Model-Based Electroencephalogram Instantaneous Frequency Tracking: Application in Automated Sleep–Wake Stage Classification
Abstract
:1. Introduction
2. A Kalman Filter-Based IF Estimation Scheme
2.1. The Concept of IE and IF
2.2. Bandpass Filtering
2.3. Amplitude Normalization
2.4. Time-Varying Auto-Regressive (TVAR) EEG Instantaneous Frequency Modeling
2.5. TVAR Model Parameter Tracking Using Kalman Filter
2.6. KF Parameter Selection and Optimization
3. Evaluation
3.1. Dataset
3.2. Implementation
3.3. Processing and Feature Extraction Pipeline
- In the pre-processing stage, the raw signal is passed through five parallel FIR band-pass filters with bandwidth ranges corresponding to the (0.5–4.0 Hz), (4.0–8 Hz), (8.0–13.0 Hz), (13.0–30.0 Hz), and (30–50 Hz) brain rhythms. The EOG channel is separately processed using a band-pass filter with a range of 0.5–20 Hz to preserve its main frequency components.
- The EEG and EOG of each subband are represented in analytic form (1), and their modulus are set as the IE.
- Signals are normalized by their analytical form modulus using (1).
- The IF of the normalized signals are estimated using the robust KF algorithm detailed in Section 2.
- Steps 2 and 4 generate five pairs of IF-IE for each EEG channel and a single pair of IF-IE for the EOG channel, all of which match the original signals in length. As hypnogram labels are provided for 30 s intervals, time-interval averaging is applied to IF-IE vectors to compute the mean IE-IF over non-overlapping 30 s windows.
3.4. Classification Pipeline
4. Results
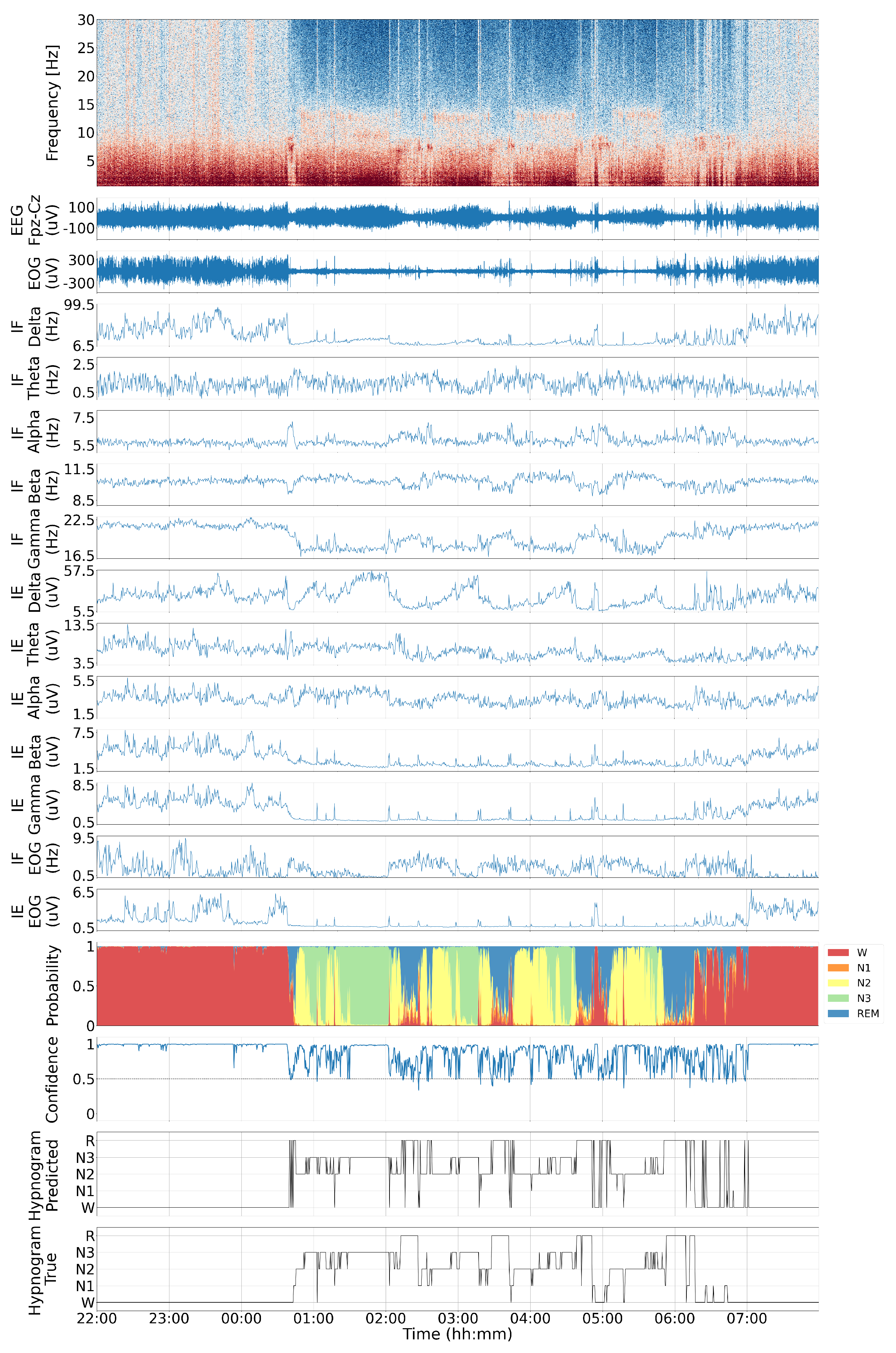
4.1. Visual Inspection
4.2. Classification Results Across All Subjects
4.3. Subject-Wise Performances
4.4. Feature Importance
5. Discussion
5.1. Enhanced Frequency Tracking
5.2. Pipeline Validation Through Sleep Stage Analysis
5.3. Limitations
5.4. Future Directions
5.4.1. IF Estimate Confidence Interval
5.4.2. Drowsiness Detection
5.4.3. Kalman Filter Versus Kalman Smoother
5.4.4. Comparison with Existing Methods and Broader Applications
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AASM | American Academy of Sleep Medicine | KF | Kalman filter |
| AR | Auto-Regressive | KS | Kalman Smoother |
| ARMA | Auto-Regressive Moving Average | LGBM | Light Gradient Boosting Machine |
| CV | Cross-Validation | LR | Logistic Regression |
| DOA | Depth of anesthesia | LSTM | Long Short-Term Memory |
| EEG | Electroencephalogram | MMSE | Minimum Mean Square Error |
| EOG | Electrooculogram | OvR | One-vs-Rest |
| FIR | Finite Impulse Response | PSD | Power Spectral Density |
| IE | Instantaneous Energy | PSG | Polysomnography |
| IF | Instantaneous Frequency | REM | Rapid Eye Movement |
| IP | Instantaneous Phase | RNN | Recurrent Neural Network |
| R&K | Rechtschaffen and Kales | SHAP | Shapley Additive Explanations |
| STFT | Short-Time Fourier Transform | SVM | Support Vector Machine |
| TVAR | Time-Varying Auto-Regressive | WGN | White Gaussian Noise |
| XGB | Extreme Gradient Boosting |
References
- Šušmáková, K. Human sleep and sleep EEG. Meas. Sci. Rev. 2004, 4, 59–74. [Google Scholar]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Lloyd, R.M.; Marcus, C.L.; Vaughn, B.V.; American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications: Version 2.3. American Academy of Sleep Medicine. 2015. Available online: https://books.google.com/books?id=SySXAQAACAAJ (accessed on 9 May 2024).
- Warby, S.C.; Wendt, S.L.; Welinder, P.; Munk, E.G.; Carrillo, O.; Sorensen, H.B.; Jennum, P.; Peppard, P.E.; Perona, P.; Mignot, E. Sleep-spindle detection: Crowdsourcing and evaluating performance of experts, non-experts and automated methods. Nat. Methods 2014, 11, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Stephansen, J.B.; Olesen, A.N.; Olsen, M.; Ambati, A.; Leary, E.B.; Moore, H.E.; Carrillo, O.; Lin, L.; Han, F.; Yan, H.; et al. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat. Commun. 2018, 9, 5229. [Google Scholar] [CrossRef] [PubMed]
- Basner, M.; Griefahn, B.; Penzel, T. Inter-rater agreement in sleep stage classification between centers with different backgrounds. Somnologie 2008, 12, 75–84. [Google Scholar] [CrossRef]
- Miller, C.B.; Bartlett, D.J.; Mullins, A.E.; Dodds, K.L.; Gordon, C.J.; Kyle, S.D.; Kim, J.W.; D’Rozario, A.L.; Lee, R.S.; Comas, M.; et al. Clusters of insomnia disorder: An exploratory cluster analysis of objective sleep parameters reveals differences in neurocognitive functioning, quantitative EEG, and heart rate variability. Sleep 2016, 39, 1993–2004. [Google Scholar] [CrossRef]
- Baandrup, L.; Christensen, J.A.; Fagerlund, B.; Jennum, P. Investigation of sleep spindle activity and morphology as predictors of neurocognitive functioning in medicated patients with schizophrenia. J. Sleep Res. 2019, 28, e12672. [Google Scholar] [CrossRef]
- Koch, H.; Jennum, P.; Christensen, J.A. Automatic sleep classification using adaptive segmentation reveals an increased number of rapid eye movement sleep transitions. J. Sleep Res. 2019, 28, e12780. [Google Scholar] [CrossRef]
- Olesen, A.N.; Cesari, M.; Christensen, J.A.E.; Sorensen, H.B.D.; Mignot, E.; Jennum, P. A comparative study of methods for automatic detection of rapid eye movement abnormal muscular activity in narcolepsy. Sleep Med. 2018, 44, 97–105. [Google Scholar] [CrossRef]
- Prinz, P.N.; Peskind, E.R.; Vitaliano, P.P.; Raskind, M.A.; Eisdorfer, C.; Zemcuznikov, H.N.; Gerber, C.J. Changes in the sleep and waking EEGs of nondemented and demented elderly subjects. J. Am. Geriatr. Soc. 1982, 30, 86–92. [Google Scholar] [CrossRef]
- Koch, H.; Schneider, L.D.; Finn, L.A.; Leary, E.B.; Peppard, P.E.; Hagen, E.; Sorensen, H.B.D.; Jennum, P.; Mignot, E. Breathing disturbances without hypoxia are associated with objective sleepiness in sleep apnea. Sleep 2017, 40, zsx152. [Google Scholar] [CrossRef]
- Doroshenkov, L.; Konyshev, V.; Selishchev, S. Classification of human sleep stages based on EEG processing using hidden Markov models. Biomed. Eng. 2007, 41, 25–28. [Google Scholar] [CrossRef]
- Zoubek, L.; Charbonnier, S.; Lesecq, S.; Buguet, A.; Chapotot, F. Feature selection for sleep/wake stages classification using data driven methods. Biomed. Signal Process. Control 2007, 2, 171–179. [Google Scholar] [CrossRef]
- Subasi, A. EEG signal classification using wavelet feature extraction and a mixture of expert model. Expert Syst. Appl. 2007, 32, 1084–1093. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Mikaeili, M.; Estrada, E.; Nazeran, H. Automatic sleep stage classification based on EEG signals by using neural networks and wavelet packet coefficients. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 20–24 August 2008; IEEE: Piscataway, NJ, USA, 2008; pp. 1151–1154. [Google Scholar]
- Sinha, R.K. Artificial neural network and wavelet based automated detection of sleep spindles, REM sleep and wake states. J. Med Syst. 2008, 32, 291–299. [Google Scholar] [CrossRef]
- Oropesa, E.; Cycon, H.L.; Jobert, M. Sleep stage classification using wavelet transform and neural network. Int. Comput. Sci. Inst. 1999, 2, 1–7. [Google Scholar]
- Fraiwan, L.; Lweesy, K.; Khasawneh, N.; Wenz, H.; Dickhaus, H. Automated sleep stage identification system based on time–frequency analysis of a single EEG channel and random forest classifier. Comput. Methods Programs Biomed. 2012, 108, 10–19. [Google Scholar] [CrossRef]
- Šušmáková, K.; Krakovská, A. Discrimination ability of individual measures used in sleep stages classification. Artif. Intell. Med. 2008, 44, 261–277. [Google Scholar] [CrossRef]
- Flexer, A.; Gruber, G.; Dorffner, G. A reliable probabilistic sleep stager based on a single EEG signal. Artif. Intell. Med. 2005, 33, 199–207. [Google Scholar] [CrossRef]
- Pan, S.T.; Kuo, C.E.; Zeng, J.H.; Liang, S.F. A transition-constrained discrete hidden Markov model for automatic sleep staging. Biomed. Eng. Online 2012, 11, 1. [Google Scholar] [CrossRef]
- Özşen, S. Classification of sleep stages using class-dependent sequential feature selection and artificial neural network. Neural Comput. Appl. 2013, 23, 1239–1250. [Google Scholar] [CrossRef]
- Chapotot, F.; Becq, G. Automated sleep–wake staging combining robust feature extraction, artificial neural network classification, and flexible decision rules. Int. J. Adapt. Control Signal Process. 2010, 24, 409–423. [Google Scholar] [CrossRef]
- Subasi, A.; Kiymik, M.K.; Akin, M.; Erogul, O. Automatic recognition of vigilance state by using a wavelet-based artificial neural network. Neural Comput. Appl. 2005, 14, 45–55. [Google Scholar] [CrossRef]
- Tagluk, M.E.; Sezgin, N.; Akin, M. Estimation of sleep stages by an artificial neural network employing EEG, EMG and EOG. J. Med. Syst. 2010, 34, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Ronzhina, M.; Janoušek, O.; Kolářová, J.; Nováková, M.; Honzík, P.; Provazník, I. Sleep scoring using artificial neural networks. Sleep Med. Rev. 2012, 16, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Şen, B.; Peker, M.; Çavuşoğlu, A.; Çelebi, F.V. A comparative study on classification of sleep stage based on EEG signals using feature selection and classification algorithms. J. Med. Syst. 2014, 38, 18. [Google Scholar] [CrossRef]
- Gudmundsson, S.; Runarsson, T.P.; Sigurdsson, S. Automatic sleep staging using support vector machines with posterior probability estimates. In Proceedings of the International Conference on Computational Intelligence for Modelling, Control and Automation and International Conference on Intelligent Agents, Web Technologies and Internet Commerce (CIMCA-IAWTIC’06), Vienna, Austria, 28–30 November 2005; IEEE: Piscataway, NJ, USA, 2005; Volume 2, pp. 366–372. [Google Scholar]
- Koley, B.; Dey, D. An ensemble system for automatic sleep stage classification using single channel EEG signal. Comput. Biol. Med. 2012, 42, 1186–1195. [Google Scholar] [CrossRef]
- Yeo, M.V.; Li, X.; Shen, K.; Wilder-Smith, E.P. Can SVM be used for automatic EEG detection of drowsiness during car driving? Saf. Sci. 2009, 47, 115–124. [Google Scholar] [CrossRef]
- Lee, Y.H.; Chen, Y.S.; Chen, L.F. Automated sleep staging using single EEG channel for REM sleep deprivation. In Proceedings of the 2009 Ninth IEEE International Conference on Bioinformatics and BioEngineering, Taichung, Taiwan, 22–24 June 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 439–442. [Google Scholar]
- Lajnef, T.; Chaibi, S.; Ruby, P.; Aguera, P.E.; Eichenlaub, J.B.; Samet, M.; Kachouri, A.; Jerbi, K. Learning machines and sleeping brains: Automatic sleep stage classification using decision-tree multi-class support vector machines. J. Neurosci. Methods 2015, 250, 94–105. [Google Scholar] [CrossRef]
- Motamedi-Fakhr, S.; Moshrefi-Torbati, M.; Hill, M.; Hill, C.M.; White, P.R. Signal processing techniques applied to human sleep EEG signals—A review. Biomed. Signal Process. Control 2014, 10, 21–33. [Google Scholar] [CrossRef]
- Cohen, L. Time-Frequency Analysis; Prentice Hall PTR: Upper Saddle River, NJ, USA, 1995; Volume 778. [Google Scholar]
- Coenen, A. Neuronal activities underlying the electroencephalogram and evoked potentials of sleeping and waking: Implications for information processing. Neurosci. Biobehav. Rev. 1995, 19, 447–463. [Google Scholar] [CrossRef]
- Patomäki, L.; Kaipio, J.; Karjalainen, P.A. Tracking of nonstationary EEG with the roots of ARMA models. In Proceedings of the 17th International Conference of the Engineering in Medicine and Biology Society, Montréal, QC, Canada, 20–23 September 1995; Volume 95. [Google Scholar]
- Rogowski, Z.; Gath, I.; Bental, E. On the prediction of epileptic seizures. Biol. Cybern. 1981, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Dahal, N.; Nandagopal, D.N.; Cocks, B.; Vijayalakshmi, R.; Dasari, N.; Gaertner, P. TVAR modeling of EEG to detect audio distraction during simulated driving. J. Neural Eng. 2014, 11, 036012. [Google Scholar] [CrossRef] [PubMed]
- Lashkari, A.; Boostani, R.; Afrasiabi, S. Estimation of the anesthetic depth based on instantaneous frequency of electroencephalogram. In Proceedings of the 2015 38th International Conference on Telecommunications and Signal Processing (TSP), Prague, Czech Republic, 9–11 July 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 403–407. [Google Scholar]
- Picinbono, B. On instantaneous amplitude and phase of signals. IEEE Trans. Signal Process. 1997, 45, 552–560. [Google Scholar] [CrossRef]
- Khosla, A.; Khandnor, P.; Chand, T. A comparative analysis of signal processing and classification methods for different applications based on EEG signals. Biocybern. Biomed. Eng. 2020, 40, 649–690. [Google Scholar] [CrossRef]
- Sameni, R.; Seraj, E. A robust statistical framework for instantaneous electroencephalogram phase and frequency estimation and analysis. Physiol. Meas. 2017, 38, 2141. [Google Scholar] [CrossRef] [PubMed]
- Seraj, E.; Sameni, R. Robust electroencephalogram phase estimation with applications in brain-computer interface systems. Physiol. Meas. 2017, 38, 501–523. [Google Scholar] [CrossRef]
- Karimzadeh, F.; Boostani, R.; Seraj, E.; Sameni, R. A Distributed Classification Procedure for Automatic Sleep Stage Scoring Based on Instantaneous Electroencephalogram Phase and Envelope Features. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 362–370. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Wilson, M.A.; Brown, E.N.; Barbieri, R. Measuring instantaneous frequency of local field potential oscillations using the Kalman smoother. J. Neurosci. Methods 2009, 184, 365–374. [Google Scholar] [CrossRef]
- Boashash, B. Estimating and interpreting the instantaneous frequency of a signal. II. Algorithms and applications. Proc. IEEE 1992, 80, 540–568. [Google Scholar] [CrossRef]
- Almeida, L.B. The fractional Fourier transform and time-frequency representations. IEEE Trans. Signal Process. 1994, 42, 3084–3091. [Google Scholar] [CrossRef]
- Kwok, H.K.; Jones, D.L. Improved instantaneous frequency estimation using an adaptive short-time Fourier transform. IEEE Trans. Signal Process. 2000, 48, 2964–2972. [Google Scholar] [CrossRef]
- Boashash, B. Estimating and interpreting the instantaneous frequency of a signal. I. Fundamentals. Proc. IEEE 1992, 80, 520–538. [Google Scholar] [CrossRef]
- Boashash, B.; O’shea, P. Use of the cross Wigner-Ville distribution for estimation of instantaneous frequency. IEEE Trans. Signal Process. 1993, 41, 1439–1445. [Google Scholar] [CrossRef]
- Grenier, Y. Time-dependent ARMA modeling of nonstationary signals. IEEE Trans. Acoust. Speech Signal Process. 1983, 31, 899–911. [Google Scholar] [CrossRef]
- Box, G.E.; Jenkins, G.M.; Reinsel, G.C.; Ljung, G.M. Time Series Analysis: Forecasting and Control; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Shumway, R.H.; Stoffer, D.S.; Stoffer, D.S. Time Series Analysis and Its Applications; Springer: Berlin/Heidelberg, Germany, 2000; Volume 3. [Google Scholar]
- Arnold, M.; Milner, X.; Witte, H.; Bauer, R.; Braun, C. Adaptive AR modeling of nonstationary time series by means of Kalman filtering. IEEE Trans. Biomed. Eng. 1998, 45, 553–562. [Google Scholar] [CrossRef]
- Aboy, M.; Márquez, O.W.; McNames, J.; Hornero, R.; Trong, T.; Goldstein, B. Adaptive modeling and spectral estimation of nonstationary biomedical signals based on Kalman filtering. IEEE Trans. Biomed. Eng. 2005, 52, 1485–1489. [Google Scholar] [CrossRef]
- Kay, S.M. Fundamentals of Statistical Signal Processing, Volume I: Estimation Theory; Prentice Hall: Englewood Cliffs, NJ, USA, 1993. [Google Scholar]
- Tarvainen, M.P.; Hiltunen, J.K.; Ranta-aho, P.O.; Karjalainen, P.A. Estimation of nonstationary EEG with Kalman smoother approach: An application to event-related synchronization (ERS). IEEE Trans. Biomed. Eng. 2004, 51, 516–524. [Google Scholar] [CrossRef]
- Haykin, S. Adaptive Filter Theory; Prentice-Hall: Englewood Cliffs, NJ, USA, 1986; Volume 607. [Google Scholar]
- Brown, R.G. Introduction to Random Signal Analysis and Kalman Filtering; John Wiley & Sons: Hoboken, NJ, USA, 1983. [Google Scholar]
- Gelb, A. Applied Optimal Estimation; MIT Press: Cambridge, MA, USA, 1974; sixteenth print 2001. [Google Scholar]
- Sameni, R.; Shamsollahi, M.B.; Jutten, C.; Clifford, G.D. A nonlinear Bayesian filtering framework for ECG denoising. IEEE Trans. Biomed. Eng. 2007, 54, 2172–2185. [Google Scholar] [CrossRef]
- Sameni, R. A linear kalman notch filter for power-line interference cancellation. In Proceedings of the 16th CSI International Symposium on Artificial Intelligence and Signal Processing (AISP 2012), Shiraz, Iran, 2–3 May 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 604–610. [Google Scholar]
- Anderson, B.D.O.; Moore, J.B. Optimal Filtering; Dover Publications, Inc.: Mineola, NY, USA, 1979. [Google Scholar]
- Grewal, M.; Weill, L.; Andrews, A. Global Positioning Systems, Inertial Navigation, and Integration; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberye, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Kemp, B.; Aeilko, Z.; Tuk, B.; Kamphuisen, H.; Oberyé, J. The Sleep-EDF Database [Expanded]. 2018. Available online: https://physionet.org/content/sleep-edfx/1.0.0/ (accessed on 9 May 2024).
- Moser, D.; Anderer, P.; Gruber, G.; Parapatics, S.; Loretz, E.; Boeck, M.; Kloesch, G.; Heller, E.; Schmidt, A.; Danker-Hopfe, H.; et al. Sleep classification according to AASM and Rechtschaffen & Kales: Effects on sleep scoring parameters. Sleep 2009, 32, 139–149. [Google Scholar]
- Rechtschaffen, A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects; Public Health Service: Washington, DC, USA, 1968. [Google Scholar]
- Van Sweden, B.; Kemp, B.; Kamphuisen, H.; Ver der Velde, A. Alternative electrode placement in (automatic) sleep scoring. Sleep 1990, 13, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Voss, L.; Sleigh, J. Monitoring consciousness: The current status of EEG-based depth of anaesthesia monitors. Best Pract. Res. Clin. Anaesthesiol. 2007, 21, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Mikkelsen, K.; Chen, O.Y.; Koch, P.; Mertins, A.; De Vos, M. SleepTransformer: Automatic Sleep Staging With Interpretability and Uncertainty Quantification. IEEE Trans. Biomed. Eng. 2022, 69, 2456–2467. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.K.; Reddy, V.; Shumway, K.R.; Araujo, J.F. Physiology, sleep stages. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Armitage, R. The distribution of EEG frequencies in REM and NREM sleep stages in healthy young adults. Sleep 1995, 18, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Delorme, A. EEG is better left alone. Sci. Rep. 2023, 13, 2372. [Google Scholar] [CrossRef] [PubMed]
- Sanei, S.; Chambers, J.A. EEG Signal Processing; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Urigüen, J.A.; Garcia-Zapirain, B. EEG artifact removal—State-of-the-art and guidelines. J. Neural Eng. 2015, 12, 031001. [Google Scholar] [CrossRef]
- Sameni, R.; Gouy-Pailler, C. An iterative subspace denoising algorithm for removing electroencephalogram ocular artifacts. J. Neurosci. Methods 2014, 225, 97–105. [Google Scholar] [CrossRef]
- Armington, J.C.; Mitnick, L.L. Electroencephalogram and sleep deprivation. J. Appl. Physiol. 1959, 14, 247–250. [Google Scholar] [CrossRef]









| Model | #Step | Metric | Wake | N1 | N2 | N3 | REM |
|---|---|---|---|---|---|---|---|
| XGBoost | 1 | SE | |||||
| SP | |||||||
| LGBM | 1 | SE | |||||
| SP | |||||||
| XGBoost | 2 | SE | |||||
| SP | |||||||
| LGBM | 2 | SE | |||||
| SP |
| Model | #Step | F1-Macro | Cohen’s Kappa | Accuracy |
|---|---|---|---|---|
| SVM | 1 | |||
| LR | 1 | |||
| XGBoost | 1 | |||
| LGBM | 1 | |||
| SVM | 2 | |||
| LR | 2 | |||
| XGBoost | 2 | |||
| LGBM | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nateghi, M.; Rahbar Alam, M.; Amiri, H.; Nasiri, S.; Sameni, R. Model-Based Electroencephalogram Instantaneous Frequency Tracking: Application in Automated Sleep–Wake Stage Classification. Sensors 2024, 24, 7881. https://doi.org/10.3390/s24247881
Nateghi M, Rahbar Alam M, Amiri H, Nasiri S, Sameni R. Model-Based Electroencephalogram Instantaneous Frequency Tracking: Application in Automated Sleep–Wake Stage Classification. Sensors. 2024; 24(24):7881. https://doi.org/10.3390/s24247881
Chicago/Turabian StyleNateghi, Masoud, Mahdi Rahbar Alam, Hossein Amiri, Samaneh Nasiri, and Reza Sameni. 2024. "Model-Based Electroencephalogram Instantaneous Frequency Tracking: Application in Automated Sleep–Wake Stage Classification" Sensors 24, no. 24: 7881. https://doi.org/10.3390/s24247881
APA StyleNateghi, M., Rahbar Alam, M., Amiri, H., Nasiri, S., & Sameni, R. (2024). Model-Based Electroencephalogram Instantaneous Frequency Tracking: Application in Automated Sleep–Wake Stage Classification. Sensors, 24(24), 7881. https://doi.org/10.3390/s24247881





