.; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2017; pp. 83–91. [Google Scholar] [CrossRef]
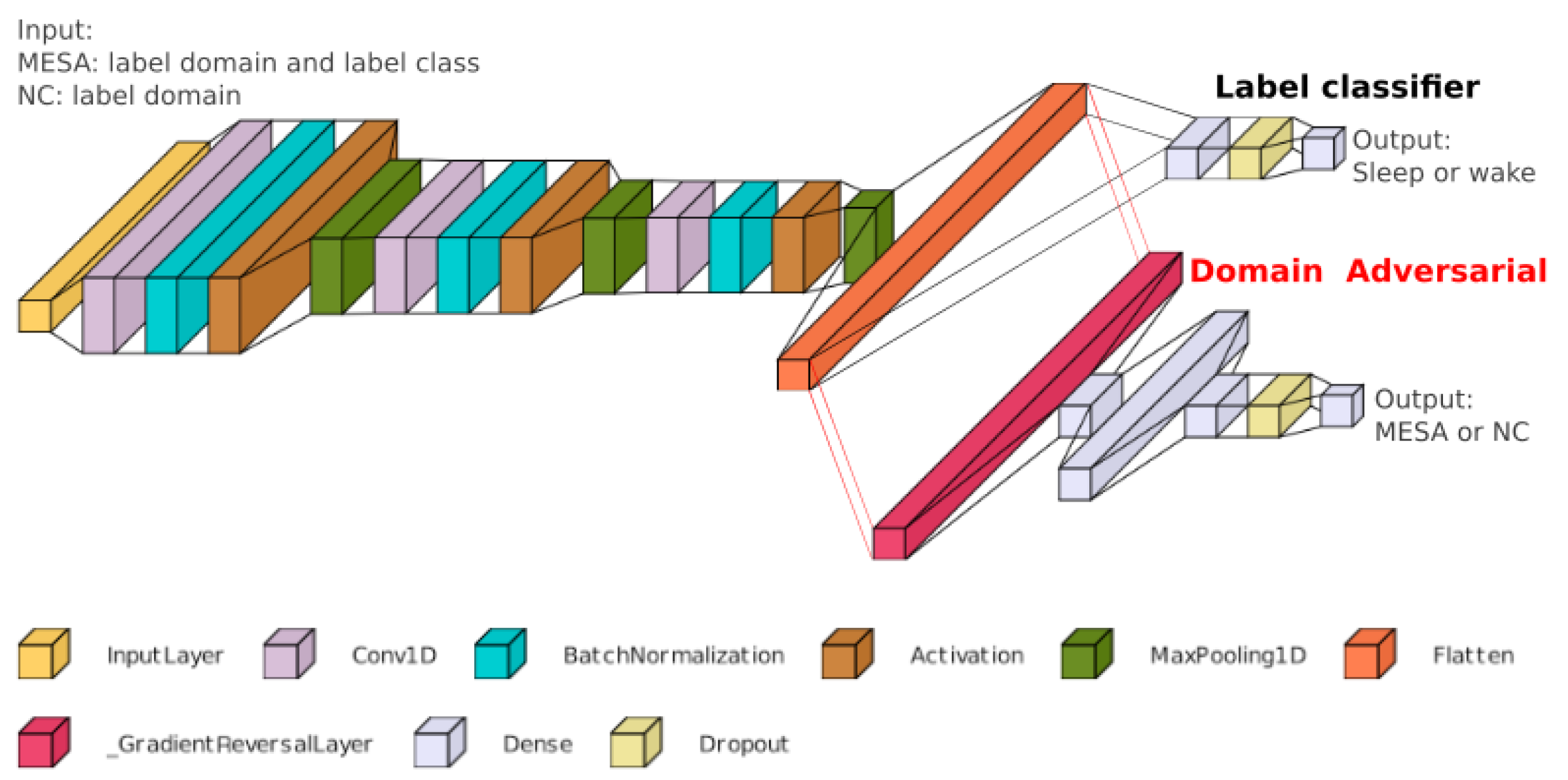
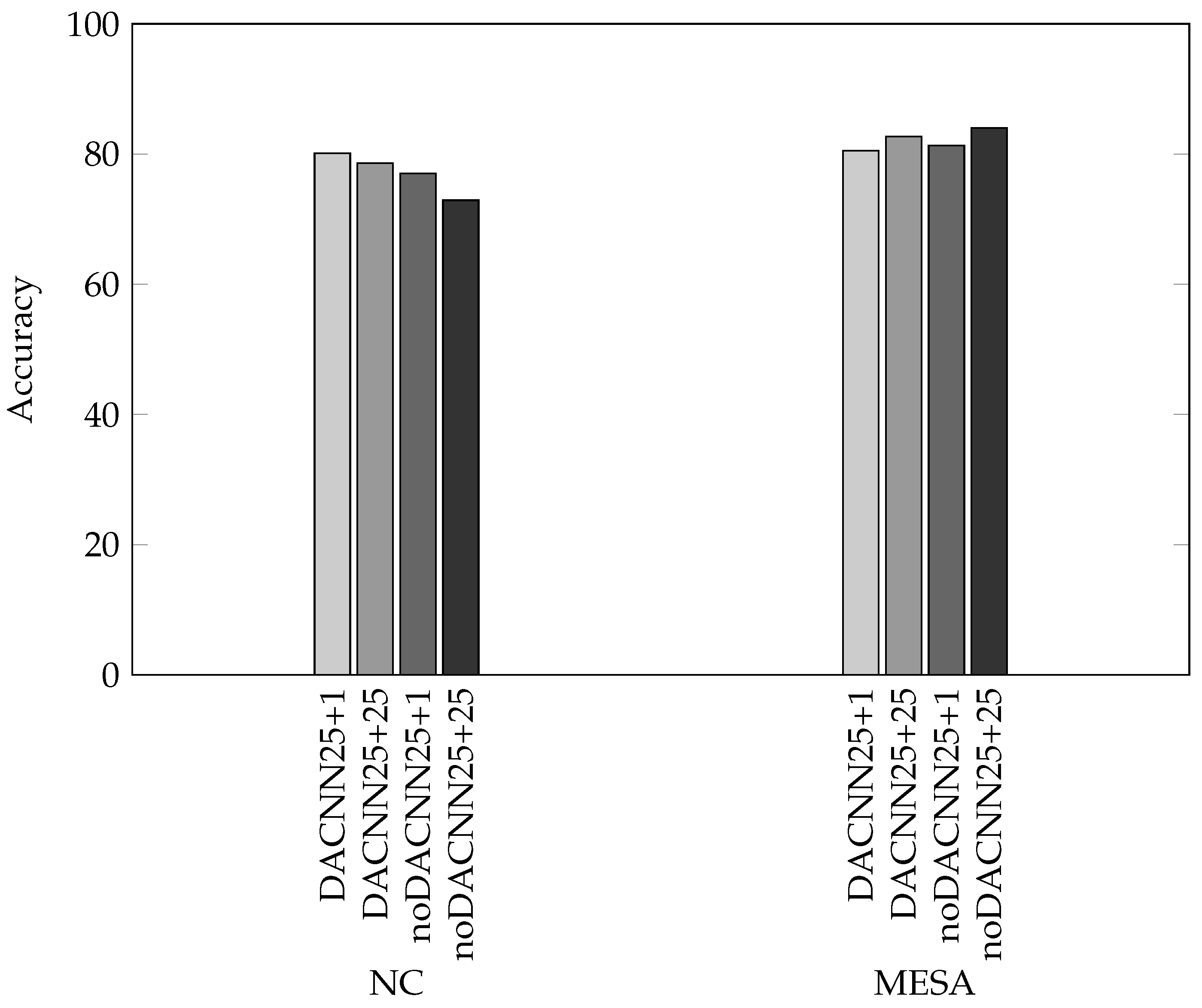

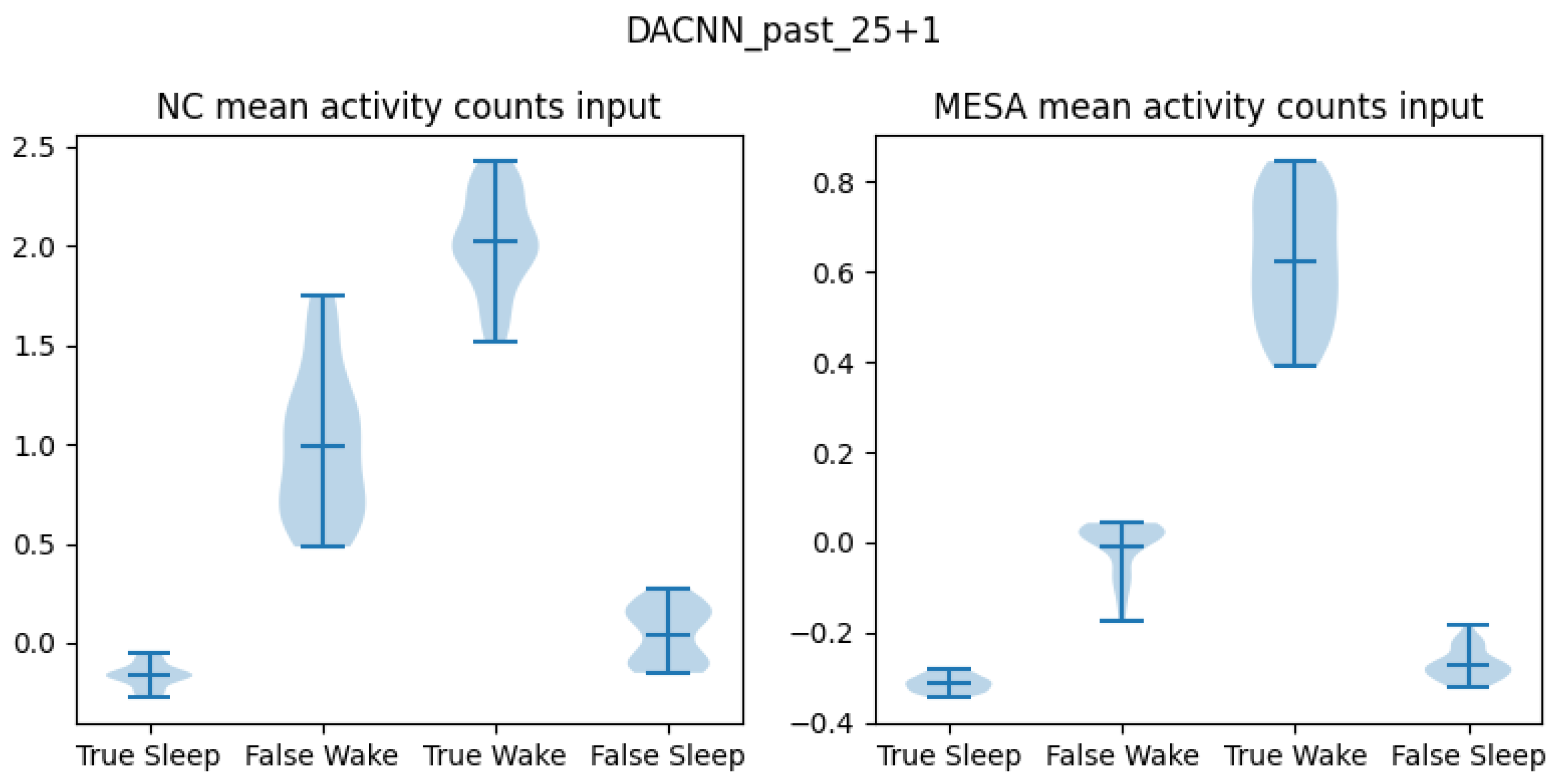
| Model | Past 25 min + Future 25 min | Past 25 min + Future 1 min |
|---|---|---|
| Without DA | noDACNN25+25 | noDACNN25+1 |
| With DA | DACNN25+25 | DACNN25+1 |
| Model | acc | sens | spec | prec | F1 | WASO | RMSE | MAE | CI Width | SE | RMSE | MAE | CI Width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 80.1 | 83.9 | 57.6 | 81 | 81.7 | 159.3 | 80.9 | 48.7 | 309.1 | 71.8 | 11.8 | 8.0 | 45.7 | |
| 78.6 | 81.1 | 55.7 | 79.9 | 79.5 | 165.2 | 83.2 | 54.8 | 323.8 | 70.6 | 12.5 | 9.1 | 49.2 | |
| 77 | 84.9 | 48.5 | 77.2 | 80.1 | 137.1 | 102.8 | 76.5 | 353.4 | 75.6 | 17.7 | 13.5 | 57.1 | |
| 72.9 | 68.9 | 68.3 | 81.3 | 73.2 | 231 | 94.1 | 57.2 | 331.5 | 59 | 13.3 | 9.1 | 46.1 | |
| Previous Algorithms | |||||||||||||
| Z-angle | 76.2 | 83.6 | 47.5 | 76.2 | 79.1 | 140.8 | 91.1 | 57.5 | 325.3 | 75 | 13.0 | 9.3 | 46.2 |
| Cole rescored | 71.9 | 66.1 | 73.6 | 82.6 | 72.2 | 247.5 | 105.8 | 86.2 | 322.5 | 56.3 | 18.4 | 15.4 | 51.7 |
| Sadeh rescored | 69.6 | 61.9 | 75.5 | 83.1 | 69 | 268.6 | 133.1 | 108.2 | 393.2 | 52.5 | 23.3 | 19.3 | 64.1 |
| Baselines | |||||||||||||
| All sleep | 69.2 | 100 | 0 | 69.2 | 79.6 | 0 | 223.9 | 180.1 | 526.1 | 100 | 37.2 | 30.8 | 83.1 |
| All wake | 30.8 | 0 | 100 | 0 | 0 | 571.4 | 409.4 | 391.3 | 476.5 | 0 | 72.4 | 69.2 | 83.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunes, A.S.; Patterson, M.R.; Gerstel, D.; Khan, S.; Guo, C.C.; Neishabouri, A. Domain Adversarial Convolutional Neural Network Improves the Accuracy and Generalizability of Wearable Sleep Assessment Technology. Sensors 2024, 24, 7982. https://doi.org/10.3390/s24247982
Nunes AS, Patterson MR, Gerstel D, Khan S, Guo CC, Neishabouri A. Domain Adversarial Convolutional Neural Network Improves the Accuracy and Generalizability of Wearable Sleep Assessment Technology. Sensors. 2024; 24(24):7982. https://doi.org/10.3390/s24247982
Chicago/Turabian StyleNunes, Adonay S., Matthew R. Patterson, Dawid Gerstel, Sheraz Khan, Christine C. Guo, and Ali Neishabouri. 2024. "Domain Adversarial Convolutional Neural Network Improves the Accuracy and Generalizability of Wearable Sleep Assessment Technology" Sensors 24, no. 24: 7982. https://doi.org/10.3390/s24247982
APA StyleNunes, A. S., Patterson, M. R., Gerstel, D., Khan, S., Guo, C. C., & Neishabouri, A. (2024). Domain Adversarial Convolutional Neural Network Improves the Accuracy and Generalizability of Wearable Sleep Assessment Technology. Sensors, 24(24), 7982. https://doi.org/10.3390/s24247982






