Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE)
Abstract
:1. Introduction
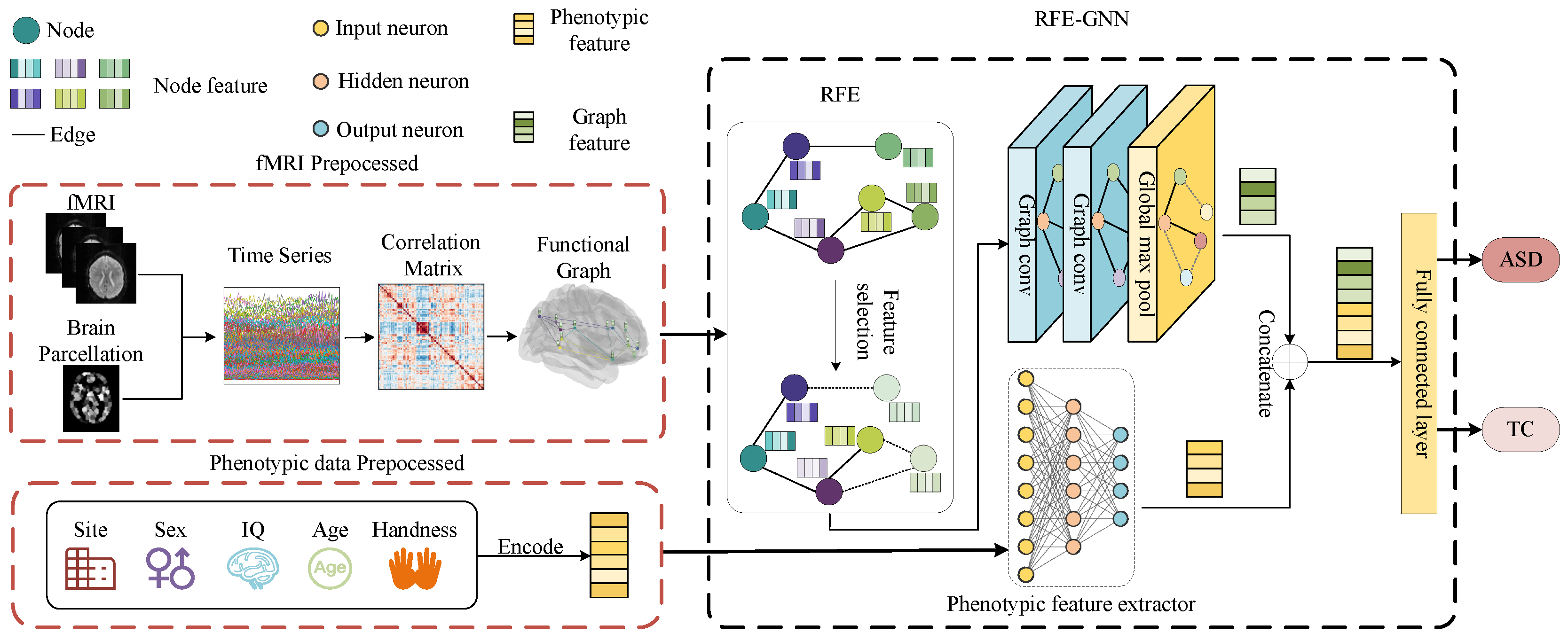
- There is a limited sample size of medical datasets and a substantial number of feature dimensions in graph data. To overcome this challenge, we leverage RFE to procure a subset of original graph features, which excludes features exhibiting lower classification scores.
- To combat the issue of overfitting in medical datasets, we incorporate phenotypic data for ASD detection in our research. We devise a novel feature extraction module, termed PFE, which is capable of extracting pertinent features from phenotypic data by continually adjusting its parameters during neural network training, thereby facilitating feature selection. By effectively capturing the underlying representations from phenotypic data, PFE augments the detection of ASD.
- Leveraging the aforementioned modules, we present a pioneering framework for detecting ASD utilizing multimodal data. Comprehensive experiments conducted on two medical datasets and comparisons with state-of-the-art techniques substantiate the efficacy of the proposed framework. By incorporating RFE and PFE, we surmount the challenge of the high-dimensionality and limited sample size of the datasets while mitigating overfitting. Our framework is endowed with exceptional processing capabilities for multimodal data and can seamlessly integrate diverse modalities to distinguish ASD patients. This has profound implications for doctors to employ computer-aided diagnosis techniques in the prevention and scientific treatment of ASD.
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
2.2. Methods
The Framework of RFE–GNN
3. Experiments and Results
3.1. Training Setup
3.2. Statistical Metrics
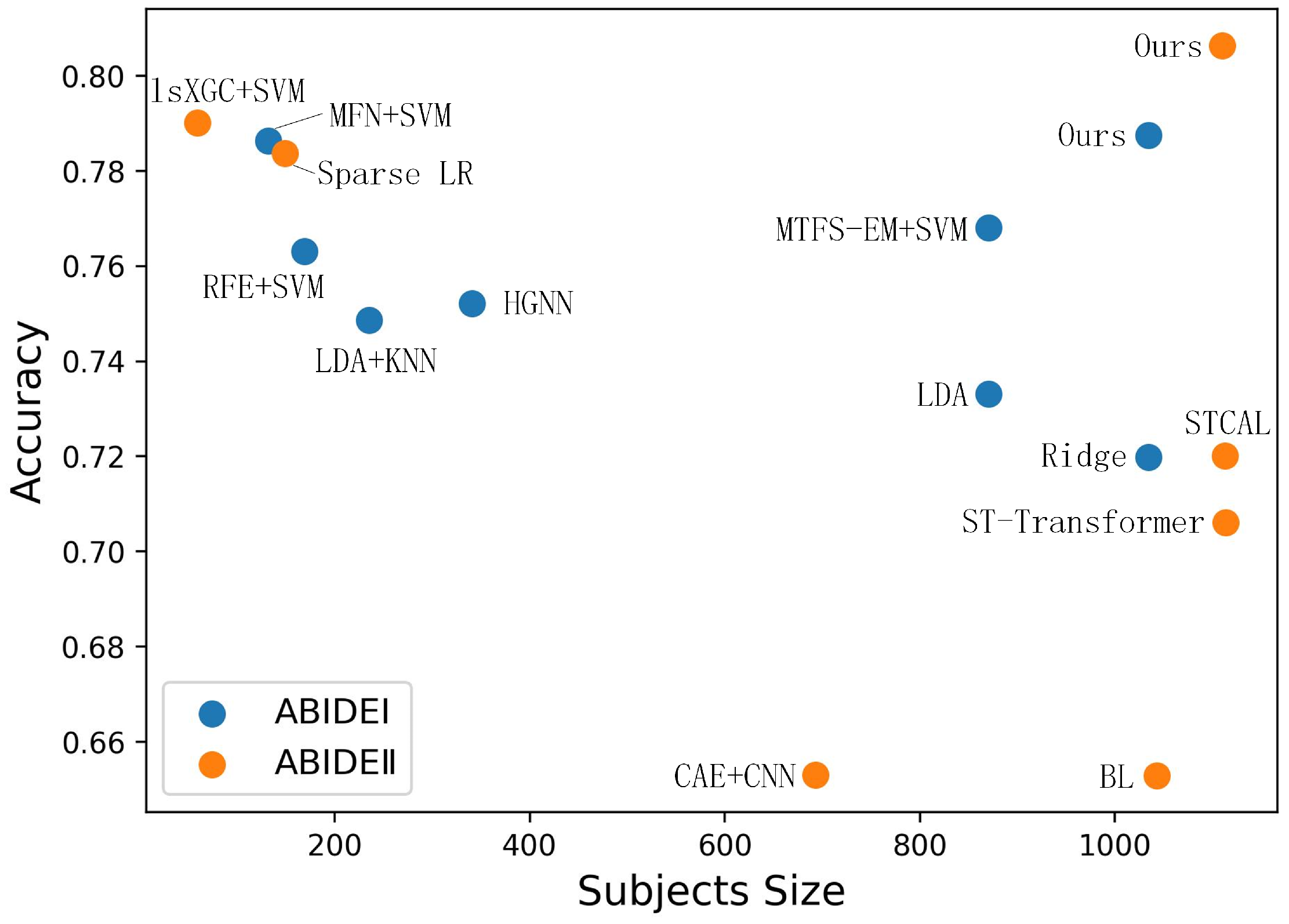
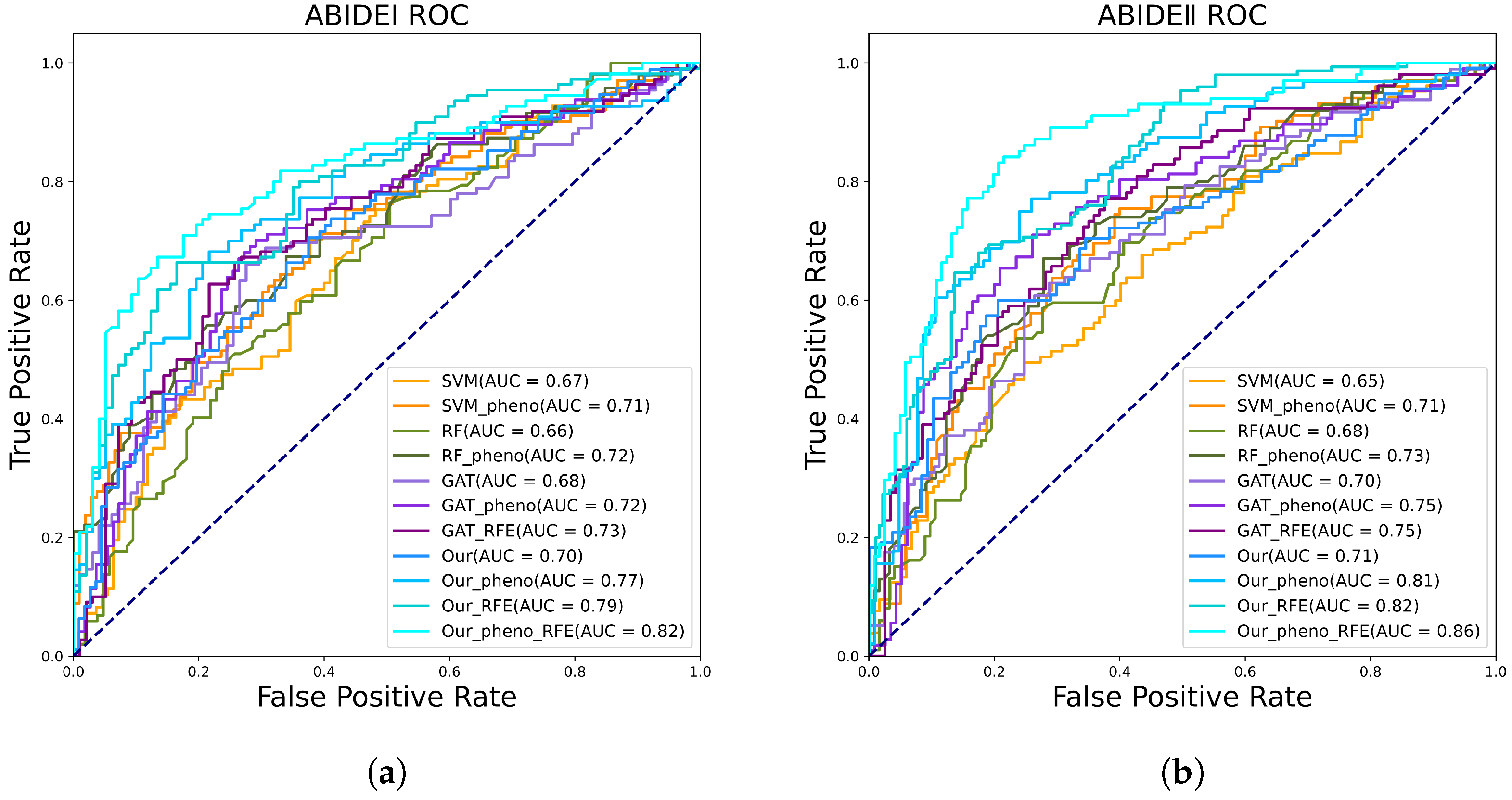
3.3. Comparison with State-of-the-Art (SOTA) Models
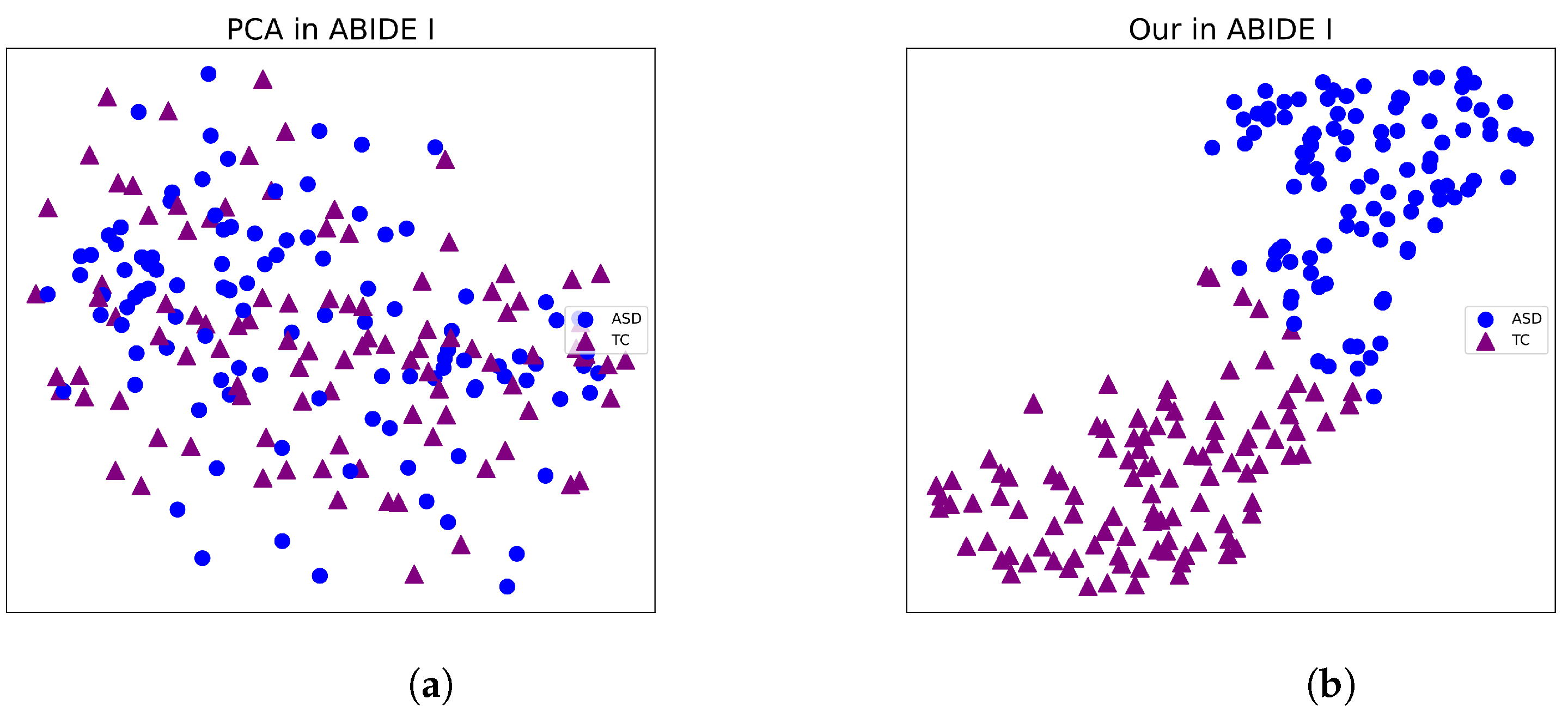
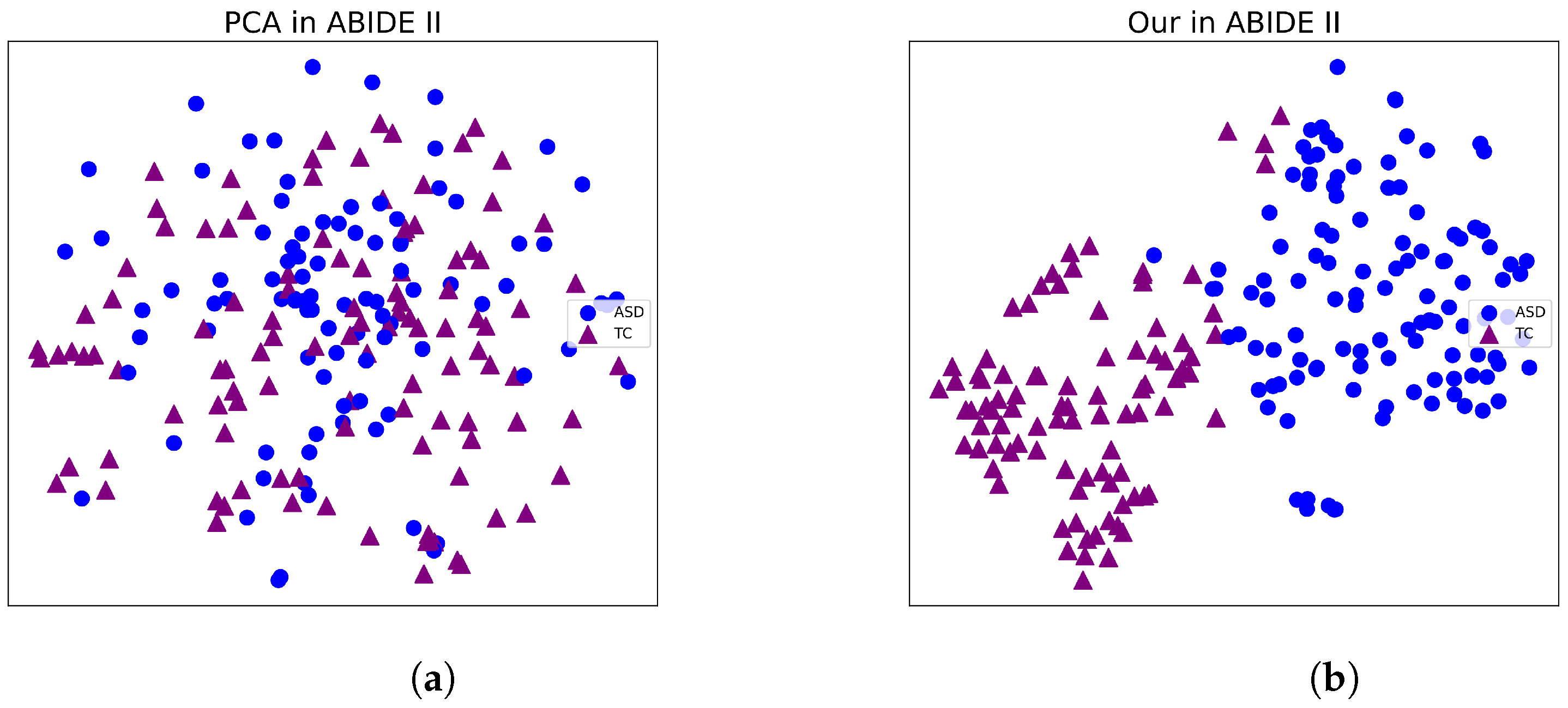
3.4. Visualization the Classification
3.5. Comparison with Different Models
- SVM: SVM finds a hyperplane in the feature space of input data such that the distance between each sample and the hyperplane is maximized, achieving the classification effect of the original data. In the subsequent experiments, we use the SVC model from the sklearn Python library. Parameters are set as regularization parameter = 1.0 and kernel = ‘linear’, and the remaining parameters are set to default.
- RF [44]: Random Forest is an ensemble algorithm that builds multiple decision trees on data and combines the results. The RF model is also found in the sklearn library; the parameter of n_estimators = 1600, and the other is set to default.
- GAT [45]: GAT is a graph convolutional layer based on GCN. It uses a self-attention mechanism to aggregate neighbor nodes and adaptively match the weights of different neighbors. The GATConv we use is in the torch_geometric library, and the layer parameter is set to default; in the GAT model, there are two layers.
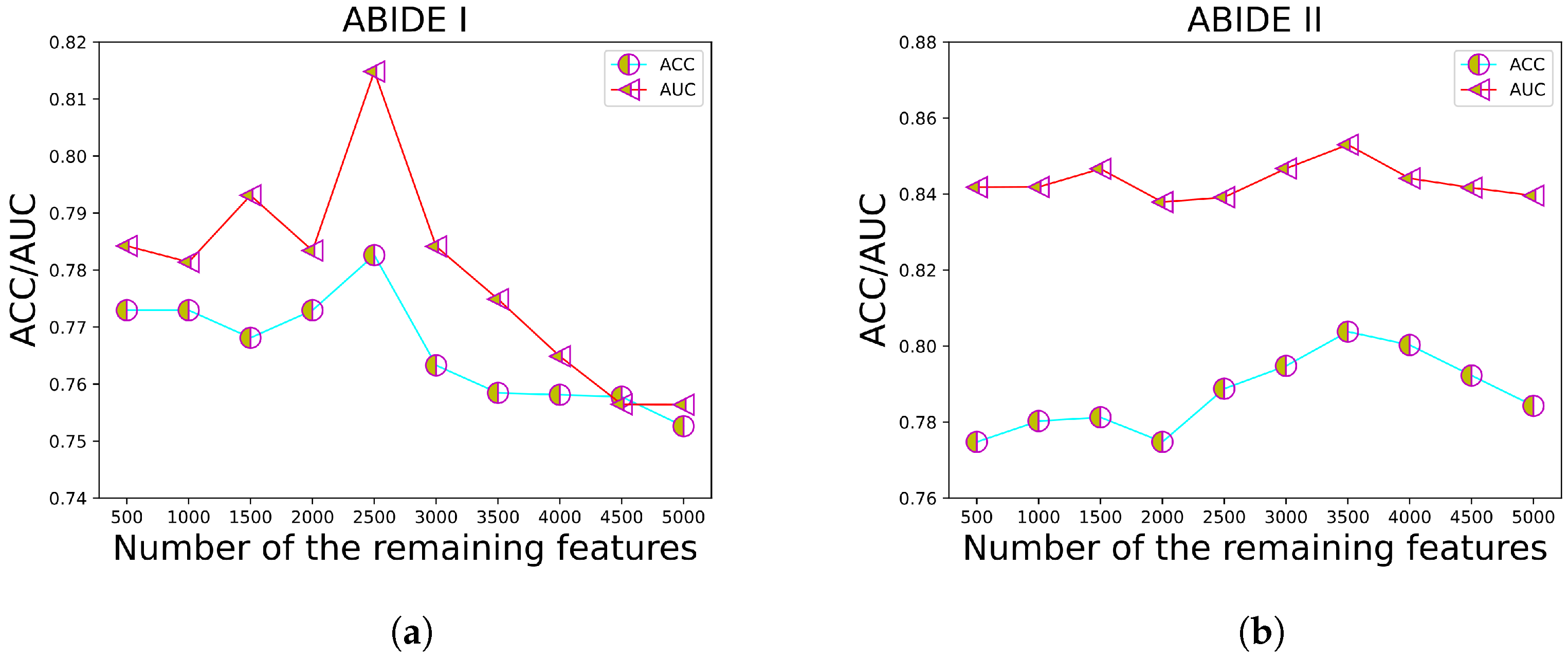
3.6. Hyperparameter Discussion
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum Disorder |
| RFE–GNN | Recursive Feature Elimination-Graph Neural Network |
| PFE | Phenotypic Feature Extractor |
| fMRI | functional Magnetic Resonance Imaging |
| ABIDE | Autism Brain Imaging Data Exchange |
| CAD | Computer-aided Diagnosis |
| ADHD | Attention Deficit Hyperactivity Disorder |
| LR | Logistic Regression |
| SVM | Support Vector Machine |
| LDA | Linear Discriminant Analysis |
| KNN | K-nearest neighbors |
| TC | Typical Control |
| GNNs | Graph Neural Networks |
| PSCR | Pearson Correlation-based Spatially Constrained Representation |
| CPAC | Configurable Pipeline for the Analysis of Connectomes |
| ROIs | Regions of Interest |
| FIQ | Full Intelligence Quotient |
| VIQ | Verbal Intelligence Quotient |
| PIQ | Performance Intelligence Quotient |
| MLP | Multilayer Perceptron |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under Curve |
| PCA | Principal Components Analysis |
| SOTA | state-of-the-art |
References
- Williams, J.B.; First, M. Diagnostic and statistical manual of mental disorders. In Encyclopedia of Social Work; National Association of Social Workers: Washington, DC, USA, 2013. [Google Scholar]
- Cao, X.; Cao, J. Commentary: Machine learning for autism spectrum disorder diagnosis—Challenges and opportunities—A commentary on Schulte-Rüther et al. (2022). J. Child Psychol. Psychiatry 2023, 64, 966–967. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, S.; Tang, L.; Wu, F.X. Diagnosis of autism spectrum disorder based on eigenvalues of brain networks. IEEE Access 2019, 7, 128474–128486. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Shao, L.; You, Y.; Du, H.; Fu, D. Classification of ADHD with fMRI data and multi-objective optimization. Comput. Methods Programs Biomed. 2020, 196, 105676. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yao, D.; Ma, L.; Liu, M. Multi-site clustering and nested feature extraction for identifying autism spectrum disorder with resting-state fMRI. Med. Image Anal. 2022, 75, 102279. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cao, L.; Li, T.; Liao, B.; Dong, S.; Li, P. Interpretable learning approaches in resting-state functional connectivity analysis: The case of autism spectrum disorder. Comput. Math. Methods Med. 2020, 2020, 1394830. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, X.; Sun, B.; Feng, K.; Kou, L.; Chen, Y.; Li, Y.; Chen, H.; Tian, E.; Ni, Q.; et al. Online Knowledge Distillation Based Multiscale Threshold Denoising Networks for Fault Diagnosis of Transmission Systems. IEEE Trans. Transp. Electrif. 2023. [Google Scholar] [CrossRef]
- Xu, Y.; Feng, K.; Yan, X.; Yan, R.; Ni, Q.; Sun, B.; Lei, Z.; Zhang, Y.; Liu, Z. CFCNN: A novel convolutional fusion framework for collaborative fault identification of rotating machinery. Inf. Fusion 2023, 95, 1–16. [Google Scholar] [CrossRef]
- Sherkatghanad, Z.; Akhondzadeh, M.; Salari, S.; Zomorodi-Moghadam, M.; Abdar, M.; Acharya, U.R.; Khosrowabadi, R.; Salari, V. Automated detection of autism spectrum disorder using a convolutional neural network. Front. Neurosci. 2020, 13, 1325. [Google Scholar] [CrossRef]
- Rathore, A.; Palande, S.; Anderson, J.S.; Zielinski, B.A.; Fletcher, P.T.; Wang, B. Autism classification using topological features and deep learning: A cautionary tale. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part III 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 736–744. [Google Scholar]
- Craddock, C.; Benhajali, Y.; Chu, C.; Chouinard, F.; Evans, A.; Jakab, A.; Khundrakpam, B.S.; Lewis, J.D.; Li, Q.; Milham, M.; et al. The neuro bureau preprocessing initiative: Open sharing of preprocessed neuroimaging data and derivatives. Front. Neuroinform. 2013, 7, 27. [Google Scholar]
- Li, X.; Dvornek, N.C.; Zhuang, J.; Ventola, P.; Duncan, J. Graph embedding using infomax for ASD classification and brain functional difference detection. In Proceedings of the Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging, Houston, TX, USA, 15–20 February 2020; SPIE: Bellingham, WA, USA, 2020; Volume 11317, p. 1131702. [Google Scholar]
- Yang, C.; Wang, P.; Tan, J.; Liu, Q.; Li, X. Autism spectrum disorder diagnosis using graph attention network based on spatial-constrained sparse functional brain networks. Comput. Biol. Med. 2021, 139, 104963. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cao, L.; Li, T.; Dong, S.; Li, P. GAT-LI: A graph attention network based learning and interpreting method for functional brain network classification. BMC Bioinform. 2021, 22, 379. [Google Scholar] [CrossRef] [PubMed]
- Anirudh, R.; Thiagarajan, J.J. Bootstrapping graph convolutional neural networks for autism spectrum disorder classification. In Proceedings of the 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP 2019), Brighton, UK, 12–17 May 2019; pp. 3197–3201. [Google Scholar]
- Ktena, S.I.; Parisot, S.; Ferrante, E.; Rajchl, M.; Lee, M.; Glocker, B.; Rueckert, D. Metric learning with spectral graph convolutions on brain connectivity networks. NeuroImage 2018, 169, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Mohan, P.; Paramasivam, I. Feature reduction using SVM-RFE technique to detect autism spectrum disorder. Evol. Intell. 2021, 14, 989–997. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, Z.; Wang, B.; Wu, J. Identification of autism based on SVM-RFE and stacked sparse auto-encoder. IEEE Access 2019, 7, 118030–118036. [Google Scholar] [CrossRef]
- Kumar, C.J.; Das, P.R. The diagnosis of ASD using multiple machine learning techniques. Int. J. Dev. Disabil. 2022, 68, 973–983. [Google Scholar] [CrossRef]
- Radhika, C.; Priya, N. Advance Hybrid RF-GBC-RFE Wrapper-Based Feature Selection Techniques for Prediction of Autistic Disorder. J. Algebr. Stat. 2022, 13, 503–510. [Google Scholar]
- Parisot, S.; Ktena, S.I.; Ferrante, E.; Lee, M.; Moreno, R.G.; Glocker, B.; Rueckert, D. Spectral graph convolutions for population-based disease prediction. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part III 20. Springer: Berlin/Heidelberg, Germany, 2017; pp. 177–185. [Google Scholar]
- Chen, H.; Zhuang, F.; Xiao, L.; Ma, L.; Liu, H.; Zhang, R.; Jiang, H.; He, Q. AMA-GCN: Adaptive multi-layer aggregation graph convolutional network for disease prediction. arXiv 2021, arXiv:2106.08732. [Google Scholar]
- Kazi, A.; Shekarforoush, S.; Arvind Krishna, S.; Burwinkel, H.; Vivar, G.; Kortüm, K.; Ahmadi, S.A.; Albarqouni, S.; Navab, N. InceptionGCN: Receptive field aware graph convolutional network for disease prediction. In Proceedings of the Information Processing in Medical Imaging: 26th International Conference (IPMI 2019), Hong Kong, China, 2–7 June 2019; Proceedings 26. Springer: Berlin/Heidelberg, Germany, 2019; pp. 73–85. [Google Scholar]
- Craddock, R.C.; James, G.A.; Holtzheimer, P.E., III; Hu, X.P.; Mayberg, H.S. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012, 33, 1914–1928. [Google Scholar] [CrossRef]
- Joseph, R.M.; Tager-Flusberg, H.; Lord, C. Cognitive profiles and social-communicative functioning in children with autism spectrum disorder. J. Child Psychol. Psychiatry 2002, 43, 807–821. [Google Scholar] [CrossRef]
- Yan, C.G.; Wang, X.D.; Zuo, X.N.; Zang, Y.F. DPABI: Data processing & analysis for (resting-state) brain imaging. Neuroinformatics 2016, 14, 339–351. [Google Scholar] [PubMed]
- Morris, C.; Ritzert, M.; Fey, M.; Hamilton, W.L.; Lenssen, J.E.; Rattan, G.; Grohe, M. Weisfeiler and leman go neural: Higher-order graph neural networks. In Proceedings of the AAAI conference on artificial intelligence, Honolulu, HI, USA, 27–28 January 2019; Volume 33, pp. 4602–4609. [Google Scholar]
- Li, Q.; Han, Z.; Wu, X.M. Deeper insights into graph convolutional networks for semi-supervised learning. In Proceedings of the AAAI conference on artificial intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Yang, X.; Islam, M.S.; Khaled, A.A. Functional connectivity magnetic resonance imaging classification of autism spectrum disorder using the multisite ABIDE dataset. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar]
- Bernas, A.; Aldenkamp, A.P.; Zinger, S. Wavelet coherence-based classifier: A resting-state functional MRI study on neurodynamics in adolescents with high-functioning autism. Comput. Methods Programs Biomed. 2018, 154, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Epalle, T.M.; Lu, H. Characterizing and predicting autism spectrum disorder by performing resting-state functional network community pattern analysis. Front. Hum. Neurosci. 2019, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Madine, M.; Rekik, I.; Werghi, N. Diagnosing autism using T1-W MRI with multi-kernel learning and hypergraph neural network. In Proceedings of the 2020 IEEE International Conference on Image Processing (ICIP), Virtual Conference, 25–28 October 2020; pp. 438–442. [Google Scholar]
- Jung, M.; Tu, Y.; Park, J.; Jorgenson, K.; Lang, C.; Song, W.; Kong, J. Surface-based shared and distinct resting functional connectivity in attention-deficit hyperactivity disorder and autism spectrum disorder. Br. J. Psychiatry 2019, 214, 339–344. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, Y.; Lan, W.; Guo, R.; Wang, Y.; Wang, J. Improved ASD classification using dynamic functional connectivity and multi-task feature selection. Pattern Recognit. Lett. 2020, 138, 82–87. [Google Scholar] [CrossRef]
- Zheng, W.; Eilam-Stock, T.; Wu, T.; Spagna, A.; Chen, C.; Hu, B.; Fan, J. Multi-feature based network revealing the structural abnormalities in autism spectrum disorder. IEEE Trans. Affect. Comput. 2019, 12, 732–742. [Google Scholar] [CrossRef]
- Liu, J.C.; Ji, J.Z. Classification method of fMRI data based on broad learning system. J. ZheJiang Univ. Eng. Sci. 2021, 55, 1270–1278. [Google Scholar]
- Zhao, Y.; Dai, H.; Zhang, W.; Ge, F.; Liu, T. Two-stage spatial temporal deep learning framework for functional brain network modeling. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1576–1580. [Google Scholar]
- Deng, X.; Zhang, J.; Liu, R.; Liu, K. Classifying ASD based on time-series fMRI using spatial—Temporal transformer. Comput. Biol. Med. 2022, 151, 106320. [Google Scholar] [CrossRef]
- Liu, R.; Huang, Z.A.; Hu, Y.; Zhu, Z.; Wong, K.C.; Tan, K.C. Spatial—Temporal Co-Attention Learning for Diagnosis of Mental Disorders From Resting-State fMRI Data. IEEE Trans. Neural Netw. Learn. Syst. 2023. [Google Scholar] [CrossRef]
- Zhan, Y.; Wei, J.; Liang, J.; Xu, X.; He, R.; Robbins, T.W.; Wang, Z. Diagnostic classification for human autism and obsessive-compulsive disorder based on machine learning from a primate genetic model. Am. J. Psychiatry 2021, 178, 65–76. [Google Scholar] [CrossRef]
- Vosoughi, M.A.; Wismüller, A. Large-scale extended Granger causality for classification of marijuana users from functional MRI. In Proceedings of the Medical Imaging 2021: Biomedical Applications in Molecular, Structural, and Functional Imaging, Online, 15–19 February 2021; SPIE: Bellingham, WA, USA, 2021; Volume 11600, pp. 72–84. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. Stat 2017, 1050, 10-48550. [Google Scholar]
| SITE | ASD | TC 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex (m/f) | Age Avg (SD) | FIQ Avg (SD) | VIQ Avg (SD) | PIQ Avg (SD) | Hand* 2 (l/m/r) | Sex (m/f) | Age Avg (SD) | FIQ Avg (SD) | VIQ Avg (SD) | PIQ Avg (SD) | Hand* (l/m/r) | |
| CALTECH | 15/4 | 27.4 (10.3) | 107.7 (12.4) | 107.3 (15.1) | 107 (11.5) | 0/5/14 | 14/4 | 28 (10.9) | 114.8 (9.6) | 114.5 (12.8) | 111.8 (9.5) | 1/3/14 |
| CMU | 11/3 | 26.4 (5.8) | 114.5 (11.6) | 111.2 (13.7) | 114.4 (10.6) | 1/1/12 | 10/3 | 26.8 (5.7) | 114.6 (9.7) | 112.8 (9.8) | 112.8 (9.8) | 0/1/12 |
| KKI | 16/4 | 10 (1.4) | 97.9 (17.5) | N/A | N/A | 1/3/16 | 20/8 | 10 (1.2) | 112.1 (9.4) | N/A | N/A | 2/3/24 |
| LEUVEN | 26/3 | 17.8 (5) | 109.4 (13.1) | 99.1 (20) | 103.7 (16.8) | 3/0/26 | 29/5 | 18.2 (5.1) | 114.8 (12.9) | 116.4 (10.8) | 108 (13) | 4/1/29 |
| MAX_MUN | 21/3 | 26.1 (14.9) | 109.1 (14.1) | N/A | 111.5 (10.8) | 2/0/22 | 27/1 | 24.6 (8.8) | 111.8 (9.3) | N/A | 110.9 (13.8) | 0/0/28 |
| NYU | 65/10 | 14.7 (7.1) | 107.1 (16.4) | 104.9 (15.9) | 108.3 (17.3) | 0/0/75 | 74/26 | 15.7 (6.2) | 113 (13.4) | 112.8 (12.7) | 110.2 (14) | 0/0/100 |
| OHSU | 12/0 | 11.4 (2.2) | 105.5 (21.1) | N/A | N/A | 1/0/11 | 14/0 | 10.1 (1.1) | 115 (11.1) | N/A | N/A | 0/0/14 |
| OLIN | 16/3 | 16.5 (3.4) | 111.3 (17.7) | N/A | N/A | 4/0/15 | 13/2 | 16.7 (3.6) | 113.9 (16.5) | N/A | N/A | 2/0/13 |
| PITT | 25/4 | 19 (7.3) | 110.2 (14.6) | 107 (13.8) | 110.8 (14.1) | 3/0/26 | 23/4 | 18.9 (6.6) | 110.1 (9.4) | 107.7 (11) | 109.6 (9) | 1/0/26 |
| SBL | 15/0 | 35 (10.4) | N/A | N/A | N/A | 1/0/14 | 15/0 | 33.7 (6.6) | N/A | N/A | N/A | 0/0/15 |
| SDSU | 13/1 | 14.7 (1.8) | 111.4 (18) | 110.1 (18.4) | 109.7 (16.4) | 1/0/13 | 16/6 | 14.2 (1.9) | 108.1 (10.5) | 106.7 (10.4) | 107.8 (12.2) | 3/0/19 |
| STANFORD | 15/4 | 10 (1.6) | 110.7 (16.1) | 108.3 (20.4) | 110.6 (12.5) | 3/1/15 | 16/4 | 10 (1.6) | 112.1 (15.4) | 111.2 (19.7) | 110.6 (15.6) | 0/2/18 |
| TRINITY | 22/0 | 16.8 (3.2) | 108.9 (15.5) | 107.9 (14.4) | 107.6 (15.7) | 0/0/22 | 25/0 | 17.1 (3.8) | 110.9 (12.2) | 109.6 (13.7) | 110.3 (10.9) | 0/0/25 |
| UCLA | 48/6 | 13 (2.5) | 100.4 (13.5) | 101.6 (14.1) | 99.8 (13.9) | 6/0/48 | 38/6 | 13 (1.9) | 106.4 (11.1) | 107.1 (11.6) | 104.3 (11.7) | 4/0/40 |
| UM | 57/9 | 13.2 (2.4) | 105.4 (17.1) | 108.7 (20) | 102.5 (19.9) | 7/1/58 | 56/18 | 14.8 (3.6) | 107.9 (9.7) | 113.6 (12.8) | 103 (12) | 9/0/65 |
| USM | 46/0 | 23.5 (8.3) | 99.7 (16.6) | 95 (19.3) | 104.7 (16.7) | 0/0/46 | 25/0 | 21.3 (8.4) | 115.4 (15.1) | 113.6 (16) | 112.8 (14.2) | 0/0/25 |
| YALE | 20/8 | 12.7 (3) | 94.6 (21.6) | 96.5 (23.1) | 92.3 (19.2) | 6/0/22 | 20/8 | 12.7 (2.8) | 105 (17.4) | 106.8 (16) | 101.3 (16.5) | 4/0/24 |
| SITE | ASD | TC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex (m/f) | Age Avg (SD) | FIQ Avg (SD) | VIQ Avg (SD) | PIQ Avg (SD) | Hand* 1 (l/m/r) | Sex (m/f) | Age Avg (SD) | FIQ Avg (SD) | VIQ Avg (SD) | PIQ Avg (SD) | Hand* (l/m/r) | |
| BNI | 29/0 | 37.4 (16.1) | 107.8 (13.7) | N/A | N/A | 0/0/29 | 29/0 | 39.6 (15.1) | 112.4 (12.1) | N/A | N/A | 0/0/29 |
| EMC | 19/5 | 8.2 (1.2) | N/A | N/A | 99.3 (14.3) | 5/0/19 | 22/5 | 8.2 (1) | N/A | N/A | 99.4 (15.4) | 6/0/21 |
| ETH | 13/0 | 20.6 (3.4) | 109 (13) | 111 (13.3) | 105.2 (14.6) | 0/0/13 | 24/0 | 23.9 (4.5) | 116.5 (9.5) | 114.2 (14.1) | 114.1 (12.4) | 0/0/24 |
| GU | 43/8 | 10.9 (1.5) | 118.3 (15.4) | 120.3 (15.2) | 110.7 (15) | 8/0/43 | 28/27 | 10.4 (1.7) | 121.5 (13.8) | 121.6 (15.2) | 116.5 (13.3) | 3/0/52 |
| IP | 14/8 | 15.1 (4.9) | 92.4 (24.5) | 98.6 (23.3) | 92 (22.8) | 1/0/21 | 12/21 | 23.7 (11.6) | 108.1 (18.6) | 111.5 (12) | 112.9 (11.2) | 4/2/27 |
| IU | 16/4 | 25 (9.3) | 116.3 (11.8) | 117.8 (15) | 110.3 (14.4) | 2/3/15 | 15/5 | 23.8 (4.9) | 117 (10.7) | 115.3 (10.4) | 115 (12.2) | 1/2/17 |
| KKI | 41/15 | 10.3 (1.5) | 103.4 (16) | 109.9 (17.1) | 105.3 (14.2) | 2/8/46 | 99/56 | 10.3 (1.2) | 114.3 (10.5) | 118 (12.3) | 110.7 (12) | 10/12/133 |
| KUL | 28/0 | 23.6 (4.8) | 106.6 (15.8) | 109.6 (11.4) | 106.3 (20.7) | 6/0/22 | N/A | N/A | N/A | N/A | N/A | N/A |
| NYU_1 | 43/5 | 10.1 (5.7) | 101.8 (18.3) | 101 (16.5) | 102.2 (19.1) | 2/8/38 | 28/2 | 9.5 (3.3) | 115.5 (15) | 116.1 (15.7) | 112.2 (15.2) | 0/1/29 |
| NYU_2 | 24/3 | 6.8 (1.1) | 107.2 (14.1) | 110.8 (17.7) | 107 (17.2) | 4/7/16 | N/A | N/A | N/A | N/A | N/A | N/A |
| OHSU | 30/7 | 11.8 (2.3) | 106 (16.7) | N/A | N/A | 1/1/35 | 27/29 | 10.4 (1.6) | 117.5 (12) | N/A | N/A | 0/1/55 |
| OILH | 20/4 | 21.8 (3.7) | 114 (16.2) | N/A | N/A | 4/4/16 | 20/15 | 24 (3.6) | 111.2 (12.8) | N/A | N/A | 0/3/32 |
| SDSU | 26/7 | 12.9 (3.3) | 99.8 (14.7) | 97.2 (15.5) | 103.1 (18.2) | 4/2/27 | 23/2 | 13.3 (3) | 103 (11.7) | 104.9 (10.5) | 101.4 (14.7) | 1/3/21 |
| SU | 19/2 | 11.2 (1.2) | 111.8 (15.7) | 111.7 (16.8) | 109.3 (15.2) | 0/0/21 | 19/2 | 11 (1.3) | 116.1 (14) | 117.6 (16.8) | 111.4 (13) | 0/3/18 |
| TCD | 21/0 | 14.8 (3.3) | 108.5 (15.3) | 108.5 (15) | 106.2 (16.4) | 0/0/21 | 21/0 | 15.6 (3.1) | 118.5 (13.2) | 117.3 (15.8) | 115.5 (11.8) | 0/0/21 |
| U_MIA | 11/2 | 9.9 (2) | 100.8 (20.1) | 97.5 (21.3) | 102.9 (20.9) | 0/1/12 | 11/4 | 9.7 (2.1) | 115.9 (14.7) | 112.2 (12.4) | 111.7 (19.3) | 0/0/15 |
| UCD | 14/4 | 14.8 (2) | 103.4 (12.2) | 101.1 (15.9) | 104.9 (12.8) | 0/1/17 | 10/4 | 14.8 (1.7) | 113 (11.2) | 112.1 (10.5) | 110.6 (12.8) | 0/0/14 |
| UCLA | 15/1 | 11.7 (2.2) | 102.1 (14) | 101.6 (17.5) | 104.4 (15.1) | 2/0/14 | 11/5 | 9.7 (2.1) | 114.5 (13.9) | 111.9 (13.6) | 114.1 (15.1) | 1/1/14 |
| USM | 15/2 | 18.3 (7) | 99.3 (20) | 99.6 (14.4) | 101.1 (17) | 0/2/15 | 13/3 | 24 (7.8) | 115.2 (16.2) | 116.1 (14.6) | 117.4 (16.4) | 0/1/15 |
| Parameter Description | Value |
|---|---|
| Train epoch | 100 |
| BatchSize | 64 |
| Learning rate | 0.001 |
| Weightdecay | 0.05 |
| Stepsize | 20 |
| Gamma | 0.5 |
| GraphConv layers | 2 |
| Optimizers | Adam |
| Methods | Classifier | Samples | Acc | Sen |
|---|---|---|---|---|
| Yang 2019 [30] | Ridge | 505 ASD, 530 HC | 0.7198 | 0.7089 |
| Bernas 2018 [31] | LDA | 403 ASD, 468 HC | 0.733 | 0.667 |
| Song 2019 [32] | LDA + KNN | 119 ASD, 116 HC | 0.7486 | 0.7167 |
| Madine 2020 [33] | HGNN | 155 ASD, 186 HC | 0.752 | N/A |
| Jung 2019 [34] | RFE + SVM | 86 ASD, 83 HC | 0.763 | 0.792 |
| Liu 2020 [35] | MTFS-EM + SVM | 403ASD, 468 HC | 0.768 | 0.725 |
| Zheng 2019 [36] | MFN + SVM | 66 ASD, 66 HC | 0.7863 | 0.8 |
| Ours | RFE + GNN + PFE | 505 ASD, 530 HC | 0.7874 | 0.7429 |
| Methods | Classifier | Samples | Acc | Sen |
|---|---|---|---|---|
| Liu 2021 [37] | BL | 487 ASD, 556 HC | 0.6529 | 0.6293 |
| Zhao 2019 [38] | CAE + CNN | 303 ASD, 390 HC | 0.653 | N/A |
| Deng 2022 [39] | ST-Transformer | 521 ASD, 593 HC | 0.7061 | 0.6875 |
| Liu 2023 [40] | STCAL | 521 ASD, 592 HC | 0.72 | 0.744 |
| Zhang 2021 [41] | Sparse LR | 60 ASD, 89 HC | 0.7836 | 0.7391 |
| Wismu 2020 [42] | lsXGC + SVM | 24 ASD, 35 HC | 0.79 | N/A |
| Ours | RFE + GNN + PFE | 518 ASD, 592 HC | 0.8036 | 0.7624 |
| Methods | Acc | Sen | Pre | F1_Score |
|---|---|---|---|---|
| SVM | 0.6232 | 0.5979 | 0.6077 | 0.6028 |
| SVM () | 0.6522 | 0.5842 | 0.6629 | 0.6211 |
| RF | 0.6232 | 0.5502 | 0.6579 | 0.5992 |
| RF (pheno) | 0.6667 | 0.6 | 0.6477 | 0.623 |
| GAT | 0.6763 | 0.6972 | 0.6909 | 0.6941 |
| GAT (pheno) | 0.7005 | 0.6995 | 0.7023 | 0.7009 |
| GAT () | 0.715 | 0.7045 | 0.7092 | 0.7068 |
| Ours | 0.6715 | 0.7579 | 0.6154 | 0.6792 |
| Ours (pheno) | 0.7291 | 0.7813 | 0.6944 | 0.7353 |
| Ours (RFE) | 0.7488 | 0.7579 | 0.7277 | 0.7425 |
| Ours (pheno, RFE) | 0.7874 | 0.7429 | 0.8041 | 0.7723 |
| Methods | Acc | Sen | Pre | F1_Score |
|---|---|---|---|---|
| SVM | 0.6216 | 0.519 | 0.6567 | 0.5798 |
| SVM (pheno) | 0.6667 | 0.549 | 0.6667 | 0.6022 |
| RF | 0.6486 | 0.5455 | 0.6207 | 0.5806 |
| RF (pheno) | 0.6712 | 0.56 | 0.7077 | 0.6252 |
| GAT | 0.6802 | 0.6276 | 0.6477 | 0.6375 |
| GAT (pheno) | 0.7207 | 0.6542 | 0.7368 | 0.6931 |
| GAT (RFE) | 0.7252 | 0.6905 | 0.745 | 0.7167 |
| Ours | 0.6937 | 0.6 | 0.7582 | 0.6699 |
| Ours (pheno) | 0.75 | 0.7604 | 0.73 | 0.7449 |
| Ours (RFE) | 0.7808 | 0.7267 | 0.7533 | 0.7398 |
| Ours (pheno, RFE) | 0.8063 | 0.7624 | 0.8021 | 0.7817 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Hu, M.; Hu, Y.; Zhang, Z.; Zhong, J. Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE). Sensors 2023, 23, 9647. https://doi.org/10.3390/s23249647
Yang J, Hu M, Hu Y, Zhang Z, Zhong J. Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE). Sensors. 2023; 23(24):9647. https://doi.org/10.3390/s23249647
Chicago/Turabian StyleYang, Jiahong, Miaojun Hu, Yao Hu, Zixi Zhang, and Jiancheng Zhong. 2023. "Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE)" Sensors 23, no. 24: 9647. https://doi.org/10.3390/s23249647
APA StyleYang, J., Hu, M., Hu, Y., Zhang, Z., & Zhong, J. (2023). Diagnosis of Autism Spectrum Disorder (ASD) Using Recursive Feature Elimination–Graph Neural Network (RFE–GNN) and Phenotypic Feature Extractor (PFE). Sensors, 23(24), 9647. https://doi.org/10.3390/s23249647






