A Two-Stage Automatic Color Thresholding Technique
Abstract
1. Introduction
- A fully automated histogram-based color thresholding approach is provided, which is invariant to natural variations in images such as varying background colors, lighting differences, and camera specifications.
- Block size determination and addressing the blocking artefacts problem during the local thresholding stage are achieved by automatically detecting the blocks from the global thresholded image.
- The method represents an unsupervised technique, as it does not require any labeled data for training, making it advantageous in situations where labeled data are limited or difficult/costly to generate.
2. Related Work
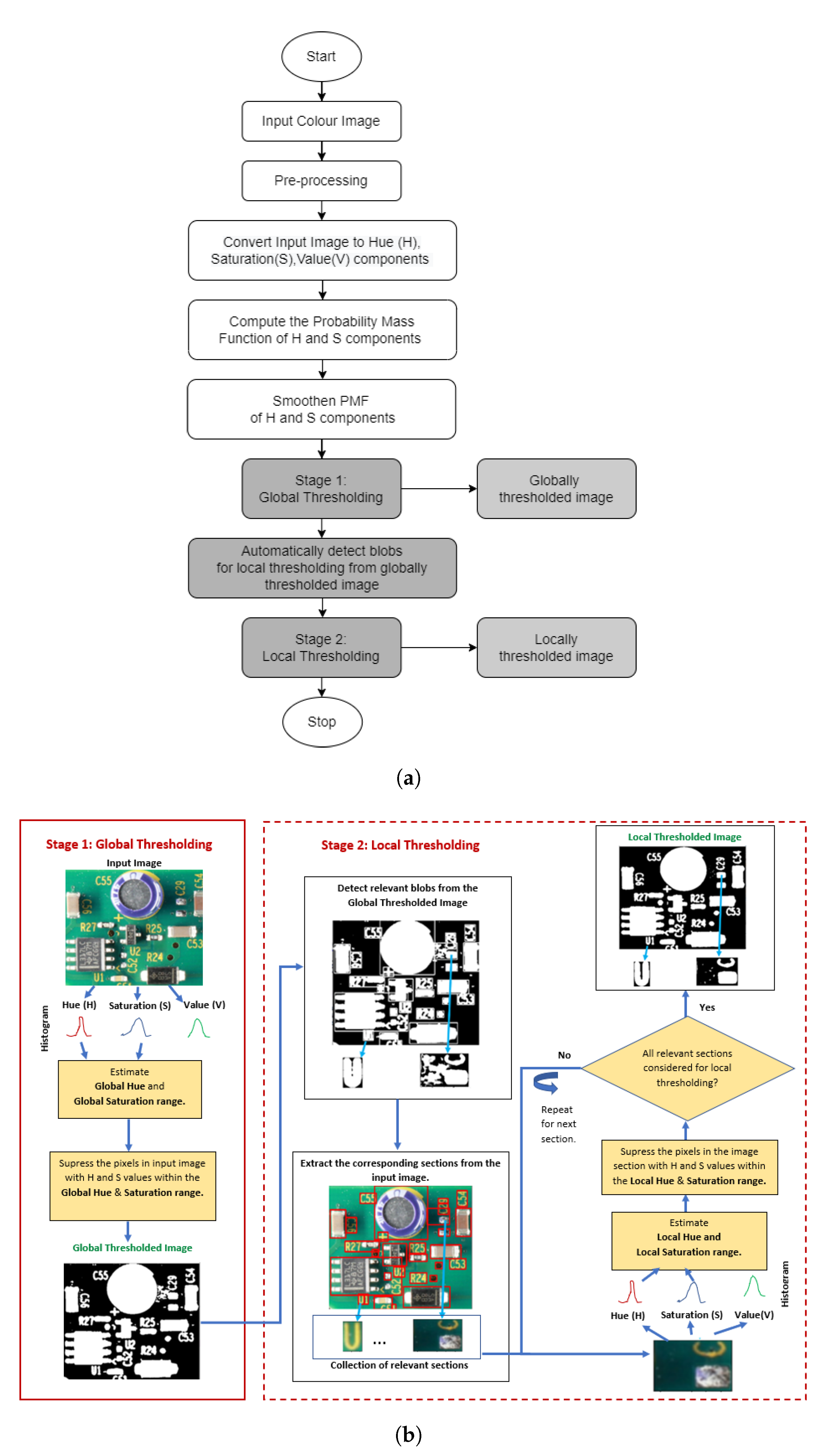
3. Methods
| Algorithm 1 Two-stage global–local thresholding | ||
| Input: | ||
| ▹ Input color image | ||
| ▹ Threshold value for histogram | ||
| ▹ Threshold value for histogram | ||
| ▹ Window size for calculating the and | ||
| ▹ The limit for maximum continuous hue range | ||
| ▹ The limit for maximum continuous saturation range | ||
| ▹ A constant that determines the degree of change from global hue to local hue | ||
| ▹ A constant that determines the degree of change from global saturation to local saturation | ||
| Output: | ||
| ▹ Globally thresholded image, background in black and foreground in white | ||
| ▹ Locally thresholded image, background in black and foreground in white | ||
| ||
| ||
3.1. Stage 1: Global Thresholding
3.2. Stage 2: Local Thresholding
4. Implementation Details
5. Evaluation Metrics
5.1. Dice Similarity Index
5.2. Matthews Correlation Coefficient
- Correctly predicted foreground pixels are considered true positives (TP)—the number of pixels segmented as foreground in both GT and T images.
- Falsely predicted foreground pixels are considered false positives (FP)—the number of pixels segmented as foreground in T and background in GT.
- Correctly predicted background pixels are considered true negatives (TN)—the number of pixels segmented as background in both GT and T images.
- Falsely predicted background pixels are considered false negatives (FN)—the number of pixels segmented as background in T and foreground in GT.
5.3. Peak Signal-to-Noise Ratio
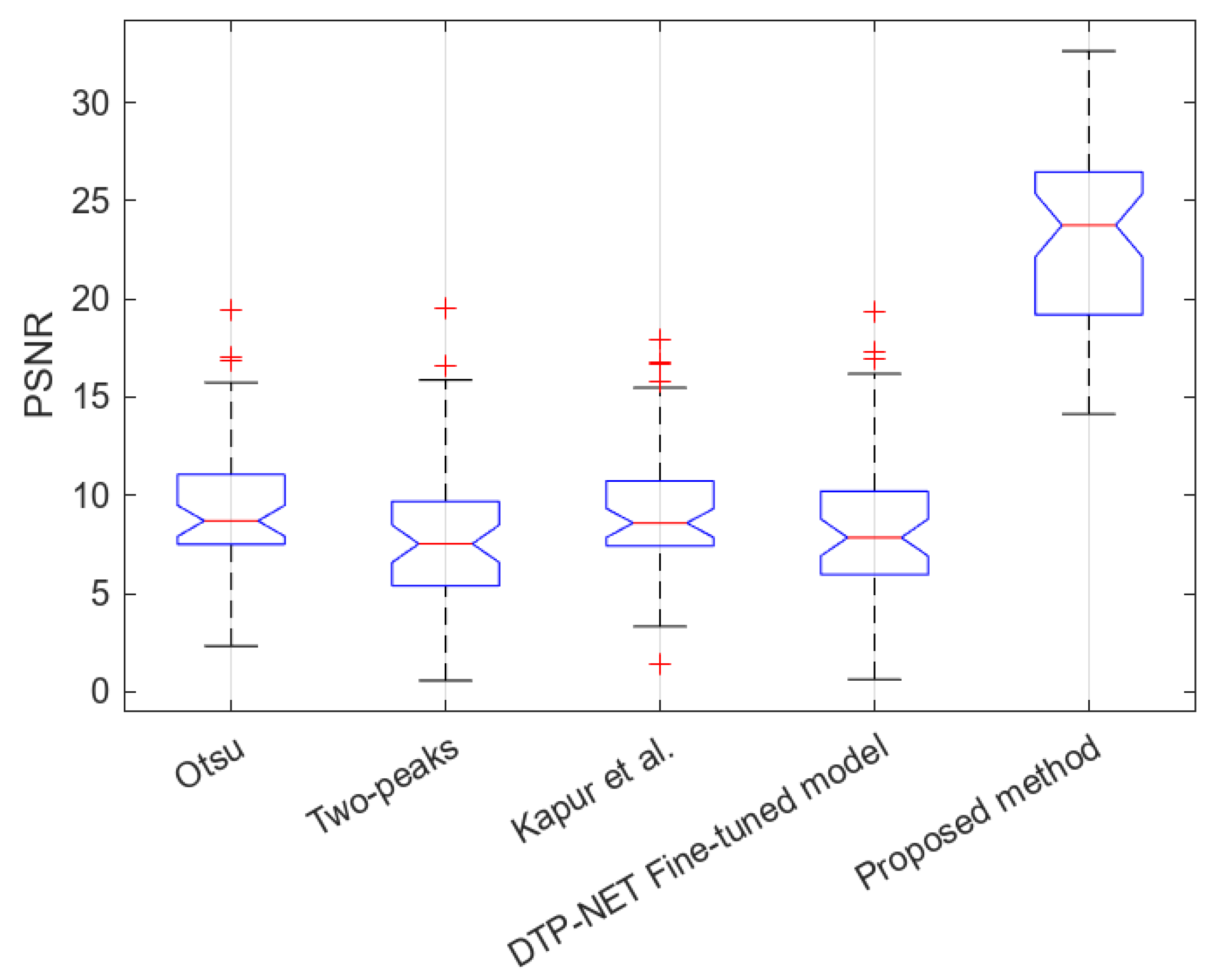
6. Results
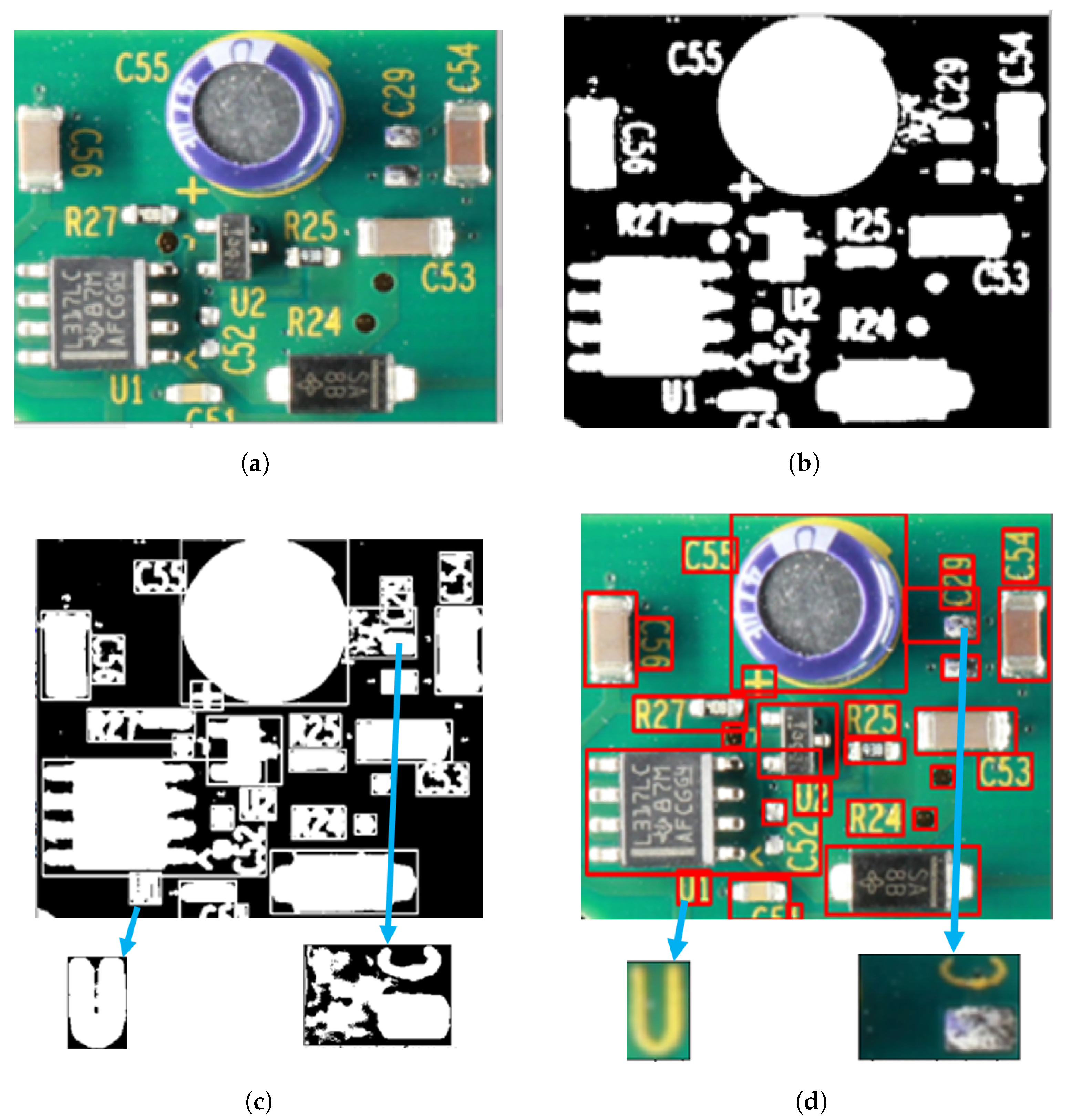
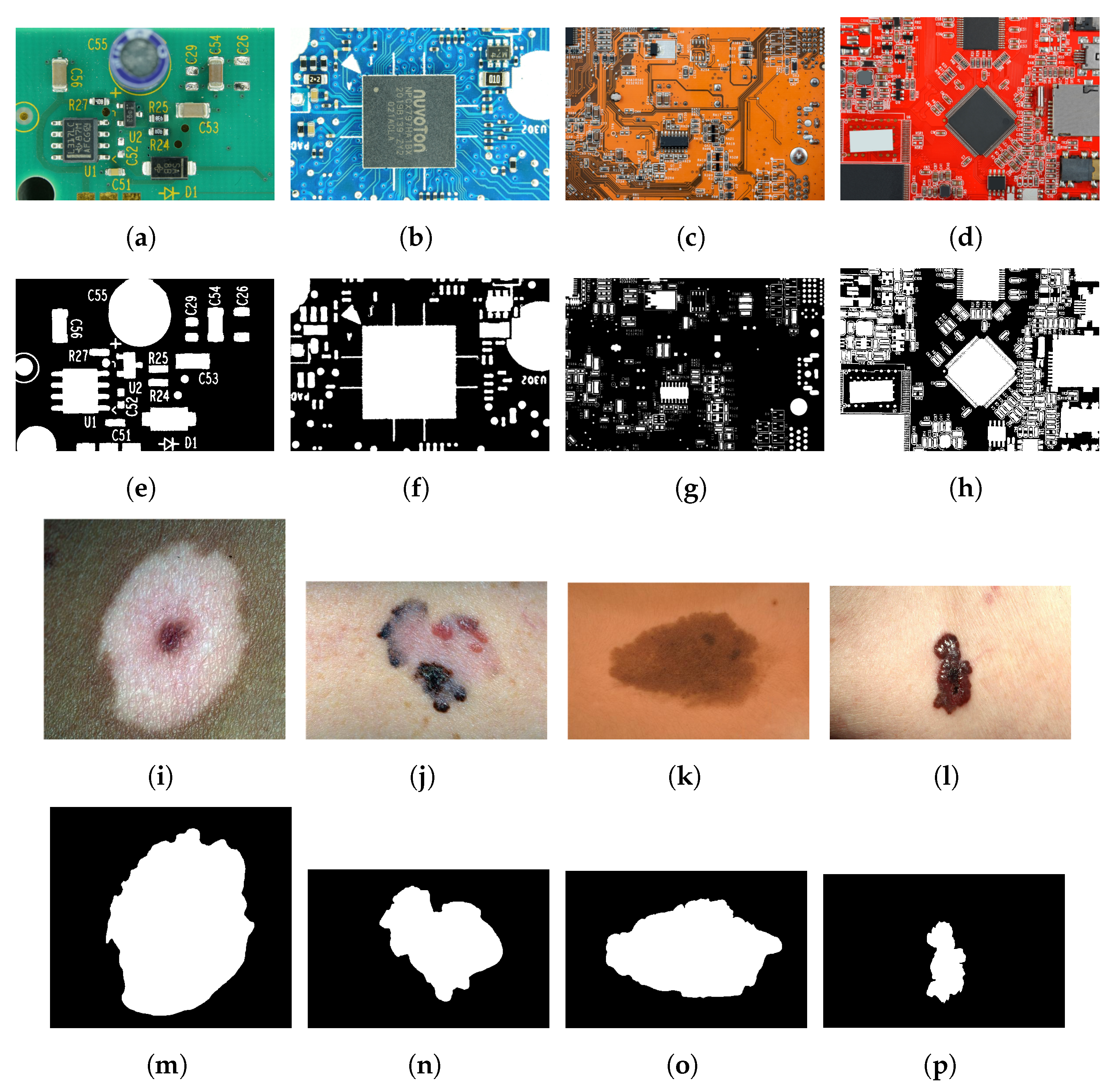
6.1. Experimental Results Using the Skin Cancer Dataset
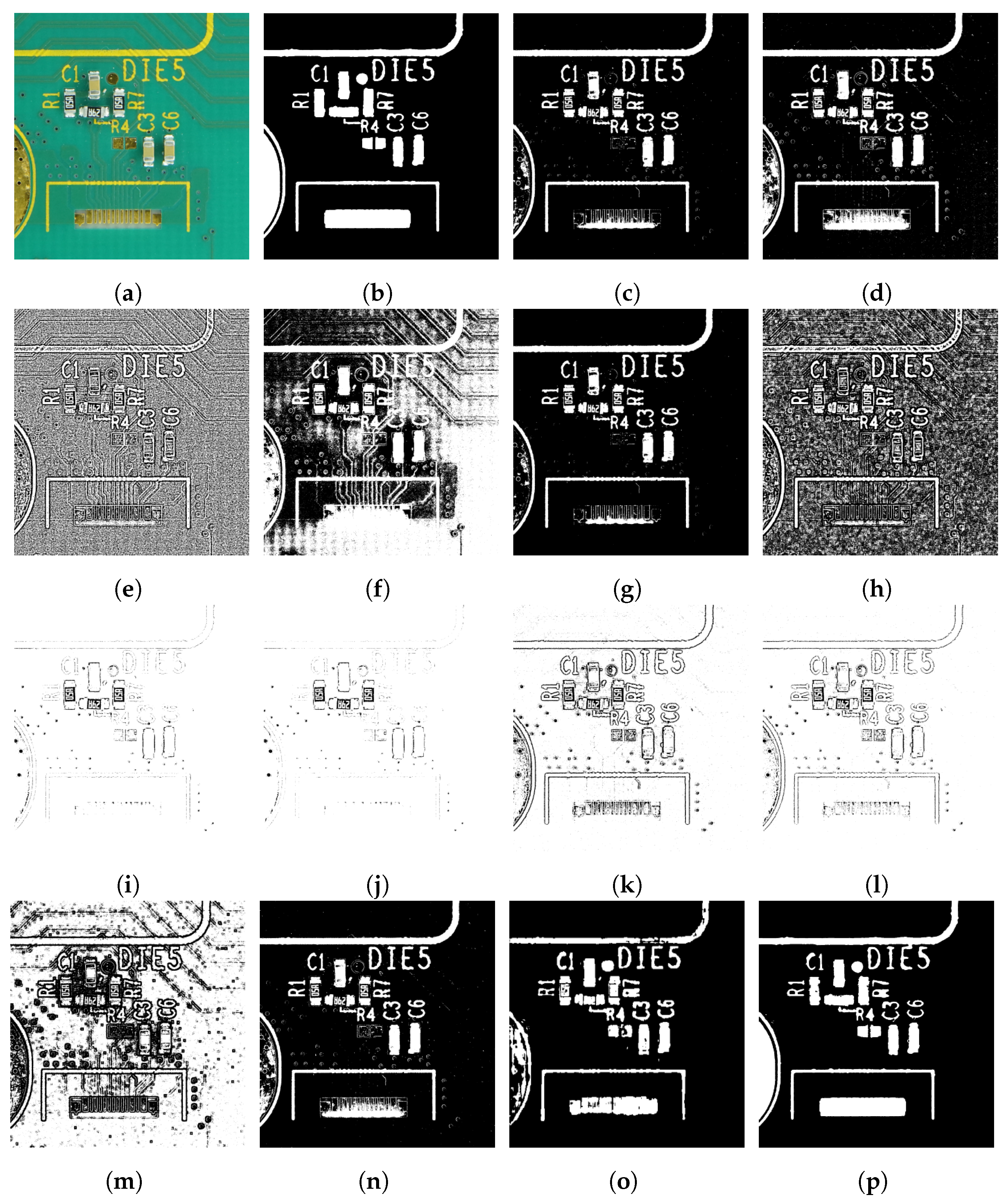
6.2. Experimental Results Using the PCA Board Dataset
7. Discussion
- An unsupervised method, as it does not require any ground-truth data;
- A robust method that is invariant to background color variations and changes in intensity;
- A fully automated color thresholding approach, as there is no need to adjust parameters based on varying image conditions;
- Able to automatically detect the block size for the local thresholding stage;
- Effective at suppressing shadow regions;
- Easily adjustable to different image qualities;
- Efficient in suppressing background pixels of images with tiny foreground components;
- Efficient in determining the threshold value for unimodal, bimodal, and multimodal histograms and also for histograms with sharp peaks and elongated shoulders;
- Effective for symmetric, skewed, or uniform histogram analysis.
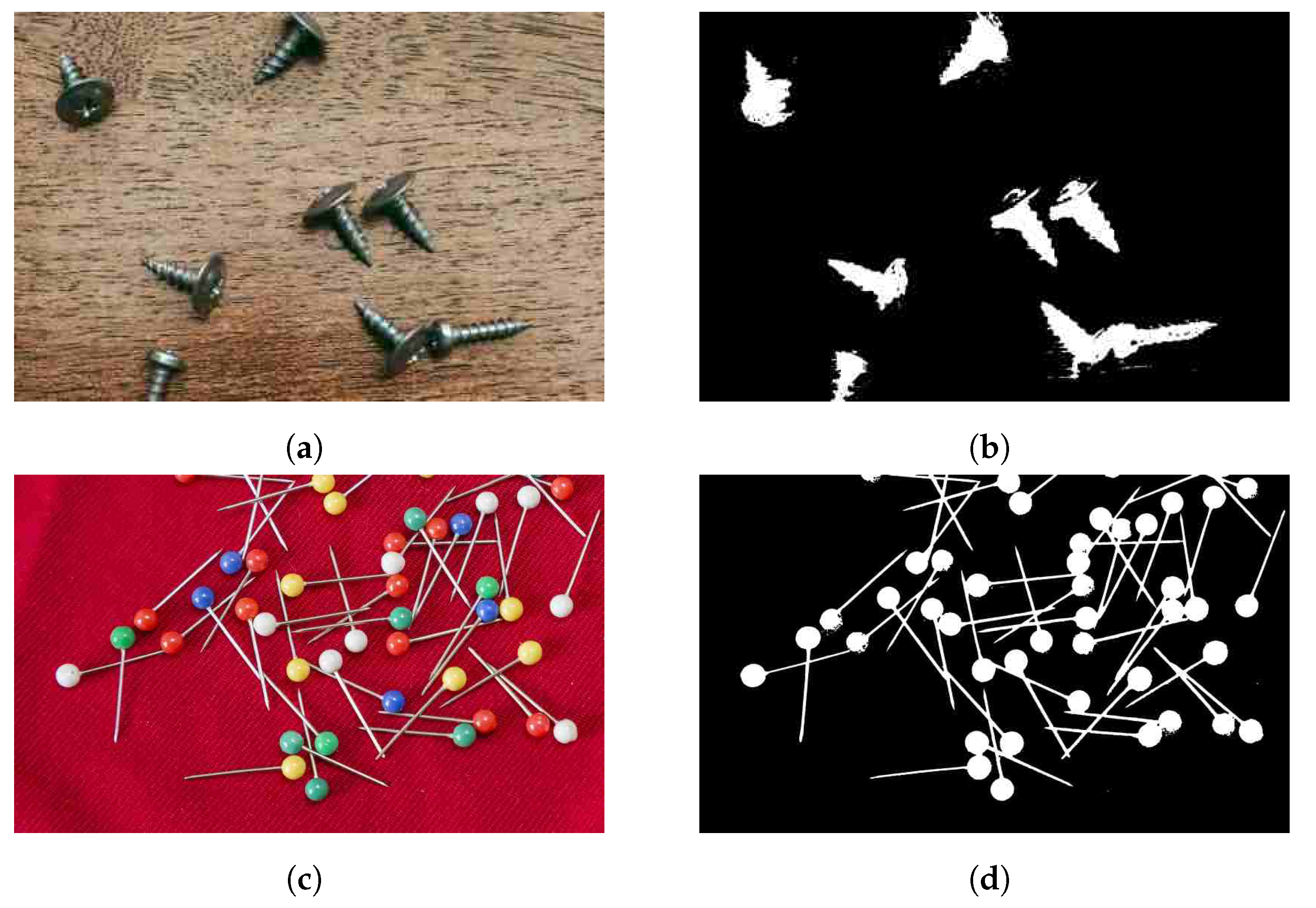
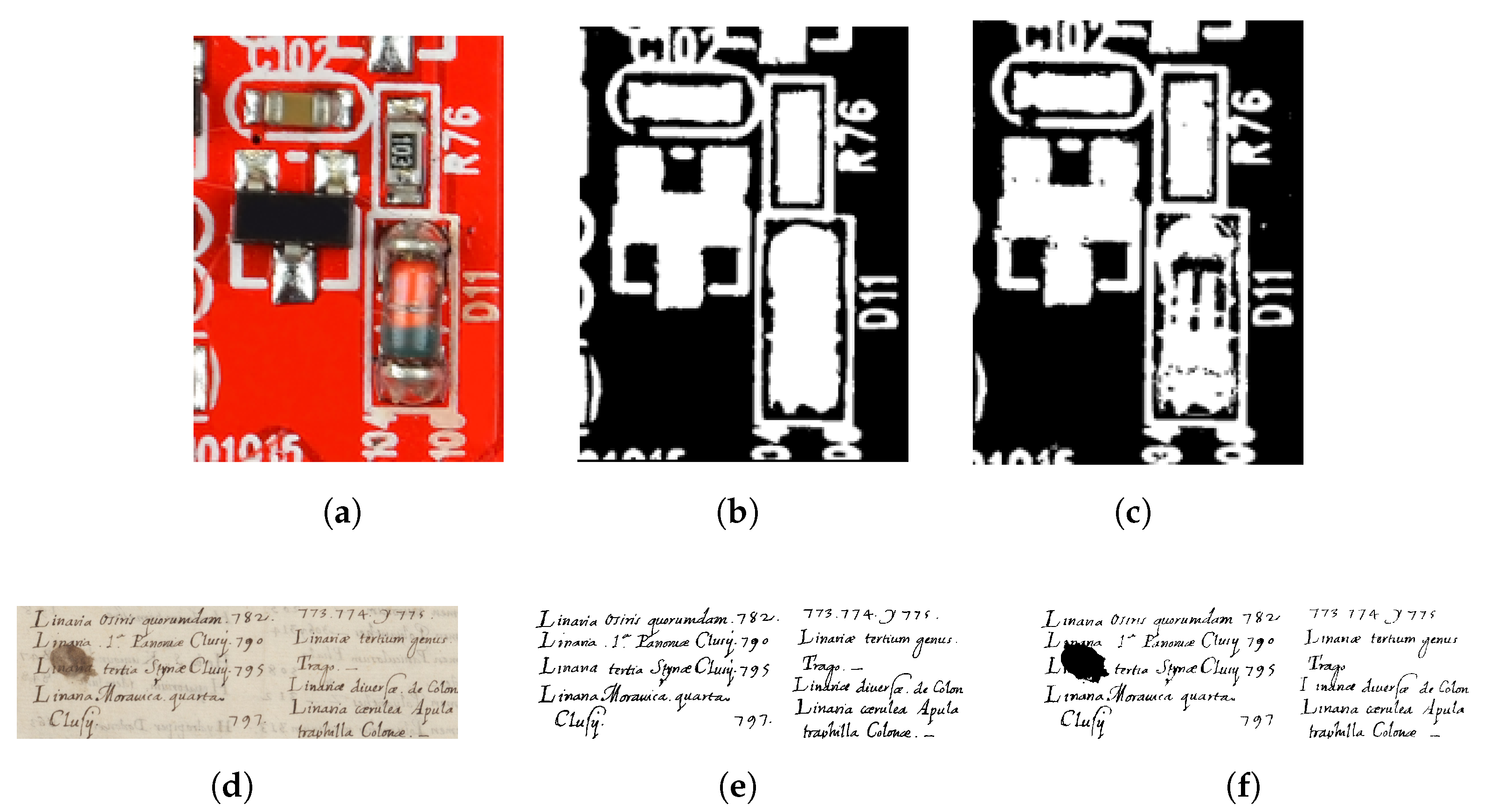
8. Application Areas
9. Limitations and Future Work
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CV | Computer vision |
| DCNN | Deep convolutional neural network |
| DSI | Dice similarity index |
| FN | False negative |
| FP | False positive |
| GT | Ground truth |
| HSV | Hue, saturation, value |
| MCC | Matthews correlation coefficient |
| MP | Megapixels |
| PCA | Printed circuit assembly |
| PCB | Printed circuit board |
| PMF | Probability mass function |
| PSNR | Peak signal-to-noise ratio |
| ROI | Region of interest |
| TN | True negative |
| TP | True positive |
Appendix A. Statistical Evaluation of the Proposed Method on the PCA Board Dataset
| p-Value | |
|---|---|
| Otsu [11] | 0.1383 |
| Two-peak [27] | 0.1404 |
| Kapur et al. [12] | 0.0501 |
| DTP-NET fine-tuned model [46] | 0.1239 |
| Proposed method | 0.4575 |
| PSNR | |
|---|---|
| p-value | 2.11 × 10−60 |
| Method 1 | Method 2 | Difference | p-Value |
|---|---|---|---|
| Proposed method | Otsu [11] | −13.8644 | 0.0000 |
| Proposed method | Two-peaks [27] | −15.4534 | 0.0000 |
| Proposed method | Kapur et al. [12] | −13.8882 | 0.0000 |
| Proposed method | DTP-NET (fine-tuned model) [46] | −14.9383 | 0.0000 |
| Otsu [11] | Two-peak [27] | 1.5889 | 0.2719 |
| Otsu [11] | Kapur et al. [12] | 0.0237 | 1.0000 |
| Otsu [11] | DTP-NET (fine-tuned model) [46] | −0.5151 | 0.6640 |
| Two-peak [27] | Kapur et al. [12] | −1.5652 | 0.2868 |
| Two-peak [27] | DTP-NET (fine-tuned model) [46] | 1.0739 | 0.9677 |
| Kapur et al. [12] | DTP-NET (fine-tuned model) [46] | 1.0502 | 0.6827 |
Appendix B. Configurations of Parameters
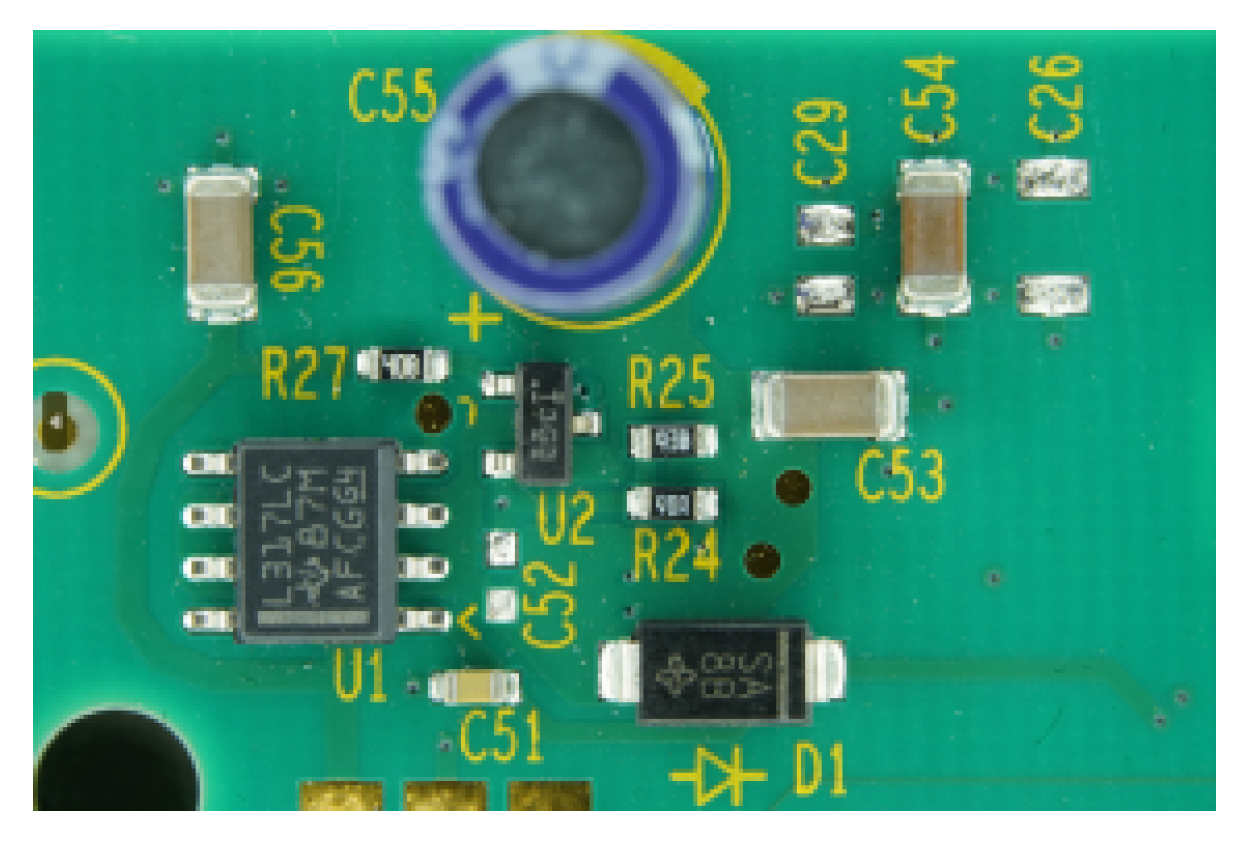
| Thresholding Result | Parameters |
|---|---|
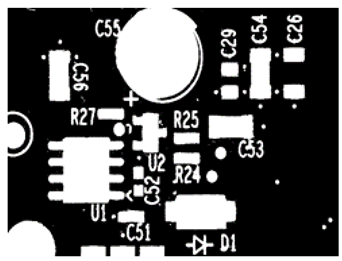 | Ideal parameters: determined heuristically. Cutoff_Gradient: 0.001 Cutoff_Area: 1/180, Window_Size: 5 Limit1: 4, Limit2: 10 C1: 5, C2: 10 |
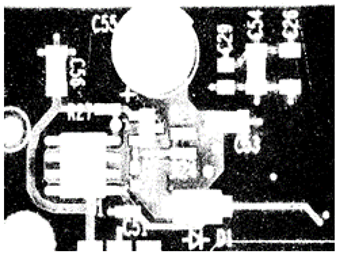 | Cutoff_Gradient: 0.1 An increase in Cutoff_Gradient by a factor of 100 to 0.1 led to a decrease in the nominated hue or saturation range, which led to the misclassification of some background pixels as foreground. |
 | Cutoff_Gradient: 0.00001 A decrease in Cutoff_Gradient by a factor of 100 to 0.00001 led to an increase in the nominated hue or saturation range, which led to the misclassification of some foreground pixels as background. A change in Cutoff_Area led to the same effect. |
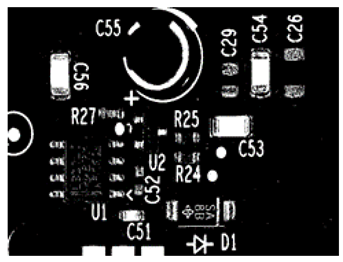 | Window_Size: 10, Limit1: 8, Limit2: 20 An increase in Window_Size, Limit1, and Limit2 by a factor of 2 led to an increase in the nominated hue or saturation range, which led to the misclassification of some foreground pixels as background pixels. Limit1 defines the allowable hue discontinuity, and Limit2 defines the allowable saturation discontinuity. |
 | Window_Size: 2, Limit1: 2, Limit2: 5 A decrease in Window_Size, Limit1, and Limit2 by a factor of 2 led to a decrease in the nominated hue or saturation range, which led to the misclassification of some background pixels as foreground. |
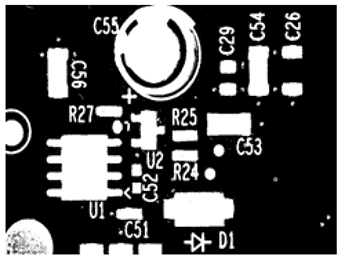 | C1: 10, C2: 20 An increase in C1 and C2 by a factor of 2 led to the misclassification of some foreground pixels as background in the local thresholding stage. |
 | C1: 2, C2: 5 A decrease in C1 and C2 by a factor of 2 led to the misclassification of background pixels as foreground in the local thresholding stage. |
References
- Fan, H.; Xie, F.; Li, Y.; Jiang, Z.; Liu, J. Automatic segmentation of dermoscopy images using saliency combined with Otsu threshold. Comput. Biol. Med. 2017, 85, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Prewitt, J.M.; Mendelsohn, M.L. The analysis of cell images. Ann. N. Y. Acad. Sci. 1966, 128, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, A.; Kumar, A.; Singh, G. Tsallis entropy based multilevel thresholding for colored satellite image segmentation using evolutionary algorithms. Expert Syst. Appl. 2015, 42, 8707–8730. [Google Scholar] [CrossRef]
- Fan, J.; Yu, J.; Fujita, G.; Onoye, T.; Wu, L.; Shirakawa, I. Spatiotemporal segmentation for compact video representation. Signal Process. Image Commun. 2001, 16, 553–566. [Google Scholar] [CrossRef]
- Bradley, D.; Roth, G. Adaptive thresholding using the integral image. J. Graph. Tools 2007, 12, 13–21. [Google Scholar] [CrossRef]
- White, J.M.; Rohrer, G.D. Image thresholding for optical character recognition and other applications requiring character image extraction. IBM J. Res. Dev. 1983, 27, 400–411. [Google Scholar] [CrossRef]
- Shaikh, S.H.; Maiti, A.K.; Chaki, N. A new image binarization method using iterative partitioning. Mach. Vis. Appl. 2013, 24, 337–350. [Google Scholar] [CrossRef]
- Garcia-Lamont, F.; Cervantes, J.; López, A.; Rodriguez, L. Segmentation of images by color features: A survey. Neurocomputing 2018, 292, 1–27. [Google Scholar] [CrossRef]
- Sahoo, P.K.; Soltani, S.A.; Wong, A.K. A survey of thresholding techniques. Comput. Vis. Graph. Image Process. 1988, 41, 233–260. [Google Scholar] [CrossRef]
- Hamuda, E.; Mc Ginley, B.; Glavin, M.; Jones, E. Automatic crop detection under field conditions using the HSV colour space and morphological operations. Comput. Electron. Agric. 2017, 133, 97–107. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Kapur, J.; Sahoo, P.; Wong, A. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Gr. Image Process 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Pare, S.; Kumar, A.; Bajaj, V.; Singh, G.K. A multilevel color image segmentation technique based on cuckoo search algorithm and energy curve. Appl. Soft Comput. 2016, 47, 76–102. [Google Scholar] [CrossRef]
- Mushrif, M.M.; Ray, A.K. A-IFS histon based multithresholding algorithm for color image segmentation. IEEE Signal Process. Lett. 2009, 16, 168–171. [Google Scholar] [CrossRef]
- Cao, Z.; Zhang, X.; Mei, X. Unsupervised segmentation for color image based on graph theory. In Proceedings of the Second International Symposium on Intelligent Information Technology Application, Shanghai, China, 20–22 December 2008; Volume 2, pp. 99–103. [Google Scholar]
- Harrabi, R.; Braiek, E.B. Color image segmentation using multi-level thresholding approach and data fusion techniques. EURASIP J. Image Video Process. 2012, 1, 1–11. [Google Scholar]
- Kang, S.D.; Yoo, H.W.; Jang, D.S. Color image segmentation based on the normal distribution and the dynamic thresholding. Comput. Sci. Appl. ICCSA 2007, 4705, 372–384. [Google Scholar]
- Mehta, D.; Lu, H.; Paradis, O.P.; MS, M.A.; Rahman, M.T.; Iskander, Y.; Chawla, P.; Woodard, D.L.; Tehranipoor, M.; Asadizanjani, N. The big hack explained: Detection and prevention of PCB supply chain implants. ACM J. Emerg. Technol. Comput. Syst. (JETC) 2020, 16, 1–25. [Google Scholar] [CrossRef]
- Mittal, H.; Pandey, A.C.; Saraswat, M.; Kumar, S.; Pal, R.; Modwel, G. A comprehensive survey of image segmentation: Clustering methods, performance parameters, and benchmark datasets. Multimed. Tools Appl. 2021, 10, 1–26. [Google Scholar] [CrossRef]
- Dirami, A.; Hammouche, K.; Diaf, M.; Siarry, P. Fast multilevel thresholding for image segmentation through a multiphase level set method, Signal Process. Signal Process. 2013, 93, 139–153. [Google Scholar] [CrossRef]
- Sezgin, M.; Sankur, B. Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 2004, 13, 146–166. [Google Scholar]
- Glasbey, C.A. An analysis of histogram-based thresholding algorithms. CVGIP Graph. Model. Image Process. 1993, 55, 532–537. [Google Scholar] [CrossRef]
- Niblack, W. An Introduction to Digital Image Processing; Prentice-Hall: Englewood Cliffs, NJ, USA, 1986; pp. 115–116. [Google Scholar]
- Goh, T.Y.; Basah, S.N.; Yazid, H.; Safar, M.J.A.; Saad, F.S.A. Performance analysis of image thresholding: Otsu technique. Measurement 2018, 114, 298–307. [Google Scholar] [CrossRef]
- Su, H.; Zhao, D.; Elmannai, H.; Heidari, A.A.; Bourouis, S.; Wu, Z.; Cai, Z.; Gui, W.; Chen, M. Multilevel threshold image segmentation for COVID-19 chest radiography: A framework using horizontal and vertical multiverse optimization. Comput. Biol. Med. 2022, 146, 105618. [Google Scholar] [CrossRef] [PubMed]
- Qi, A.; Zhao, D.; Yu, F.; Heidari, A.A.; Wu, Z.; Cai, Z.; Alenezi, F.; Mansour, R.F.; Chen, H.; Chen, M. Directional mutation and crossover boosted ant colony optimization with application to COVID-19 X-ray image segmentation. Comput. Biol. Med. 2023, 148, 105810. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.R. Algorithms for Image Processing and Computer Vision; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Sauvola, J.; Seppanen, T.; Haapakoski, S.; Pietikainen, M. Adaptive document binarization. In Proceedings of the IEEE Proceedings of the Fourth International Conference on Document Analysis and Recognition, Ulm, Germany, 18–20 August 1997; Volume 1, pp. 147–152.
- Rosenfeld, A.; Kak, A.C. Digital Picture Processing, 2nd ed.; Academic Press: New York, NY, USA, 1982. [Google Scholar]
- Rosenfeld, A.; De La Torre, P. Histogram concavity analysis as an aid in threshold selection. IEEE Trans. Syst. Man Cybern. 1983, 2, 231–235. [Google Scholar] [CrossRef]
- Mason, D.; Lauder, I.; Rutovitz, D.; Spowart, G. Measurement of C-bands in human chromosomes. Comput. Biol. Med. 1975, 5, 179–201. [Google Scholar] [CrossRef]
- Tseng, D.C.; Li, Y.F.; Tung, C.T. Circular histogram thresholding for color image segmentation. In Proceedings of the 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; Volume 2, pp. 673–676. [Google Scholar]
- Doyle, W. Operations useful for similarity-invariant pattern recognition. J. ACM (JACM) 1962, 9, 259–267. [Google Scholar] [CrossRef]
- Pratikakis, I.; Zagoris, K.; Barlas, G.; Gatos, B. ICDAR2017 competition on document image binarization (DIBCO 2017). In Proceedings of the 2017 14th IAPR International Conference on Document Analysis and Recognition (ICDAR), Kyoto, Japan, 9–15 November 2017; Volume 1, pp. 1395–1403. [Google Scholar]
- Sulaiman, A.; Omar, K.; Nasrudin, M.F. Degraded historical document binarization: A review on issues, challenges, techniques, and future directions. J. Imaging 2019, 5, 48. [Google Scholar] [CrossRef]
- Wolf, C.; Jolion, J.M. Extraction and recognition of artificial text in multimedia documents. Form. Pattern Anal. Appl. 2004, 6, 309–326. [Google Scholar] [CrossRef]
- Feng, M.L.; Tan, Y.P. Contrast adaptive binarization of low quality document images. IEICE Electron. Express 2004, 1, 501–506. [Google Scholar] [CrossRef]
- Singh, O.I.; Sinam, T.; James, O.; Singh, T.R. Local contrast and mean thresholding in image binarization. Int. J. Comput. Appl. 2012, 51, 4–10. [Google Scholar]
- Sukesh, R.; Seuret, M.; Nicolaou, A.; Mayr, M.; Christlein, V. A Fair Evaluation of Various Deep Learning-Based Document Image Binarization Approaches. In Document Analysis Systems, Proceedings of the 15th IAPR International Workshop, La Rochelle, France, 22–25 May 2022; Springer International Publishing: Cham, Switzerland, 2022; Volume 1, pp. 771–785. [Google Scholar]
- Bankman, I. Handbook of Medical Image PROCESSING and Analysis; Elsevier: Amsterdam, The Netherlands, 2008; Volume 1, p. 69. [Google Scholar]
- Feng, Y.; Zhao, H.; Li, X.; Zhang, X.; Li, H. A multi-scale 3D Otsu thresholding algorithm for medical image segmentation. Digit. Signal Process. 2017, 60, 186–199. [Google Scholar] [CrossRef]
- Fazilov, S.K.; Yusupov, O.R.; Abdiyeva, K.S. Mammography image segmentation in breast cancer identification using the otsu method. Web Sci. Int. Sci. Res. J. 2022, 3, 196–205. [Google Scholar]
- Ramadas, M.; Abraham, A. Detecting tumours by segmenting MRI images using transformed differential evolution algorithm with Kapur’s thresholding. Neural Comput. Appl. 2020, 32, 6139–6149. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 3, pp. 234–241. [Google Scholar]
- Punn, N.S.; Agarwal, S. Modality specific U-Net variants for biomedical image segmentation: A survey. Artif. Intell. Rev. 2022, 55, 5845–5889. [Google Scholar] [CrossRef]
- Venugopal, V.; Joseph, J.; Das, M.V.; Nath, M.K. DTP-Net: A convolutional neural network model to predict threshold for localizing the lesions on dermatological macro-images. Comput. Biol. Med. 2022, 148, 105852. [Google Scholar] [CrossRef]
- Han, Q.; Wang, H.; Hou, M.; Weng, T.; Pei, Y.; Li, Z.; Chen, G.; Tian, Y.; Qiu, Z. HWA-SegNet: Multi-channel skin lesion image segmentation network with hierarchical analysis and weight adjustment. Comput. Biol. Med. 2023, 152, 106343. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhong, L.; Qiu, C.; Zhang, Z.; Zhang, X. Transformer-based multilevel region and edge aggregation network for magnetic resonance image segmentation. Comput. Biol. Med. 2023, 152, 106427. [Google Scholar] [CrossRef]
- Uslu, F.; Bharath, A.A. TMS-Net: A segmentation network coupled with a run-time quality control method for robust cardiac image segmentation. Comput. Biol. Med. 2023, 152, 106422. [Google Scholar] [CrossRef] [PubMed]
- Borjigin, S.; Sahoo, P.K. Color image segmentation based on multi-level Tsallis–Havrda–Charvát entropy and 2D histogram using PSO algorithms. Pattern Recognit. 2019, 92, 107–118. [Google Scholar] [CrossRef]
- Fan, P.; Lang, G.; Yan, B.; Lei, X.; Guo, P.; Liu, Z.; Yang, F. A method of segmenting apples based on gray-centered RGB color space. Remote Sens. 2021, 13, 1211. [Google Scholar] [CrossRef]
- Naik, M.K.; Panda, R.; Abraham, A. An entropy minimization based multilevel colour thresholding technique for analysis of breast thermograms using equilibrium slime mould algorithm. Appl. Soft Comput. 2021, 113, 107955. [Google Scholar] [CrossRef]
- Ito, Y.; Premachandra, C.; Sumathipala, S.; Premachandra, H.W.H.; Sudantha, B.S. Tactile paving detection by dynamic thresholding based on HSV space analysis for developing a walking support system. IEEE Access 2021, 9, 20358–20367. [Google Scholar] [CrossRef]
- Rahimi, W.N.S.; Ali, M.S.A.M. Ananas comosus crown image thresholding and crop counting using a colour space transformation scheme. Telkomnika 2020, 18, 2472–2479. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image segmentation using deep learning: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 3523–3542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.D.; Do, T.; Ngo, T.D.; Le, D.D. An evaluation of deep learning methods for small object detection. J. Electr. Comput. Eng. 2020, 2020. [Google Scholar] [CrossRef]
- OpenCV. Available online: https://docs.opencv.org/4.x/df/d9d/tutorial_py_colorspaces.html (accessed on 20 June 2022).
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells, W.M., III; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Manglem, K.; Chanu, Y.J. Image segmentation using K-means clustering algorithm and subtractive clustering algorithm. Procedia Comput. Sci. 2015, 54, 764–771. [Google Scholar] [CrossRef]
- University of Waterloo. Vision and Image Processing Lab. Skin Cancer Detection. 2022. Available online: https://uwaterloo.ca/vision-image-processing-lab/research-demos/skin-cancer-detection (accessed on 27 November 2022).
- Shutterstock. 2022. Available online: https://www.shutterstock.com (accessed on 20 October 2022).
- Giotis, I.; Molders, N.; Land, S.; Biehl, M.; Jonkman, M.F.; Petkov, N. MED-NODE: A computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst. Appl. 2015, 42, 6578–6585. [Google Scholar] [CrossRef]
- Yang, J.; Wu, X.; Liang, J.; Sun, X.; Cheng, M.M.; Rosin, P.L.; Wang, L. Self-paced balance learning for clinical skin disease recognition. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 2832–2846. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Guo, Z.; Li, D.; Hu, X.; Zhang, Y. Automatic classification of clinical skin disease images with additional high-level position information. In Proceedings of the 2019 IEEE Chinese Control Conference (CCC), Guangzhou, China, 27–30 July 2019; pp. 8606–8610. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; Volume 1, pp. 770–778. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Girden, E.R. ANOVA: Repeated Measures; Sage University Paper Series on Quantitative Applications in the Social Sciences; Sage: Thousand Oaks, CA, USA, 1992; Volume 7. [Google Scholar]
- Hochberg, Y.; Tamhane, A.C. Multiple Comparison Procedures; John Wiley & Sons, Inc.: New York, NY, USA, 1987. [Google Scholar]
- Zhao, W.; Gurudu, S.R.; Taheri, S.; Ghosh, S.; Mallaiyan Sathiaseelan, M.A.; Asadizanjani, N. PCB Component Detection Using Computer Vision for Hardware Assurance. Big Data Cogn. Comput. 2022, 6, 39. [Google Scholar] [CrossRef]
- Ghosh, S.; Basak, A.; Bhunia, S. How secure are printed circuit boards against trojan attacks? IEEE Des. Test 2014, 32, 7–16. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, S.; Lian, X.; Su, X.; Chen, E. A dummy-based user privacy protection approach for text information retrieval. Knowl.-Based Syst. 2020, 195, 105679. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, S.; Li, H.; Zhou, H.; Lu, C. A basic framework for privacy protection in personalized information retrieval: An effective framework for user privacy protection. J. Organ. End User Comput. (JOEUC) 2021, 33, 1–26. [Google Scholar] [CrossRef]
- Mustafa, W.A.; Khairunizam, W.; Zunaidi, I.; Razlan, Z.M.; Shahriman, A.B. A comprehensive review on document image (DIBCO) database. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2019; Volume 557, p. 012006. [Google Scholar]
- Zhang, S.; Zhang, C. Modified U-Net for plant diseased leaf image segmentation. Comput. Electron. Agric. 2023, 204, 107511. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q.; Wang, Y. Road extraction by deep residual U-Net. IEEE Geosci. Remote Sens. Lett. 2018, 15, 749–753. [Google Scholar] [CrossRef]







| Method | DSI | MCC | PSNR |
|---|---|---|---|
| Otsu [11] | |||
| Kapur et al. [12] | |||
| Niblack [23] | |||
| p-tile [27] | |||
| Two-peak [27] | |||
| Local contrast [27] | |||
| Sauvola et al. [28] | |||
| Wolf and Jolion [36] | |||
| Feng and Tan [37] | |||
| Bradley and Roth [5] | |||
| Singh et al. [38] | |||
| DTP-NET pre-trained model [46] | |||
| DTP-NET training from scratch [46] | |||
| DTP-NET fine-tuned model [46] | |||
| U-Net (Resnet-152) pre-trained [44] | |||
| U-Net (Resnet-152) fine-tuned [44] | |||
| Proposed method |
| Method | DSI | MCC | PSNR |
|---|---|---|---|
| Otsu [11] | |||
| Kapur et al. [12] | |||
| Niblack [23] | |||
| p-tile [27] | |||
| Two-peak [27] | |||
| Local contrast [27] | |||
| Sauvola et al. [28] | |||
| Wolf and Jolion [36] | |||
| Feng and Tan [37] | |||
| Bradley and Roth [5] | |||
| Singh et al. [38] | |||
| DTP-NET pre-trained [46] | |||
| DTP-NET training from scratch [46] | |||
| DTP-NET fine-tuned [46] | |||
| U-Net (Resnet-152) pre-trained [44] | |||
| U-Net (Resnet-152) training f. s. [44] | |||
| U-Net (Resnet-152) fine-tuned [44] | |||
| Proposed method |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pootheri, S.; Ellam, D.; Grübl, T.; Liu, Y. A Two-Stage Automatic Color Thresholding Technique. Sensors 2023, 23, 3361. https://doi.org/10.3390/s23063361
Pootheri S, Ellam D, Grübl T, Liu Y. A Two-Stage Automatic Color Thresholding Technique. Sensors. 2023; 23(6):3361. https://doi.org/10.3390/s23063361
Chicago/Turabian StylePootheri, Shamna, Daniel Ellam, Thomas Grübl, and Yang Liu. 2023. "A Two-Stage Automatic Color Thresholding Technique" Sensors 23, no. 6: 3361. https://doi.org/10.3390/s23063361
APA StylePootheri, S., Ellam, D., Grübl, T., & Liu, Y. (2023). A Two-Stage Automatic Color Thresholding Technique. Sensors, 23(6), 3361. https://doi.org/10.3390/s23063361






