BCG Signal Quality Assessment Based on Time-Series Imaging Methods
Abstract
:1. Introduction
2. Related Works
3. Materials and Methods
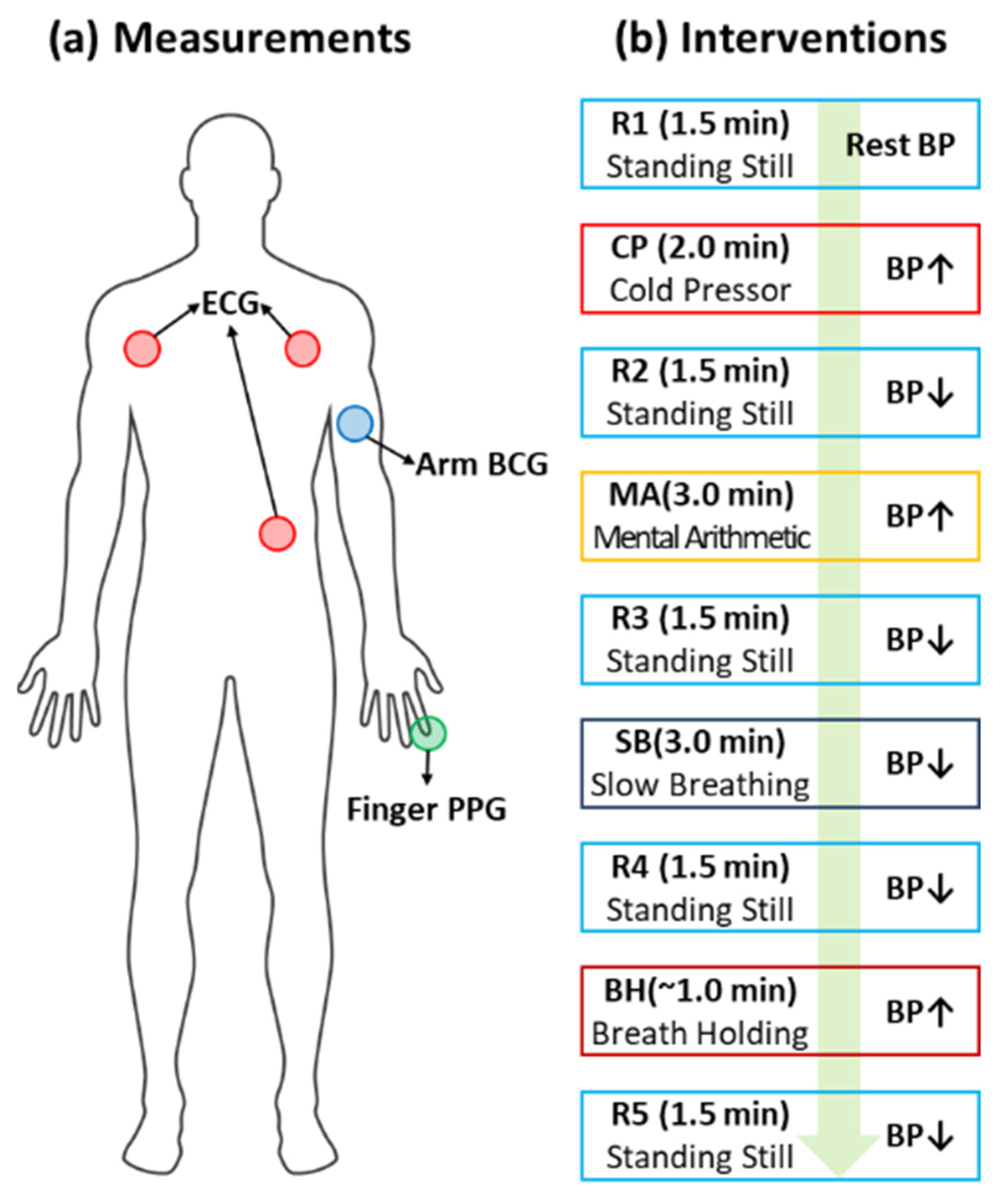
3.1. BCG Dataset [8]
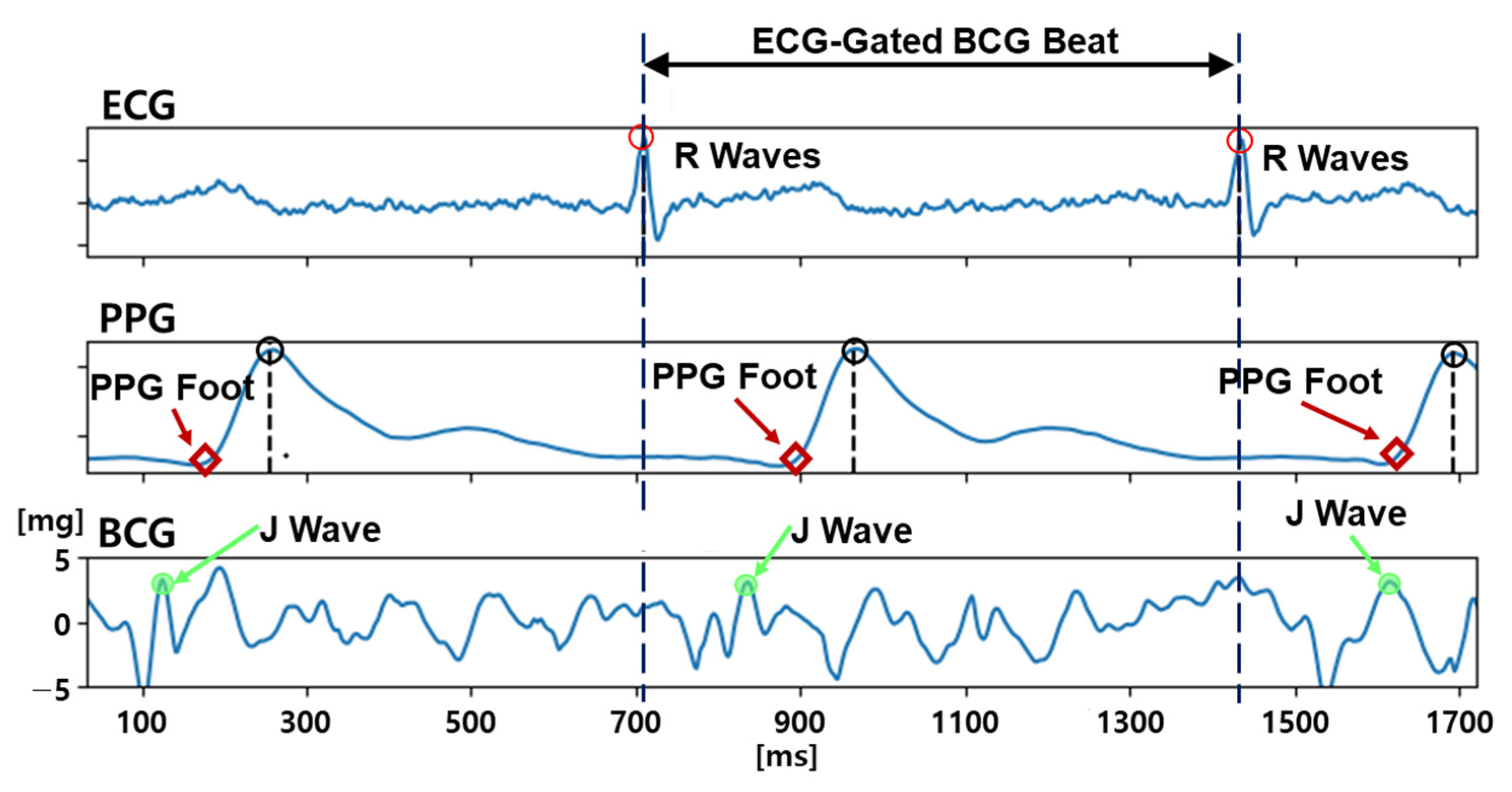
3.2. BCG Quality Labeling Procedure
- (1)
- The existence of a distinguishable and sharp J-wave candidate before the PPG foot,
- (2)
- The magnitude of the J-wave candidate being more than 3 mg (micro-gravity), and
- (3)
- The width of the J-wave candidate being less than 100 ms.
3.3. Time-Series Imaging Methods
3.3.1. Recurrence Plot
3.3.2. Gramian Angular Field
3.3.3. Markov Transition Field
3.3.4. CNN-Based Binary Classification for BCG Signal Quality Assessment
- (1)
- Use 1 × 1 filters instead of 3 × 3 filters for fewer parameters.
- (2)
- Decrease the number of input channels to reduce the number of parameters.
- (3)
- Conduct down-sampling at the latter part of the network to gain large activation maps (maximizing the performance with a reduced number of parameters).
4. Results
- SqueezeNet with GADF resulted in the highest accuracy (87.5%); however, there was no statistically significant difference (p < 0.05) with the independent samples t-test (p-value = 0.64).
- Relatively, the GADF imaging approach outperformed the others in all the 2D CNN classifiers.
- RP and GASF showed similar performance in all the 2D CNN classifiers.
- MTF produced the lowest accuracy.
- The variance of the accuracy alongside the same imaging approach was less than the variance alongside the same 2D CNN classifier.
- The accuracy of all 2D CNN classifiers, except LeNet_Tanh and SqueezeNet with MTF, exceeded that of the 1D CNN approach (baseline).
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Drawz, P.E.; Abdalla, M.; Rahman, M. Blood Pressure Measurement: Clinic, Home, Ambulatory, and Beyond. Am. J. Kidney Dis. 2012, 60, 449–462. [Google Scholar] [CrossRef]
- George, J.; MacDonald, T. Home Blood Pressure Monitoring. Eur. Cardiol. Rev. 2015, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Ogedegbe, G.; Pickering, T. Principles and Techniques of Blood Pressure Measurement. Cardiol. Clin. 2010, 28, 571–586. [Google Scholar] [CrossRef]
- Alpert, B.S.; Quinn, D.; Gallick, D. Oscillometric blood pressure: A review for clinicians. J. Am. Soc. Hypertens. 2014, 8, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhar, A.; Yavarimanesh, M.; Hahn, J.-O.; Sung, S.-H.; Chen, C.-H.; Cheng, H.-M.; Mukkamala, R. Formulas to Explain Popular Oscillometric Blood Pressure Estimation Algorithms. Front. Physiol. 2019, 10, 1415. [Google Scholar] [CrossRef] [PubMed]
- Beevers, G. ABC of hypertension: Blood pressure measurement. BMJ 2001, 322, 1043–1047. [Google Scholar] [CrossRef]
- Williams, B.; Mancia, G.; Spiering, W.; Agabiti Rosei, E.; Azizi, M.; Burnier, M.; Clement, D.L.; Coca, A.; de Simone, G.; Dominiczak, A.; et al. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur. Heart J. 2018, 39, 3021–3104. [Google Scholar] [CrossRef]
- Shin, S.; Yousefian, P.; Mousavi, A.S.; Kim, C.-S.; Mukkamala, R.; Jang, D.-G.; Ko, B.-H.; Lee, J.; Kwon, U.-K.; Kim, Y.H.; et al. A Unified Approach to Wearable Ballistocardiogram Gating and Wave Localization. IEEE Trans. Biomed. Eng. 2021, 68, 1115–1122. [Google Scholar] [CrossRef]
- Ghamari, M. A review on wearable photoplethysmography sensors and their potential future applications in health care. Int. J. Biosens. Bioelectron. 2018, 4, 195–202. [Google Scholar] [CrossRef]
- Eckmann, J.-P.; Kamphorst, S.O.; Ruelle, D. Recurrence Plots of Dynamical Systems. Europhys. Lett. 1987, 4, 973–977. [Google Scholar] [CrossRef]
- Wang, Z.; Oates, T. Encoding time series as images for visual inspection and classification using tiled convolutional neural networks. In Proceedings of the Workshops at the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 1. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Wang, Z.; Yan, W.; Oates, T. Time series classification from scratch with deep neural networks: A strong baseline. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1578–1585. [Google Scholar]
- Zeng, L.; Wang, R.; Yang, S.; Zeng, X.; Guo, Z. Classification of cardiac abnormality based on BCG signal using 1-D convolutional neural network. In Proceedings of the Third International Conference on Computer Science and Communication Technology (ICCSCT 2022), Beijing, China, 30–31 July 2022; p. 241. [Google Scholar]
- Hong, S.; Heo, J.; Park, K.S. Signal Quality Index Based on Template Cross-Correlation in Multimodal Biosignal Chair for Smart Healthcare. Sensors 2021, 21, 7564. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Meng, Y.; Zeng, L.; Liu, Z.; Lan, R. Shapelet Feature Learning Method of BCG Signal Based on ESOINN. In Proceedings of the 2019 Tenth International Conference on Intelligent Control and Information Processing (ICICIP), Marrakesh, Morocco, 11–16 December 2019; pp. 226–232. [Google Scholar]
- Bicen, A.O.; Whittingslow, D.C.; Inan, O.T. Template-Based Statistical Modeling and Synthesis for Noise Analysis of Ballistocardiogram Signals: A Cycle-Averaged Approach. IEEE J. Biomed. Heal. Informatics 2019, 23, 1516–1525. [Google Scholar] [CrossRef]
- Rahman, S.; Karmakar, C.; Natgunanathan, I.; Yearwood, J.; Palaniswami, M. Robustness of electrocardiogram signal quality indices. J. R. Soc. Interface 2022, 19, 20220012. [Google Scholar] [CrossRef] [PubMed]
- Smital, L.; Haider, C.R.; Vitek, M.; Leinveber, P.; Jurak, P.; Nemcova, A.; Smisek, R.; Marsanova, L.; Provaznik, I.; Felton, C.L.; et al. Real-Time Quality Assessment of Long-Term ECG Signals Recorded by Wearables in Free-Living Conditions. IEEE Trans. Biomed. Eng. 2020, 67, 2721–2734. [Google Scholar] [CrossRef]
- Satija, U.; Ramkumar, B.; Manikandan, M.S. A New Automated Signal Quality-Aware ECG Beat Classification Method for Unsupervised ECG Diagnosis Environments. IEEE Sens. J. 2019, 19, 277–286. [Google Scholar] [CrossRef]
- Yaghmaie, N.; Maddah-Ali, M.A.; Jelinek, H.F.; Mazrbanrad, F. Dynamic signal quality index for electrocardiograms. Physiol. Meas. 2018, 39, 105008. [Google Scholar] [CrossRef] [PubMed]
- Satija, U.; Ramkumar, B.; Manikandan, M.S. A Review of Signal Processing Techniques for Electrocardiogram Signal Quality Assessment. IEEE Rev. Biomed. Eng. 2018, 11, 36–52. [Google Scholar] [CrossRef]
- Orphanidou, C. Quality Assessment for the Photoplethysmogram (PPG). In Signal Quality Assessment in Physiological Monitoring; Springer: Cham, Switzerland, 2018; pp. 41–63. ISBN 978-3-319-68414-7. [Google Scholar]
- Pradhan, N.; Rajan, S.; Adler, A. Evaluation of the signal quality of wrist-based photoplethysmography. Physiol. Meas. 2019, 40, 065008. [Google Scholar] [CrossRef] [PubMed]
- Mohagheghian, F.; Han, D.; Peitzsch, A.; Nishita, N.; Ding, E.; Dickson, E.L.; DiMezza, D.; Otabil, E.M.; Noorishirazi, K.; Scott, J.; et al. Optimized Signal Quality Assessment for Photoplethysmogram Signals Using Feature Selection. IEEE Trans. Biomed. Eng. 2022, 69, 2982–2993. [Google Scholar] [CrossRef]
- Moscato, S.; Lo Giudice, S.; Massaro, G.; Chiari, L. Wrist Photoplethysmography Signal Quality Assessment for Reliable Heart Rate Estimate and Morphological Analysis. Sensors 2022, 22, 5831. [Google Scholar] [CrossRef]
- Roh, D.; Shin, H. Recurrence Plot and Machine Learning for Signal Quality Assessment of Photoplethysmogram in Mobile Environment. Sensors 2021, 21, 2188. [Google Scholar] [CrossRef]
- Zhang, O.; Ding, C.; Pereira, T.; Xiao, R.; Gadhoumi, K.; Meisel, K.; Lee, R.J.; Chen, Y.; Hu, X. Explainability Metrics of Deep Convolutional Networks for Photoplethysmography Quality Assessment. IEEE Access 2021, 9, 29736–29745. [Google Scholar] [CrossRef]
- Makowski, D.; Pham, T.; Lau, Z.J.; Brammer, J.C.; Lespinasse, F.; Pham, H.; Schölzel, C.; Chen, S.H.A. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav. Res. Methods 2021, 53, 1689–1696. [Google Scholar] [CrossRef] [PubMed]
- Hermeling, E.; Reesink, K.D.; Reneman, R.S.; Hoeks, A.P.G. Measurement of Local Pulse Wave Velocity: Effects of Signal Processing on Precision. Ultrasound Med. Biol. 2007, 33, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Keogh, E.; Chakrabarti, K.; Pazzani, M.; Mehrotra, S. Dimensionality Reduction for Fast Similarity Search in Large Time Series Databases. Knowl. Inf. Syst. 2001, 3, 263–286. [Google Scholar] [CrossRef]
- Yousefian, P.; Shin, S.; Mousavi, A.S.; Tivay, A.; Kim, C.S.; Mukkamala, R.; Jang, D.G.; Ko, B.H.; Lee, J.; Kwon, U.K.; et al. Pulse Transit Time-Pulse Wave Analysis Fusion Based on Wearable Wrist Ballistocardiogram for Cuff-Less Blood Pressure Trend Tracking. IEEE Access 2020, 8, 138077–138087. [Google Scholar] [CrossRef]




| 2D CNN-Based Classifier | (Baseline) * | ||||||
|---|---|---|---|---|---|---|---|
| ResNet | SqueezeNet | DenseNet | LeNet (Tanh) | LeNet (ReLU) | FCN | ||
| Time-series Image | RP | 83.1% | 82.9% | 84.0% | 82.9% | 83.7% | 78.1% (±1.5%) |
| (±1.4%) | (±1.3%) | (±0.4%) | (±0.6%) | (±0.6%) | |||
| GASF | 84.1% | 82.3% | 84.0% | 81.6% | 83.3% | ||
| (±0.7%) | (±0.7%) | (±0.8%) | (±0.9%) | (±0.6%) | |||
| GADF | 86.7% | 87.5% | 87.3% | 85.6% | 86.9% | ||
| (±0.9%) | (±0.7%) | (±0.9%) | (±0.5%) | (±0.9%) | |||
| MTF | 79.6% | 76.7% | 78.5% | 75.6% | 78.8% | ||
| (±1.2%) | (±1.4%) | (±1.4%) | (±0.4%) | (±0.6%) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.; Choi, S.; Kim, C.; Mousavi, A.S.; Hahn, J.-O.; Jeong, S.; Jeong, H. BCG Signal Quality Assessment Based on Time-Series Imaging Methods. Sensors 2023, 23, 9382. https://doi.org/10.3390/s23239382
Shin S, Choi S, Kim C, Mousavi AS, Hahn J-O, Jeong S, Jeong H. BCG Signal Quality Assessment Based on Time-Series Imaging Methods. Sensors. 2023; 23(23):9382. https://doi.org/10.3390/s23239382
Chicago/Turabian StyleShin, Sungtae, Soonyoung Choi, Chaeyoung Kim, Azin Sadat Mousavi, Jin-Oh Hahn, Sehoon Jeong, and Hyundoo Jeong. 2023. "BCG Signal Quality Assessment Based on Time-Series Imaging Methods" Sensors 23, no. 23: 9382. https://doi.org/10.3390/s23239382
APA StyleShin, S., Choi, S., Kim, C., Mousavi, A. S., Hahn, J.-O., Jeong, S., & Jeong, H. (2023). BCG Signal Quality Assessment Based on Time-Series Imaging Methods. Sensors, 23(23), 9382. https://doi.org/10.3390/s23239382







