Efficient Sleep Stage Identification Using Piecewise Linear EEG Signal Reduction: A Novel Algorithm for Sleep Disorder Diagnosis
Abstract
1. Introduction
2. Related Work
3. Dataset Used
Dataset
| Subject File | Classes | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|
| slp01a | 1,2,3,4,W,R | 92.59 | 93.84 | 93.21 |
| slp01b | 1,2,W,R | 96.86 | 98.78 | 97.82 |
| alp2a | 1,2,3,4,W,R | 92.51 | 94.68 | 93.59 |
| slp2b | 1,2,W,R,M | 94.34 | 95.77 | 95.05 |
| slp03 | 1,2,3,W,R | 96.42 | 97.73 | 97.075 |
| slp04 | 1,2,3,W,R | 94.59 | 97.73 | 96.16 |
| slp14 | 1,2,3,4,W,R | 94.42 | 96.75 | 95.58 |
| slp16 | 1,2,3,4,W,R | 96.84 | 97.92 | 97.38 |
| Avg | 94.82 | 96.65 | 95.73 |
| Author and Year | Records Used | Classifier | Avg Accuracy (%) |
|---|---|---|---|
| Redmond et al. [28], 2003 | 17 | QDA | 76.75 |
| Adnane et al. [18], 2012 | 17 | SVM | 79.99 |
| Hayet et al. [29] 2012 | 09 | ELM | 83.59 |
| Werteni et al. [30], 2015 | 17 | SVM | 56.81 |
| Tripathy et al. [14], 2018 | 17 | DNN | 85.51, 94.0, 95.71 |
| Taran et al. [17], 2018 | 16 | ELM | 92.28 |
| Budak et al. [16], 2019 | 16 | LSTM | 94.31 |
| An et al. [31],2019 | 06 | W-SVM | 85.29 |
| Zhang et al. [32], 2020 | 18 | CNN | 87.6 |
| Surantha et al. [33], 2021 | 18 | SVM/ELM | 76.77, 82.1 |
| Rashidi et al. [1], 2023 | 18 | DT | 95.6, 92.72, 85.64 |
| Wang et al. [34], 2023 | 18 | GBDT | 87.15, 82.02 |
| Proposed method | 8 | KNN | 95.73% |
| Author and Year | Number of Records | Features | Classes Used | Classifier Used | Average Accuracy (%) |
|---|---|---|---|---|---|
| Redmond et al. [28], 2003 | 17 | HRV and EEG | W vs. REM vs. NREM | QDA | 76,75 |
| Adnane et al. [18], 2012 | 17 | HRV, DFA, and WDFA | Sleep vs. wake | SVM | 79.99 |
| Hayet et al. [29], 2012 | 09 | RR-time series and HRV | Sleep vs. wake | ELM | 83.59 |
| Warteni et al. [30], 2015 | 17 | HRV | Sleep vs. wake REM | SVM | 56.81 |
| Tripathy et al. [14], 2018 | 17 | Dispersion entropy and variance | wake vs. light, sleep vs. deep, sleep vs. REM | Neural network | 91.71 |
| Taran et al. [17], 2018 | 16 | Hermite coefficients | alert (w) and drowsiness (s1) | ELM | 92.28 |
| Budak et al. [16], 2019 | 16 | Spectrogram images and instanious frequencies | alert and drowsiness | LSTM | 94.31 |
| Proposed method, 2 classes | 8 | Halfwave | 2 random classes | KNN | 96.6 |
| An et al. [31], 2019 | 06 | Statistical features | NREM (s1–s4), REM, Wake | W-SVM | 85.29 |
| Zhang et al. [32], 2020 | 18 | Hilbert Huang coefficients | REM, NREM, wake | CNN | 87.6 |
| Proposed method, random 4 classes | 8 | Halfwave features | random 4 classes | KNN | 95.96 |
| Proposed method, all 6 classes | 8 | Halfwave | Wake, Sleep (all), REM | KNN | 95.73 |
4. Proposed Method
4.1. Time Domain: Halfwave Method
4.1.1. Mathematical Formalization of Proposed Halfwave Method
4.1.2. Advantages of Proposed Halfwave Method
- 1.
- Efficiency in Processing: The method simplifies EEG and other signals by treating them as piecewise linear functions, which reduces data complexity while preserving essential information. This makes the Halfwave method efficient in processing lengthy signals.
- 2.
- Low Complexity: By computing the extremal points of the original signal and constructing a piecewise linear function, the method reduces unnecessary data, thereby lowering the complexity of the analysis. This simplification helps in faster processing and real-time application.
- 3.
- Accuracy: The Halfwave method has shown high accuracy in various applications, such as sleep state detection. It is particularly effective in distinguishing normal and abnormal patterns within signals, which is crucial for applications like epileptic seizure detection and other brain disorders.
- 4.
- Adaptability: The method’s flexibility allows for iterative decomposition, which can be adjusted to find the most suitable level of detail for a specific application. This adaptability is essential for tasks like sleep detection, where different levels of signal detail may be required.
5. Features Extraction and Classification
5.1. Feature Extraction
5.2. Classification
5.2.1. K-Nearest Neighbors Classification
Data Representation
- is the i-th feature vector with d dimensions;
- is the class label corresponding to ;
- N is the total number of training samples;
- C is the number of distinct classes.
Distance Metric
Algorithm Steps
Formal Definition
Example Illustration
- Traditional computation of Euclidean Distance Matrix
- 1.
- .
- 2.
- .
- 3.
- .
- 4.
- .
| Basic steps | Storage | MACs |
| Input: | - | |
| - | - | |
| - | - | |
| Total |
- 1.
- .
- 2.
- .
| Basic steps | Storage | MACs |
| Input: | - | |
| Total |
| Algorithm 1 Naive Computation of Euclidean Distance Matrix |
|
| Algorithm 2 Expanded Computation of Euclidean Distance Matrix |
|
5.3. Class Balancing
6. Experimental Results
7. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rashidi, S.; Asl, B. Strength of ensemble learning in automatic sleep stages classification using single-channel EEG and ECG signals. Med. Biol. Eng. Comput. 2023, 62, 997–1015. [Google Scholar] [CrossRef] [PubMed]
- Hogervorst, M.; Brouwer, A.; van Erp, J.B. Combining and comparing EEG peripheral physiology and eye-related measures for the assessment of mental workload. Front. Neurosci 2014, 8, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Rechtschaffen, A.; Kales, A. A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects; Public Health Service, US Government Printing Office: Washington, DC, USA, 1968. [CrossRef]
- Kevin, S.; Michaiel, T.; Rosemary, H. The use of actigraphy for assessment of the development of sleep-wake patterns in infants during the first 12 months of life. J. Sleep Res. 2007, 16, 181–187. [Google Scholar] [CrossRef]
- Correa, A.G.; Orosco, L.; Laciar, E. Automatic detection of drowsiness in EEG records based on multimodal analysis. Med. Eng. Phys. 2014, 36, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, R. Application of intrinsic band function technique for automated detection of sleep apnea using HRV and EDR signals. Biocybern. Biomed. Eng. 2018, 38, 136–144. [Google Scholar] [CrossRef]
- Alshammari, T.; Sarheed, T. Applying Machine Learning Algorithms for the Classification of Sleep Disorders. IEEE Access 2024, 12, 36110–36121. [Google Scholar] [CrossRef]
- Satapathy, S.K.; Patel, V.; Gandhi, M.; Mohapatra, R.K. Comparative Study of Brain Signals for Early Detection of Sleep Disorder Using Machine and Deep Learning Algorithm. In Proceedings of the 2024 IEEE International Conference on Interdisciplinary Approaches in Technology and Management for Social Innovation (IATMSI), Gwalior, India, 14–16 March 2024; Volume 2, pp. 1–6. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, Y.; Wang, Q.; Wang, X. Comparative analysis of different characteristics of automatic sleep stages. Comput. Methods Programs Biomed. 2019, 175, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, S.F.; Abbasi, Q.H.; Saeed, F.; Alghamdi, N.S. A convolutional neural network-based decision support system for neonatal quiet sleep detection. Math. Biosci. Eng. 2023, 20, 17018–17036. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gu, T.; Yao, W. Research on the application of the Sleep EEG Net model based on domain adaptation transfer in the detection of driving fatigue. Biomed. Signal Process. Control 2024, 90, 105832. [Google Scholar] [CrossRef]
- Jiang, D.; Ma, Y.; Wang, Y. Sleep stage classification using covariance features of multi-channel physiological signals on Riemannian manifolds. Comput. Methods Programs Biomed. 2019, 178, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Sci. 2019, 106, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, R.; Acharya, U.R. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern. Biomed. Eng. 2018, 38, 890–902. [Google Scholar] [CrossRef]
- da Silveira, T.; Kozakevicius, A.; Rodrigues, C. Single channel EEG sleep stage classification based on a streamlined set of statistical features in wavelet domain. Med. Biol. Eng. Comput. 2017, 55, 343–352. [Google Scholar] [CrossRef]
- Budak, U.; Bajaj, V.; Akbulut, Y.; Atila, O.; Sengur, A. An Effective Hybrid Model for EEG-Based Drowsiness Detection. IEEE Sens. J. 2019, 19, 7624–7631. [Google Scholar] [CrossRef]
- Taran, S.; Bajaj, V. Drowsiness Detection Using Adaptive Hermite Decomposition and Extreme Learning Machine for Electroencephalogram Signals. IEEE Sens. J. 2018, 18, 8855–8862. [Google Scholar] [CrossRef]
- Adnane, M.; Jiang, Z.; Yan, Z. Sleep-wake stages classification and sleep efficiency estimation using single-lead electrocardiogram. Expert Syst. Appl. 2012, 39, 1401–1413. [Google Scholar] [CrossRef]
- Barnes, L.; Lee, K.; Kempa-Liehr, A.; Hallum, L. Detection of sleep apnea from single-channel electroencephalogram (EEG) using an explainable convolutional neural network (CNN). PLoS ONE 2022, 17, e0272167. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.; Bhat, S.; Faust, O.; Adeli, H.; Chua, E.; Lim, W.; Koh, J. Nonlinear dynamics measures for automated EEG-based sleep stage detection. Eur. Neurol. 2015, 74, 268–287. [Google Scholar] [PubMed]
- Acharya, U.; Chua, E.; Chua, K.; Lim, C.; Tamura, T. Analysis and automatic identification of sleep stages using higher order spectra. Int. J. Neural Syst. 2010, 20, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Paul, Y. Various epileptic seizure detection techniques using biomedical signals: A review. Brain Inform. 2018, 5, 1–19. [Google Scholar] [CrossRef]
- Paul, Y.; Fridli, S. A Hybrid Approach for Sleep States Detection Using Blood Pressure and EEG Signal. Lect. Notes Electr. Eng. LNEE 2022, 832, 119–132. [Google Scholar] [CrossRef]
- Paul, Y.; Fridli, S. Epileptic Seizure Detection Using Piecewise Linear Reduction. Lect. Notes Comput. Sci. LNTCS 2020, 12014, 364–371. [Google Scholar] [CrossRef]
- Paul, Y.; Kumar, D.N. A Comparative Study of Famous Classification Techniques and Data Mining Tools. Lect. Notes Electr. Eng. LNEE 2020, 597, 627–644. [Google Scholar] [CrossRef]
- Paul, Y.; Fridli, S. Sleep states detection using halfwave and Franklin transformation. Ann. Univ. Sci. Math. Bp. 2021; LXIV, 157–177. [Google Scholar]
- Ichimaru, Y.; Moody, G. Development of the polysomnographic database on CD-ROM. PCN Psychiatry Clin. Neurosci. 1999, 53, 175–177. [Google Scholar] [CrossRef]
- Redmond, S.; Heneghan, C. Electrocardiogram-based automatic sleep staging in sleep disordered breathing. Comput. Cardiol. IEEE 2003, 30, 609–612. [Google Scholar]
- Hayet, W.; Yacoub, S. Sleep-wake stages classification based on heart rate variability. In Proceedings of the Biomedical Engineering and Informatics (BMEI), 5th International Conference, Chongqing, China, 16–18 October 2012; pp. 996–999. [Google Scholar] [CrossRef]
- Werteni, H.; Yacoub, S.; Ellouze, N. Classification of Sleep Stages Based on EEG Signals. Int. Rev. Comput. Softw. (IRECOS) 2015, 10, 174. [Google Scholar] [CrossRef]
- An, P.; Si, W.; Ding, S.; Xue, G.; Yuan, Z. A Novel EEG Sleep Staging Method for Wearable Devices Based on Amplitude-time Mapping. In Proceedings of the 2019 IEEE 4th International Conference on Advanced Robotics and Mechatronics (ICARM), Toyonaka, Japan, 3–5 July 2019; pp. 124–129. [Google Scholar]
- Zhang, J.; Yao, R.; Ge, W.; Gao, J. Orthogonal convolutional neural networks for automatic sleep stage classification based on single-channel EEG. Comput. Methods Programs Biomed. 2020, 183, 105089. [Google Scholar] [CrossRef] [PubMed]
- Surantha, N.; Lesmana, T.F.; Isa, S.M. Sleep stage classification using extreme learning machine and particle swarm optimization for healthcare big data. J. Big Data 2021, 8, 14. [Google Scholar] [CrossRef]
- Wang, W.; Qin, D.; Fang, Y.; Zhou, C.; Zheng, Y. Automatic Multi-class Sleep Staging Method Based on Novel Hybrid Features. J. Electr. Eng. Technol. 2023, 19, 709–722. [Google Scholar] [CrossRef]
- Motamedi-Fakhr, S.; Moshrefi-Torbati, M.; Hill, M.; Hill, C.; White, P. Signal processing techniques applied to human sleep EEG signals—A review. Biomed. Signal Process. Control 2014, 10, 21–33. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Huang, Q.; Lv, Y.; Chen, F. A review of automated sleep stage based on EEG signals. Biocybern. Biomed. Eng. 2024. [Google Scholar] [CrossRef]
- Gotman, J.; Gloor, P. Automatic recognition and quantification of interictal epileptic activity in the human scalp EEG. Electroencephalogr. Clin. Neurophysiol. 1976, 41, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Albanie, S. Euclidean Distance Matrix Trick. 2019. Available online: https://samuelalbanie.com/files/Euclidean_distance_trick.pdf (accessed on 9 July 2024).
- Ren, P.; Tang, S.; Fang, F.; Luo, L.; Xu, L.; Bringas-Vega, M.L.; Yao, D.; Kendrick, K.M.; Valdes-Sosa, P.A. Gait rhythm fluctuation analysis for neurodegenerative diseases by empirical mode decomposition. IEEE Trans. Biomed. Eng. 2016, 99, 1. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.; Bowyer, K.; Hall, L.; Kegelmeyer, W. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Haibo, H.; Yang, B.; Edwardo, G.; Li, S. ADASYN: Adaptive Synthetic Sampling Approach for Imbalanced Learning. In Proceedings of the International Joint Conference on Neural Networks, Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar]
- López, V.; Fernández, A.; Moreno-Torres, J.; Herrera, F. Analysis of preprocessing vs. cost-sensitive learning for imbalanced classification. Open problems on intrinsic data characteristics. Expert Syst. Appl. 2012, 39, 6585–6608. [Google Scholar] [CrossRef]
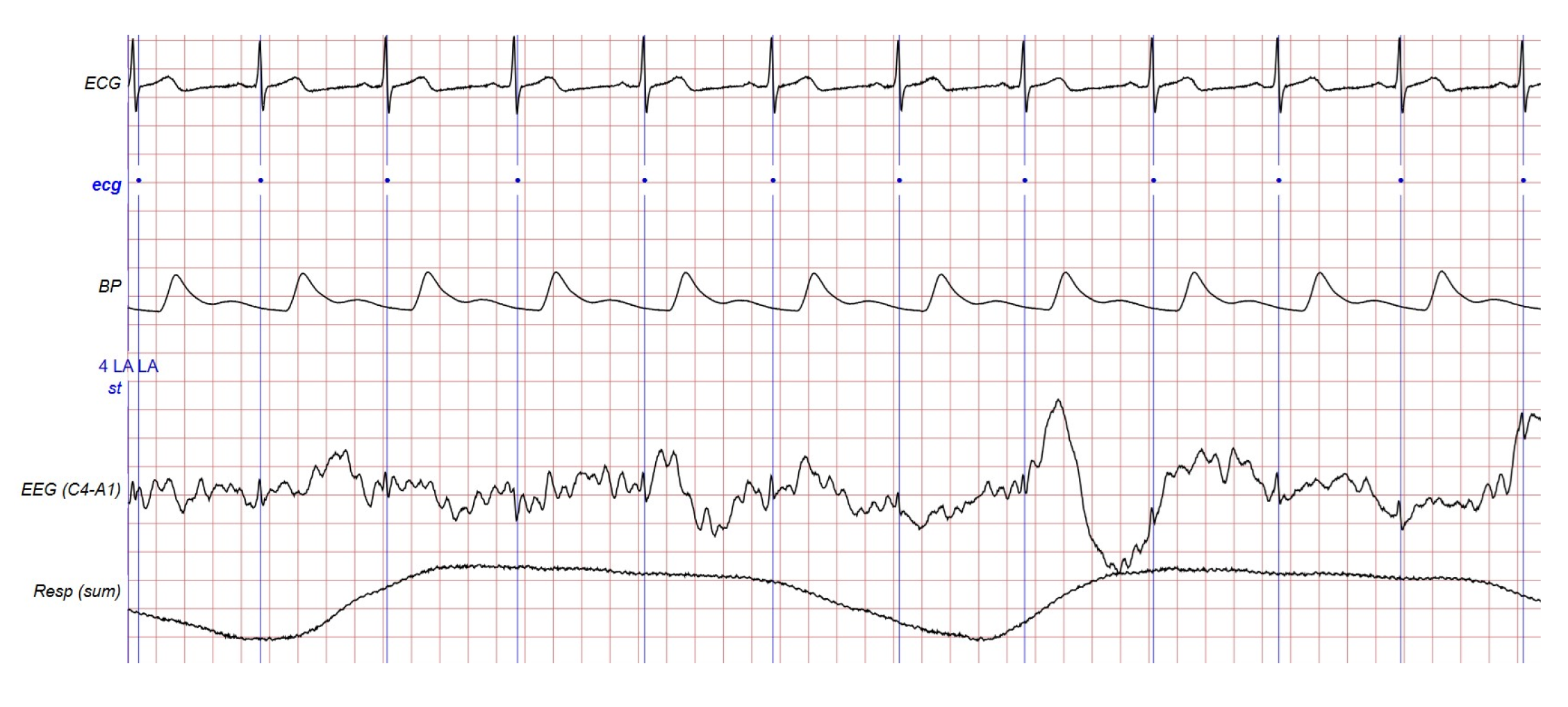
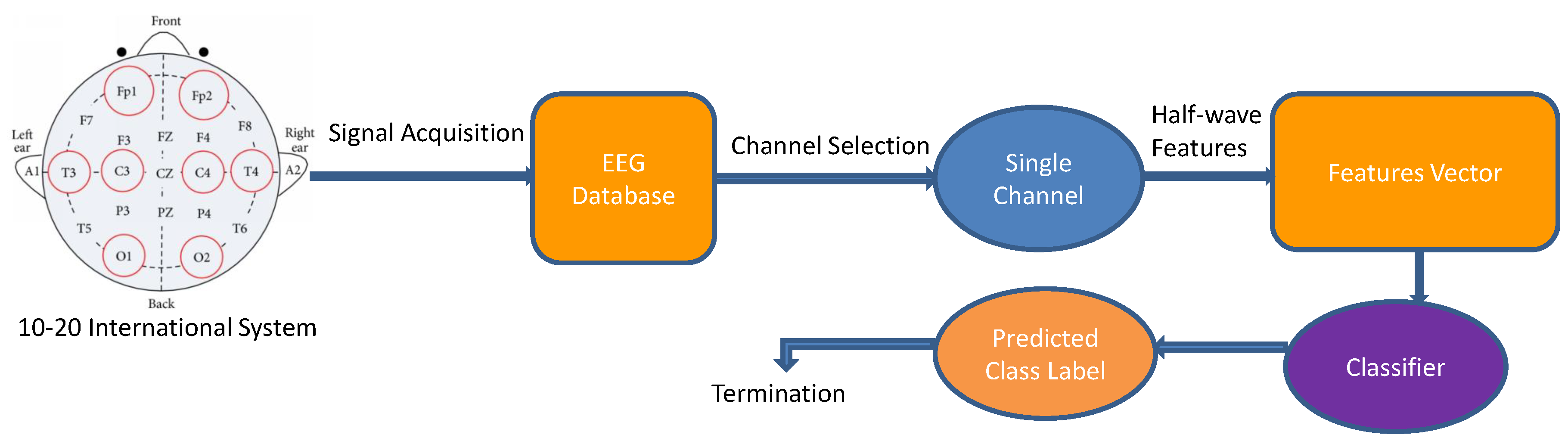
| Signal, Record and Duration | Classifier | Features | Train and Test | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| EEG, slp01a, 90 mnts | SVM | Halfwave | 60–40 | 92.36 | 91.42 | 91.89 |
| EEG, slp01b, 60 mnts | SVM | Halfwave | 60–40 | 60.87 | 89.79 | 75.33 |
| EEG, slp2a, 90 mnts | SVM | Halfwave | 60–40 | 39.60 | 80.60 | 63.1 |
| EEG, slp2b, 60 mnts | SVM | Halfwave | 60–40 | 80.36 | 90.30 | 79.21 |
| EEG, slp03, 90 mnts | SVM | Halfwave | 60–40 | 80.36 | 90.30 | 79.25 |
| Signal, Record and Duration | Classifier | Features | Train and Test | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| EEG, slp01a, 90 mnts | KNN | Halfwave | 60–40 | 97.12 | 96.63 | 97.61 |
| EEG, slp01b, 60 mnts | KNN | Halfwave | 60–40 | 90.64 | 95.40 | 93.02 |
| EEG, slp2a, 90 mnts | KNN | Halfwave | 60–40 | 94.86 | 97.72 | 96.29 |
| EEG, slp2b, 60 mnts | KNN | Halfwave | 60–40 | 96.33 | 97.75 | 97.04 |
| EEG, slp03, 90 mnts | KNN | Halfwave | 60–40 | 95.73 | 96.48 | 96.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, Y.; Singh, R.; Sharma, S.; Singh, S.; Ra, I.-H. Efficient Sleep Stage Identification Using Piecewise Linear EEG Signal Reduction: A Novel Algorithm for Sleep Disorder Diagnosis. Sensors 2024, 24, 5265. https://doi.org/10.3390/s24165265
Paul Y, Singh R, Sharma S, Singh S, Ra I-H. Efficient Sleep Stage Identification Using Piecewise Linear EEG Signal Reduction: A Novel Algorithm for Sleep Disorder Diagnosis. Sensors. 2024; 24(16):5265. https://doi.org/10.3390/s24165265
Chicago/Turabian StylePaul, Yash, Rajesh Singh, Surbhi Sharma, Saurabh Singh, and In-Ho Ra. 2024. "Efficient Sleep Stage Identification Using Piecewise Linear EEG Signal Reduction: A Novel Algorithm for Sleep Disorder Diagnosis" Sensors 24, no. 16: 5265. https://doi.org/10.3390/s24165265
APA StylePaul, Y., Singh, R., Sharma, S., Singh, S., & Ra, I.-H. (2024). Efficient Sleep Stage Identification Using Piecewise Linear EEG Signal Reduction: A Novel Algorithm for Sleep Disorder Diagnosis. Sensors, 24(16), 5265. https://doi.org/10.3390/s24165265







