Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI
Abstract
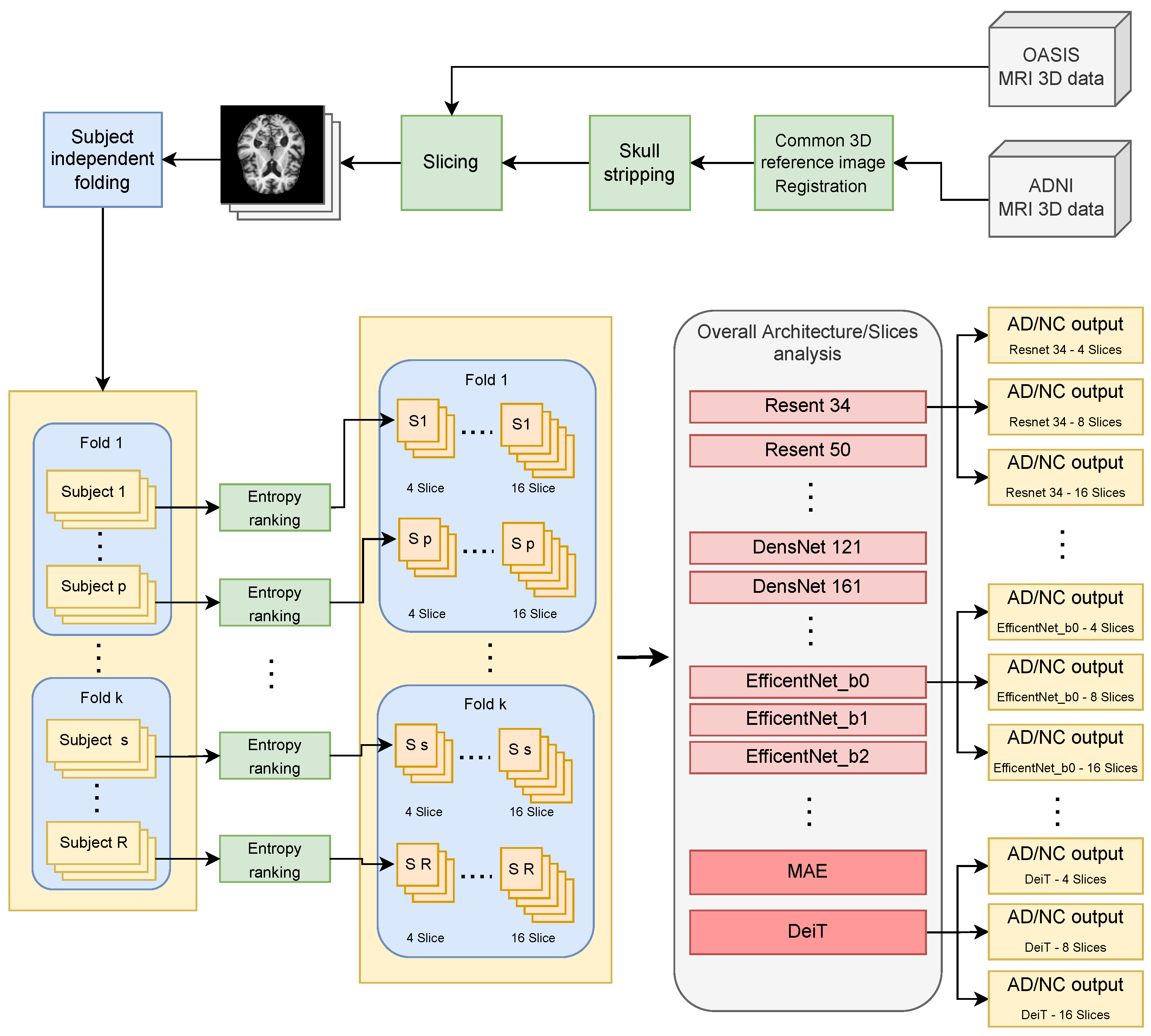
:1. Introduction
- 1.
- Most existing approaches for binary classification (dementia vs. normal) were tested on 2D MRI slices randomly sampled from the available 3D data without considering to which subject they belonged (slice-level data split strategy) [18,19]. This means that slices belonging to the same subject can occur in both the training and testing processes; in this way, the test data can have a distribution more similar to that of the training set than would be expected from new data belonging to new subjects. This is the well-known data leakage problem in machine learning [20] that has called into question the validity of many previous MRI-based CAD studies and made their use in actual clinical screenings uncertain [21]. The few studies that perform classification of neurologic diseases using MRI and with no data leakage are listed and discussed in [11,22] where it emerges that automatic classification ability is still unsatisfactory to make MRI-based CAD useful in clinical practice.
- 2.
- How many 2D slices should be extracted from the available 3D MRI volumes is an open question. Increasing the number of slices per subject may add a little discriminatory information hidden in a larger amount of useless data. The only benchmarks found in the literature are those where the number of slices per subject was fixed a priori (usually 8). The ability of the classifiers to handle this has therefore not been studied at all.
- 3.
- Deep-learning models and, in particular, convolutional neural networks (CNN) have revolutionized computer vision, but the most powerful recent CNN models have not yet been explored for AD diagnosis from MRI data. This may be due to the complexity of models’ implementation, data preparation, and validation techniques used in the machine learning community [23].
- 4.
- Recent findings in machine learning beyond Convolutional Neural Networks have also not been tested. It has been shown that the mechanism of self-attention can be a viable alternative for building image recognition models [24]. It can be used to direct attention to key areas in the image to obtain high-level information, but as far as we know, this research direction has been less explored in CAD. Recently, there have been some groundbreaking experiments with 3D brain data, but there is no work addressing AD-related issues [25].
2. Materials and Methods
2.1. Convolutional Neural Networks
2.1.1. ResNet
2.1.2. DenseNet
2.1.3. EfficientNet
2.2. Visual Transformers
2.2.1. MAE
2.2.2. DeiT
3. Results
3.1. Results by CNN
3.2. Results by Visual Transformers
3.3. Comparisons to Leading Approaches
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’ Dementia |
| DL | Deep Learning |
| NC | normal controls |
| MRI | Magnetic resonance imaging |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| OASIS | Open Access Series of Imaging Studies |
| CAD | Computer-aided diagnosis |
| MAE | Masked AutoEncoders |
| MCI | Mild Cognitive Impairment |
| MRI | Magnetic resonance imaging |
| CNN | Convolutional Neural Networks |
| DeiT | Data-efficient image Transformers |
| ViT | Visual Transformers |
References
- World Health Organization. Global Action Plan on the Public Health Response to Dementia 2017–2025; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- US National Institute on Aging: What Happens to the Brain in Alzheimer’s Disease. Available online: https://www.nia.nih.gov/health/what-happens-brain-alzheimers-disease (accessed on 27 September 2022).
- Inglese, M.; Patel, N.; Linton-Reid, K.; Loreto, F.; Win, Z.; Perry, R.J.; Carswell, C.; Grech-Sollars, M.; Crum, W.R.; Lu, H.; et al. A predictive model using the mesoscopic architecture of the living brain to detect Alzheimer’s disease. Commun. Med. 2022, 2, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef]
- Long, X.; Chen, L.; Jiang, C.; Zhang, L.; Initiative, A.D.N. Prediction and classification of Alzheimer disease based on quantification of MRI deformation. PLoS ONE 2017, 12, e0173372. [Google Scholar] [CrossRef]
- Tang, Z.; Chuang, K.V.; DeCarli, C.; Jin, L.W.; Beckett, L.; Keiser, M.J.; Dugger, B.N. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ouchicha, C.; Ammor, O.; Meknassi, M. A Novel Deep Convolutional Neural Network Model for Alzheimer’s Disease Classification Using Brain MRI. Autom. Control Comput. Sci. 2022, 56, 261–271. [Google Scholar] [CrossRef]
- Qiu, S.; Miller, M.I.; Joshi, P.S.; Lee, J.C.; Xue, C.; Ni, Y.; Wang, Y.; Anda-Duran, D.; Hwang, P.H.; Cramer, J.A.; et al. Multimodal deep learning for Alzheimer’s disease dementia assessment. Nat. Commun. 2022, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.; Nichols, K.; Wang, H.; Shen, L.; Huang, H. Joint multi-modal longitudinal regression and classification for alzheimer’s disease prediction. IEEE Trans. Med. Imaging 2019, 39, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qiao, H.; Zhu, F. Alzheimer’s Disease Diagnosis With Brain Structural MRI Using Multiview-Slice Attention and 3D Convolution Neural Network. Front. Aging Neurosci. 2022, 14, 871706. [Google Scholar] [CrossRef]
- Wen, J.; Thibeau-Sutre, E.; Diaz-Melo, M.; Samper-González, J.; Routier, A.; Bottani, S.; Dormont, D.; Durrleman, S.; Burgos, N.; Colliot, O.; et al. Convolutional neural networks for classification of Alzheimer’s disease: Overview and reproducible evaluation. Med. Image Anal. 2020, 63, 101694. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Habes, M.; Iftikhar, M.A.; Shacklett, A.; Davatzikos, C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer’s disease and its prodromal stages. NeuroImage 2017, 155, 530–548. [Google Scholar] [CrossRef]
- Altinkaya, E.; Polat, K.; Barakli, B. Detection of Alzheimer’s disease and dementia states based on deep learning from MRI images: A comprehensive review. J. Inst. Electron. Comput. 2020, 1, 39–53. [Google Scholar]
- Yamanakkanavar, N.; Choi, J.Y.; Lee, B. MRI segmentation and classification of human brain using deep learning for diagnosis of Alzheimer’s disease: A survey. Sensors 2020, 20, 3243. [Google Scholar] [CrossRef]
- Ebrahimighahnavieh, M.A.; Luo, S.; Chiong, R. Deep learning to detect Alzheimer’s disease from neuroimaging: A systematic literature review. Comput. Methods Programs Biomed. 2020, 187, 105242. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Masurkar, A.V.; Rusinek, H.; Chen, J.; Zhang, B.; Zhu, W.; Fernandez-Granda, C.; Razavian, N. Generalizable deep learning model for early Alzheimer’s disease detection from structural MRIs. Sci. Rep. 2022, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D deep learning on medical images: A review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef] [PubMed]
- Orouskhani, M.; Rostamian, S.; Zadeh, F.S.; Shafiei, M.; Orouskhani, Y. Alzheimer’s Disease Detection from Structural MRI Using Conditional Deep Triplet Network. Neurosci. Inform. 2022, 2, 100066. [Google Scholar] [CrossRef]
- AlSaeed, D.; Omar, S.F. Brain MRI Analysis for Alzheimer’s Disease Diagnosis Using CNN-Based Feature Extraction and Machine Learning. Sensors 2022, 22, 2911. [Google Scholar] [CrossRef]
- Kaufman, S.; Rosset, S.; Perlich, C.; Stitelman, O. Leakage in data mining: Formulation, detection, and avoidance. ACM Trans. Knowl. Discov. Data (TKDD) 2012, 6, 1–21. [Google Scholar] [CrossRef]
- Yagis, E.; De Herrera, A.G.S.; Citi, L. Generalization performance of deep learning models in neurodegenerative disease classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1692–1698. [Google Scholar]
- Yagis, E.; Atnafu, S.W.; García Seco de Herrera, A.; Marzi, C.; Scheda, R.; Giannelli, M.; Tessa, C.; Citi, L.; Diciotti, S. Effect of data leakage in brain MRI classification using 2D convolutional neural networks. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Thibeau-Sutre, E.; Diaz, M.; Hassanaly, R.; Routier, A.; Dormont, D.; Colliot, O.; Burgos, N. ClinicaDL: An open-source deep learning software for reproducible neuroimaging processing. Comput. Methods Programs Biomed. 2022, 220, 106818. [Google Scholar] [CrossRef]
- Zhao, H.; Jia, J.; Koltun, V. Exploring Self-Attention for Image Recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Jiang, M.; Yan, B.; Li, Y.; Zhang, J.; Li, T.; Ke, W. Image Classification of Alzheimer’s Disease Based on External-Attention Mechanism and Fully Convolutional Network. Brain Sci. 2022, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Wang, Y.; Hou, F.; Yuan, J.; Tian, J.; Zhang, Y.; Shi, Z.; Fan, J.; He, Z. A survey of visual transformers. arXiv 2021, arXiv:2111.06091. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Suganyadevi, S.; Seethalakshmi, V.; Balasamy, K. A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retr. 2022, 11, 19–38. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Touvron, H.; Cord, M.; Jégou, H. DeiT III: Revenge of the ViT. In Proceedings of the Computer Vision–ECCV 2022, Tel Aviv, Israel, 23–27 October 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 516–533. [Google Scholar]
- Yang, C.; Wang, Y.; Zhang, J.; Zhang, H.; Wei, Z.; Lin, Z.; Yuille, A. Lite vision transformer with enhanced self-attention. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11998–12008. [Google Scholar]
- Wang, Z.; Li, Y.; Childress, A.R.; Detre, J.A. Brain entropy mapping using fMRI. PLoS ONE 2014, 9, e89948. [Google Scholar] [CrossRef]
- Wang, Z.; Initiative, A.D.N. Brain entropy mapping in healthy aging and Alzheimer’s disease. Front. Aging Neurosci. 2020, 12, 596122. [Google Scholar] [CrossRef]
- Manera, A.L.; Dadar, M.; Fonov, V.; Collins, D.L. CerebrA, registration and manual label correction of Mindboggle-101 atlas for MNI-ICBM152 template. Sci. Data 2020, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Aisen, P.S.; Petersen, R.C.; Donohue, M.; Weiner, M.W.; Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s disease neuroimaging initiative 2 clinical core: Progress and plans. Alzheimer’s Dement. 2015, 11, 734–739. [Google Scholar] [CrossRef]
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. Fsl. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Kumar, S.S.; Nandhini, M. Entropy slicing extraction and transfer learning classification for early diagnosis of Alzheimer diseases with sMRI. ACM Trans. Multimed. Comput. Commun. Appl. (TOMM) 2021, 17, 1–22. [Google Scholar] [CrossRef]
- Hon, M.; Khan, N.M. Towards Alzheimer’s disease classification through transfer learning. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 1166–1169. [Google Scholar]
- Li, Y.; Hao, Z.; Lei, H. Survey of convolutional neural network. J. Comput. Appl. 2016, 36, 2508. [Google Scholar]
- Leo, M.; Farinella, G.M. Computer Vision for Assistive Healthcare; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Sarvamangala, D.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems 2019, Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Atnafu, S.W.; Diciotti, S. Development of an interpretable deep learning system for the identification of patients with Alzheimer’s disease. Res. Sq. 2022. [Google Scholar] [CrossRef]



| Dataset | Classes | Subjects | Gender | Age (in Years) |
|---|---|---|---|---|
| (Women/Men) | (Range; Mean ± SD) | |||
| ADNI-2 | AD | 100 | 44/56 | 56–89; |
| NC | 100 | 52/48 | 58–95; | |
| OASIS-1 | AD | 100 | 44/56 | 62–96; |
| NC | 100 | 52/48 | 59–94; |
| Method | Cross-Validation Accuracy (%) | |||||
|---|---|---|---|---|---|---|
| ADNI-2 | OASIS-1 | |||||
| 4 Slices | 8 Slices | 16 Slices | 4 Slices | 8 Slices | 16 Slices | |
| ResNet34 | 65.500 | 66.874 | 67.470 | 68.126 | 67.190 | 67.409 |
| ResNet50 | 65.752 | 66.374 | 65.624 | 68.502 | 68.000 | 68.653 |
| ResNet101 | 68.751 | 69.500 | 69.531 | 70.626 | 69.875 | 67.842 |
| ResNet152 | 67.876 | 65.937 | 65.594 | 72.127 | 71.124 | 69.000 |
| DenseNet121 | 66.378 | 65.438 | 66.219 | 70.252 | 70.312 | 67.654 |
| DenseNet161 | 69.375 | 70.190 | 69.253 | 71.001 | 68.190 | 68.470 |
| DenseNet169 | 66.873 | 66.564 | 65.468 | 73.501 | 70.814 | 67.780 |
| DenseNet201 | 69.751 | 69.626 | 68.626 | 69.754 | 70.190 | 66.690 |
| EfficientNet_b0 | 69.002 | 69.502 | 69.534 | 66.002 | 66.502 | 67.341 |
| EfficientNet_b1 | 66.001 | 66.066 | 66.218 | 69.502 | 66.314 | 66.470 |
| EfficientNet_b2 | 66.124 | 65.687 | 65.594 | 66.752 | 65.435 | 64.562 |
| EfficientNet_b3 | 64.626 | 63.626 | 63.408 | 66.878 | 64.690 | 64.126 |
| EfficientNet_b4 | 61.376 | 60.502 | 61.905 | 62.002 | 66.938 | 62.624 |
| EfficientNet_b5 | 60.249 | 58.500 | 59.624 | 66.004 | 65.814 | 65.034 |
| EfficientNet_b6 | 65.877 | 63.626 | 63.970 | 63.252 | 63.624 | 62.656 |
| EfficientNet_b7 | 63.243 | 61.874 | 62.534 | 68.004 | 65.124 | 63.470 |
| Method | Cross-Validation Accuracy (%) | |||||
|---|---|---|---|---|---|---|
| ADNI-2 | OASIS-1 | |||||
| 4 Slices | 8 Slices | 16 Slices | 4 Slices | 8 Slices | 16 Slices | |
| MAE | 73.555 | 73.125 | 71.875 | 72.375 | 71.875 | 70.937 |
| DeiT | 77.000 | 75.937 | 75.625 | 74.375 | 74.562 | 72.562 |
| Method | Cross-Validation Accuracy (%) | ||
|---|---|---|---|
| ADNI-2 | OASIS-1 | ||
| Previous | VGG16-v1 (8 slices) [22] | 70.1 | 66 |
| VGG16-v2 (8 slices) [22] | 66.4 | 66.1 | |
| ResNet-18 (8 slices) [22] | 68.6 | 68.8 | |
| VGG16 (10 slices) [50] | 71.6 | ||
| Proposed | ResNet152 (4 slices) | 67.2 | 72 |
| DenseNet201 (4 slices) | 69.8 | 69.8 | |
| DenseNet161 (8 slices) | 70.2 | 68.2 | |
| MAE (4 slices) | 73.6 | 72.4 | |
| DeiT (4 slices) | 77 | 74.4 | |
| DeiT (8 slices) | 76 | 75.6 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carcagnì, P.; Leo, M.; Del Coco, M.; Distante, C.; De Salve, A. Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI. Sensors 2023, 23, 1694. https://doi.org/10.3390/s23031694
Carcagnì P, Leo M, Del Coco M, Distante C, De Salve A. Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI. Sensors. 2023; 23(3):1694. https://doi.org/10.3390/s23031694
Chicago/Turabian StyleCarcagnì, Pierluigi, Marco Leo, Marco Del Coco, Cosimo Distante, and Andrea De Salve. 2023. "Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI" Sensors 23, no. 3: 1694. https://doi.org/10.3390/s23031694
APA StyleCarcagnì, P., Leo, M., Del Coco, M., Distante, C., & De Salve, A. (2023). Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI. Sensors, 23(3), 1694. https://doi.org/10.3390/s23031694









