Inversion of Forest Biomass Based on Multi-Source Remote Sensing Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overview of the Study Area
2.2. Data Source
2.3. Data Preprocessing
2.3.1. Landsat 8 OLI Data Processing Method
2.3.2. Sentinel-1A Data Preprocessing Method
2.3.3. Calculation of Tree Biomass in Sample Plot
2.3.4. Correlation Analysis of Remote Sensing Factors and Biomass
2.3.5. Build a Linear Regression Model
2.3.6. Establish BP Neuron Network Model
- , where k is the set size of input data, M and n are the number of hidden layer and input layer nodes, respectively. If i > M, specify = 0;
- , where m is the number of network output layer nodes, n is the number of network input layer nodes and a is a constant between [0 and 10];
- , where n is the number of input layer nodes.
2.3.7. Establishment of BP Neural Network Model Improved by Particle Swarm Optimization
- (1)
- Initialize a group of particles (the group size is N), including random positions and velocities;
- (2)
- Evaluate the fitness of each particle;
- (3)
- For each particle, compare its fitness value with the best position pbest it has passed, and if it is better, use it as the current best position pbest;
- (4)
- For each particle, compare its fitness value with its best position gbest, and if it is better, use it as the current best position gbest;
- (5)
- Adjust particle velocity and position;
- (6)
- If the end condition is not met, go to step 2.
2.3.8. Spatial Biomass Mapping
3. Results
3.1. Plot Data Processing
3.2. Correlation Analysis of Remote Sensing Factors and Biomass
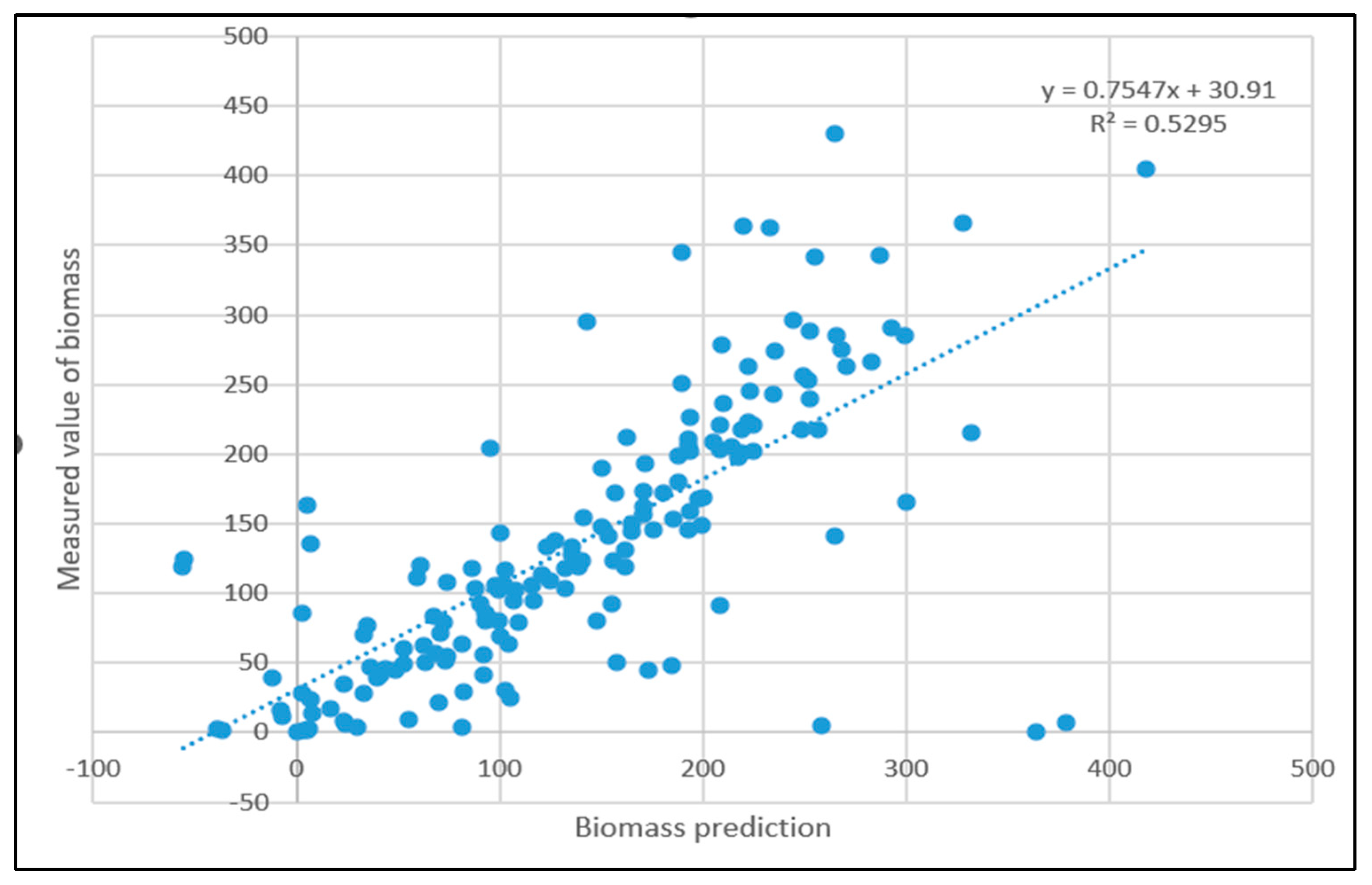
3.3. Linear Regression Model Processing Results
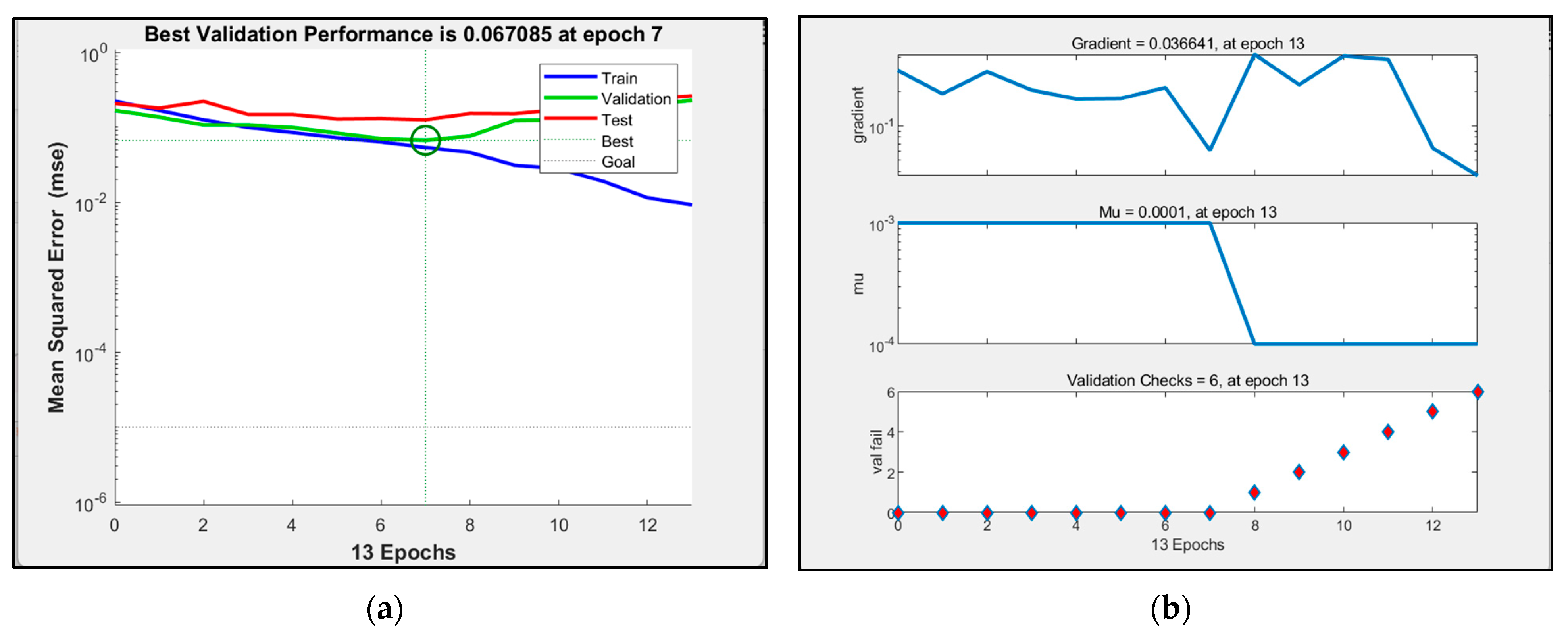
3.4. BP Neuron Network Model Processing Results
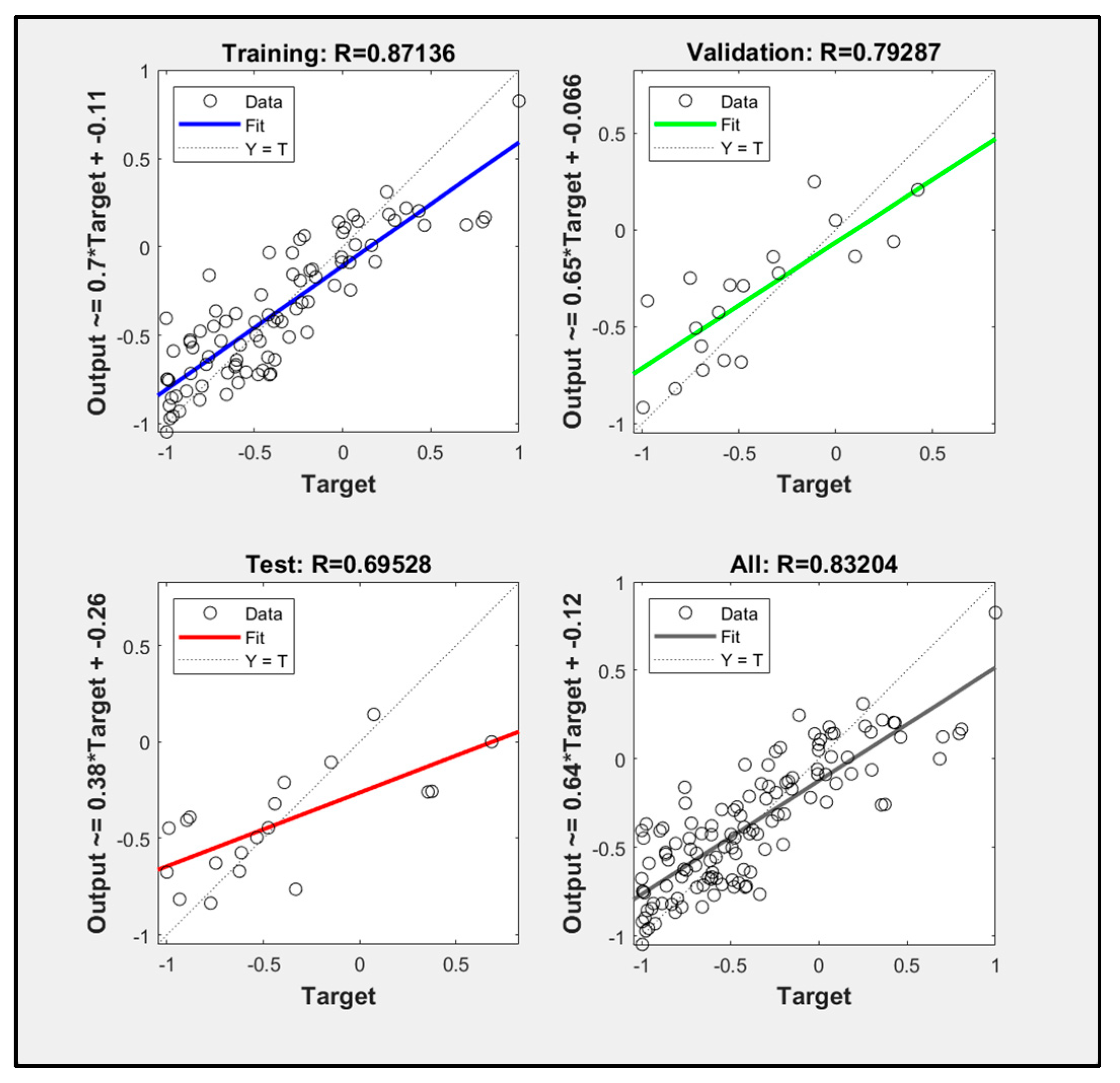
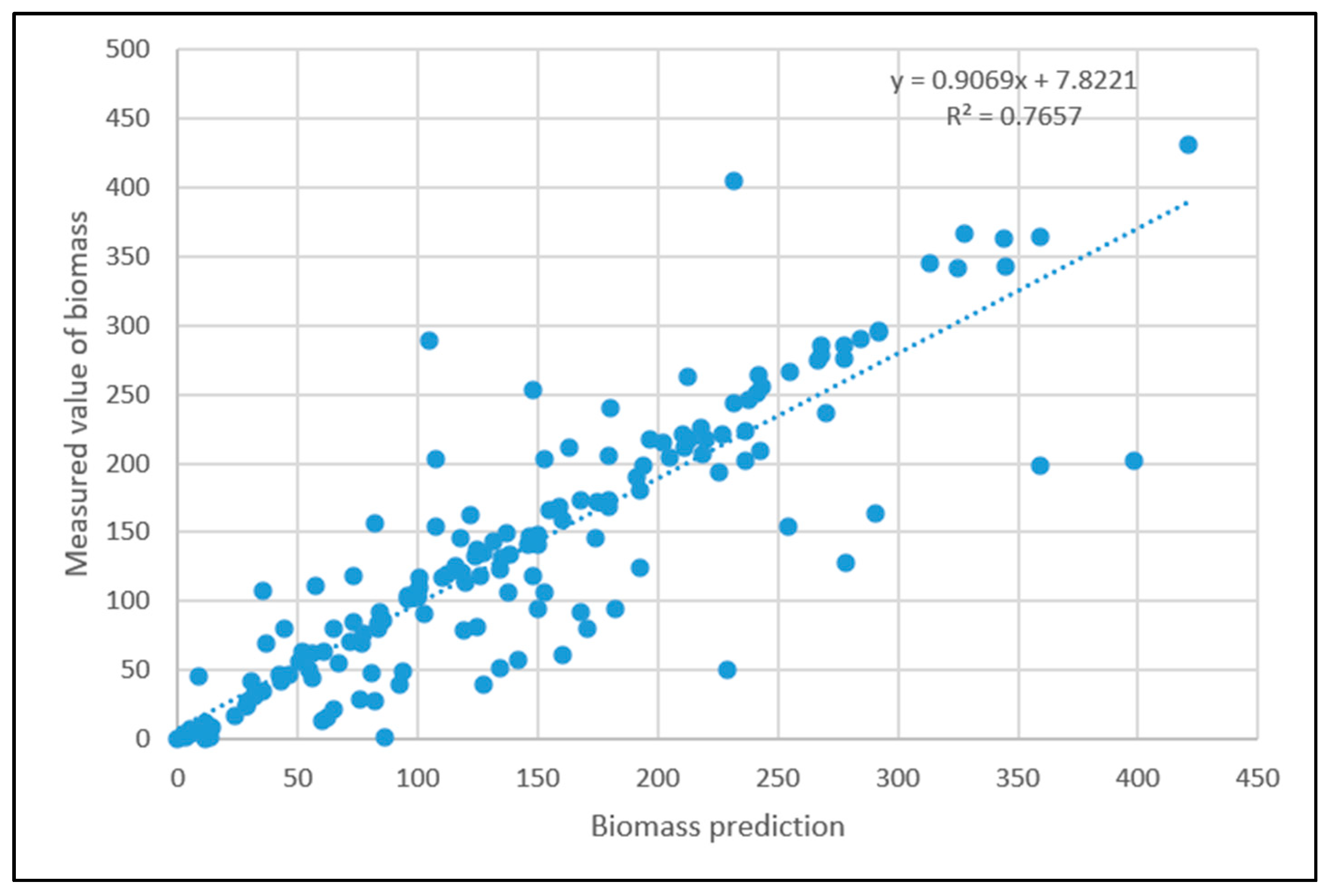
3.5. Particle Swarm Optimization Algorithm Improves BP Neural Network Model Processing Results
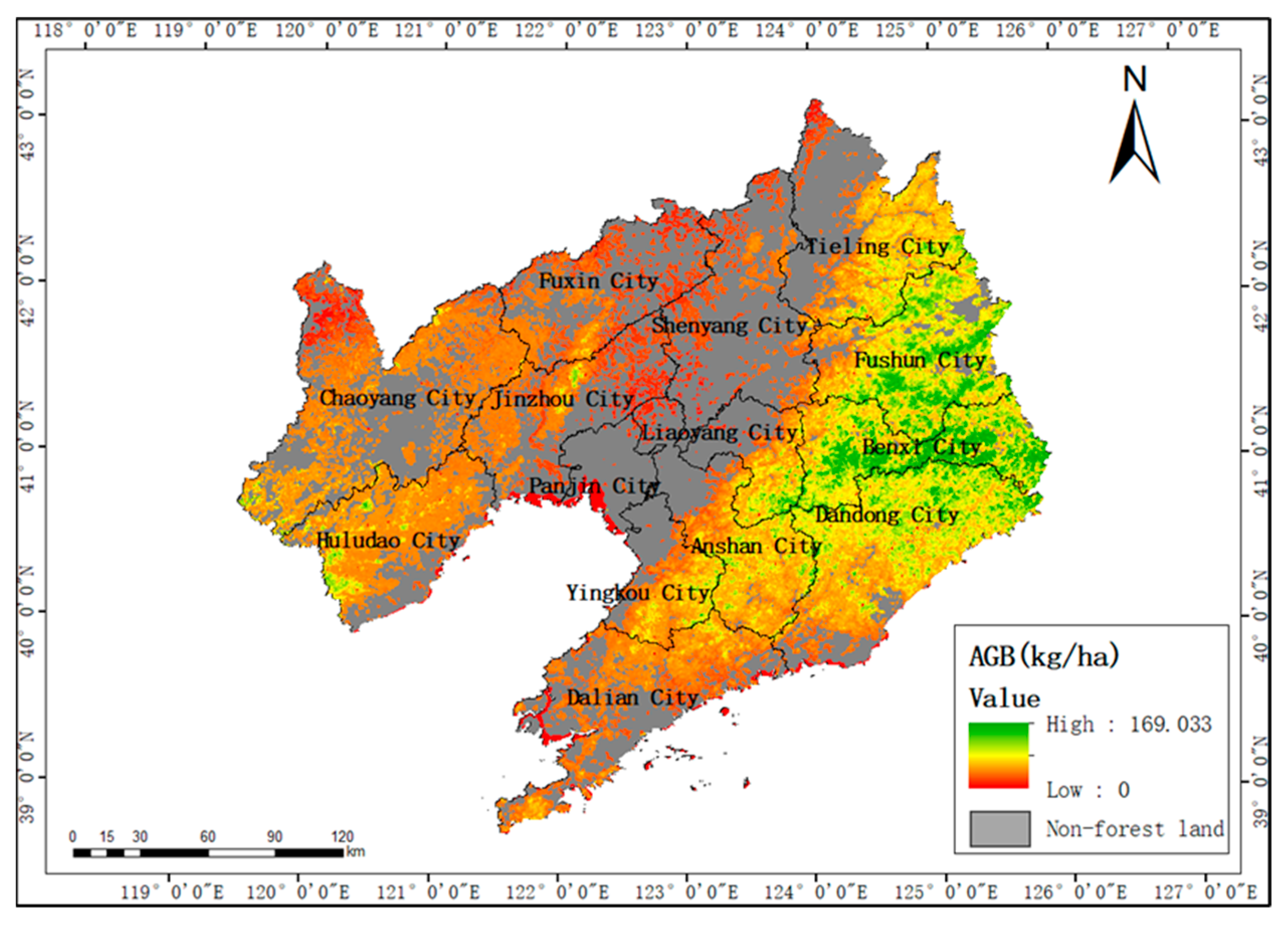
3.6. Results of Spatial Biomass Mapping
4. Discussion
- (1)
- This experiment was only a preliminary study on a forest estimation model, and there were still some shortcomings and deficiencies in the specific technical processing. More parameter characteristics would be needed for further analysis in order to better understand the impact of biological factors on factors of quantitative inversion accuracy;
- (2)
- Due to limited conditions, the number of training data samples used in this experiment was small, which made the prediction network not stable enough and could cause certain errors in the results;
- (3)
- The data collected with remote sensing technology were greatly affected by factors such as sensors, shooting angles and atmosphere, which may cause the inversion errors.
5. Conclusions
- (1)
- The correlation between altitude and biomass was the highest, and the correlation coefficient was 0.404. The B2 band of Landsat 8 and the NDPI characteristic quantity of the vegetation index have important correlations with forest biomass inversion, and the correlation coefficients are −0.342 and 0.323, respectively. Among the characteristic factors extracted from Sentinel-1A radar images, seven factors were correlated with biomass (p < 0.05), and five factors were negatively correlated, except entropy.
- (2)
- Comparing the inversion accuracy and training speed of the three models, the model based on the linear stepwise regression method has the fastest training speed, but the lowest model accuracy. The BP neural network has a strong fitting ability with complex data, a short training time and high model accuracy. Although the training time of the PSO improved neural network model is longer, the coefficient of determination between the predicted value and the measured value is the highest. The results show that there is a nonlinear relationship between the biomass and the strong correlation factors. The neural network model based on the particle swarm optimization algorithm is the best model for forest biomass inversion in the Liaoning region.
- (3)
- According to the spatial distribution map of biomass, the areas with high forest biomass in Liaoning Province are mainly distributed in areas with high altitudes and steep slopes in the east and southwest, while the areas with low biomass are mainly concentrated in the plain areas with low altitudes and gentle slopes.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Babbar, D.; Areendran, G.; Sahana, M.; Sarma, K.; Raj, K.; Sivadas, A. Assessment and prediction of carbon sequestration using Markov chain and InVEST model in Sariska Tiger Reserve, India. J. Clean. Prod. 2021, 278, 123333. [Google Scholar] [CrossRef]
- Hajima, T.; Tachiiri, K.; Ito, A.; Kawamiya, M. Uncertainty of Concentration-Terrestrial Carbon Feedback in Earth System Models. J. Clim. 2014, 27, 3425–3445. [Google Scholar] [CrossRef]
- Lu, M.; Zhou, X.; Yang, Q.; Li, H.; Luo, Y.; Fang, C.; Chen, J.; Yang, X.; Li, B.O. Responses of ecosystem carbon cycle to experimental warming: A meta-analysis. Ecology 2013, 94, 726–738. [Google Scholar] [CrossRef]
- Zhang, F.; Tian, X.; Zhang, H.; Jiang, M. Estimation of Aboveground Carbon Density of Forests Using Deep Learning and Multisource Remote Sensing. Remote Sens. 2022, 14, 3022. [Google Scholar] [CrossRef]
- Mohd Zaki, N.A.; Latif, A.Z. Carbon sinks and tropical forest biomass estimation: A review on role of remote sensing in aboveground-biomass modelling. Geocarto Int. 2016, 32, 701–716. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, X.H.; Ouyang, Z.T.; Avitabile, V.; Qi, J.G.; Chen, J.Q.; Giannico, V. Estimating aboveground biomass in subtropical forests of China by integrating multisource remote sensing and ground data. Remote Sens. Environ. 2019, 232, 111341. [Google Scholar] [CrossRef]
- Ye, Z. Research progress on photosynthesis response models to light and CO2. Chin. J. Plant Ecol. 2010, 34, 727–740. [Google Scholar]
- Chi, H.; Huang, J.; Qiu, J.; Sun, G.; Fu, A. Estimation of forest biomass from GLAS spaceborne lidar and Landsat/ETM+ data. Sci. Surv. Mapp. 2018, 43, 9–23. [Google Scholar]
- Yang, H.; Wu, B.; Zhang, J.; Lin, D.; Chang, S. Research Progress on Carbon Sequestration Function and Carbon Storage of Forest Ecosystem. J. Beijing Norm. Univ. (Nat. Sci. Ed.) 2005, 2, 172–177. [Google Scholar]
- Zhang, Y. Estimation of Aboveground Biomass in Forests in Greater Khingan Mountains Based on High-Resolution Remote Sensing and Polarimetric Radar Data. Master’s Thesis, Beijing Forestry University, Beijing, China, 2016. [Google Scholar]
- Li, D.R.; Wang, C.W.; Hu, Y.M.; Liu, S.G. Research progress of forest biomass inversion by remote sensing technology. Geomat. Inf. Sci. Wuhan Univ. 2012, 37, 631–635. [Google Scholar] [CrossRef]
- Lou, X.; Zeng, Y.; Wu, B. Advances in remote sensing estimation of forest aboveground biomass. Remote Sens. Land Resour. 2011, 1, 1–8. [Google Scholar]
- Hyde, P.; Dubayah, R.; Peterson, B.; Blair, J.B.; Hofton, M.; Hunsaker, C.; Knox, R.; Walker, W. Mapping forest structure for wildlife habitat analysis using waveform lidar: Validation of montane ecosystems. Remote Sens. Environ. 2005, 96, 427–437. [Google Scholar] [CrossRef]
- Kankare, V.; Vauhkonen, J.; Holopainen, M.; Vastaranta, M.; Hyyppä, J.; Hyyppä, H.; Alho, P. Sparse Density, Leaf-Off Airborne Laser Scanning Data in Aboveground Biomass Component Prediction. Forests 2015, 6, 1839–1857. [Google Scholar] [CrossRef]
- Kattenborn, T.; Maack, J.; Fassnacht, F.; Enßle, F.; Ermert, J.; Koch, B. Mapping forest biomass from space-Fusion of hyperspectral EO1-hyperion data and Tandem-X and WorldView-2 canopy height models. Int. J. Appl. Earth Obs. Geoinf. 2015, 35, 359–367. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Xing, Y.; Hu, X.; Gong, J. Estimation of the forest stand mean height and aboveground biomass in Northeast China using SAR Sentinel-1B, multispectral Sentinel-2A, and DEM imagery. Isprs J. Photogramm. Remote Sens. 2019, 151, 277–289. [Google Scholar] [CrossRef]
- Sadeghi, Y.; St-Onge, B.; Leblon, B.; Prieur, J.F.; Simard, M. SRTM and TanDEM-X mapping boreal forest biomass based on canopy height model and Landsat spectral index. Int. J. Appl. Earth Obs. Geogr. Inf. 2018, 68, 202–213. [Google Scholar]
- Domingues, G.F.; Soares, V.P.; Leite, H.G.; Ferraz, A.S.; Ribeiro, C.A.A.S.; Lorenzon, A.S.; Marcatti, G.E.; Teixeira, T.R.; de Castro, N.L.M.; Mota, P.H.S.; et al. High performance prediction of eucalyptus biomass based on multispectral and SAR data by artificial neural network. Agric. Comput. Electron. 2020, 168, 169–182. [Google Scholar] [CrossRef]
- Deng, S.; Katoh, M.; Guan, Q.; Yin, N.; Li, M. Estimating forest aboveground biomass by combining alos palsar and worldview-2 data: A case study at purple mountain national park, Nanjing, China. Remote Sens. 2014, 6, 7878–7910. [Google Scholar] [CrossRef]
- Bhatti, U.A.; Yu, Z.; Chanussot, J.; Zeeshan, Z.; Yuan, L.; Luo, W.; Nawaz, S.A.; Bhatti, M.A.; Ain, Q.U.; Mehmood, A. Local similarity-based spatial–spectral fusion hyperspectral image classification with deep CNN and Gabor filtering. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–15. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of herbaceous vegetation using airborne hyperspectral imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef]
- Kwak, G.; Park, N. Impact of texture information on crop classification with machine learning and UAV images. Appl. Sci. 2019, 9, 643. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Bhatti, U.A.; Tang, H.; Wu, G.; Marjan, S.; Hussain, A. Deep learning with graph convolutional networks: An overview and latest applications in computational intelligence. Int. J. Intell. Syst. 2023, 2023, 8342104. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep learning classification of land cover and crop types using remote sensing data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, J.; Ma, X.; Wang, G.; Bhatti, U.A.; Huang, M. Interactive medical image annotation using improved Attention U-net with compound geodesic distance. Expert Syst. Appl. 2024, 237, 121282. [Google Scholar] [CrossRef]
- Bhatti, U.A.; Huang, M.; Neira-Molina, H.; Marjan, S.; Baryalai, M.; Tang, H.; Wu, G.; Bazai, S.U. MFFCG–Multi feature fusion for hyperspectral image classification using graph attention network. Expert Syst. Appl. 2023, 229, 120496. [Google Scholar] [CrossRef]
- Solihin, I.M.; Tack, F.L.; Kean, L.M. Tuning of PID Controller Using Particle Swarm Optimization (PSO). Int. J. Adv. Sci. Eng. Inf. Technol. 2011, 1, 458–461. [Google Scholar] [CrossRef]
- Deng, W.; Yao, R.; Zhao, H.; Yang, X.; Li, G. A novel intelligent diagnosis method using optimal LS-SVM with improved PSO algorithm. Soft Comput. 2019, 23, 2445–2462. [Google Scholar] [CrossRef]
- Mohandes, A.M. Modeling global solar radiation using Particle Swarm Optimization (PSO). Sol. Energy 2012, 86, 3137–3145. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Y.; Liu, W.; Sangaiah, A.K.; Kim, H.-J. An Improved Routing Schema with Special Clustering Using PSO Algorithm for Heterogeneous Wireless Sensor Network. Sensors 2019, 19, 671. [Google Scholar] [CrossRef]
- She, X.; Zhang, L.; Cen, Y.; Wu, T.; Huang, C.; Baig, M.H.A. Comparison of the Continuity of Vegetation Indices Derived from Landsat 8 OLI and Landsat 7 ETM+ Data among Different Vegetation Types. Remote Sens. 2015, 7, 13485–13506. [Google Scholar] [CrossRef]
- Dong, L. Research on the Biomass Model of Main Tree Species and Stand Types in the Northeast Forest Region. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2015. [Google Scholar]
- Liu, S. Estimation of Forest Biomass in Nanchuan District, Chongqing City Based on Sentinel-1/2. Master’s Thesis, Chengdu University of Technology, Chengdu, China, 2020. [Google Scholar]
- Vogelmann, J.E.; Howard, S.M.; Yang, L.; Larson, C.R.; Wylie, B.K.; Van Driel, N. Completion of the Native American National Land Cover Dataset for the 1990s from LANDSAT Thematic Mapper Data and Auxiliary Data Sources. Photogramm. Eng. Remote Sens. 2001, 67, 650–662. [Google Scholar]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Ohmann, J.L.; Gregory, M.J. Predictive Mapping of Forest Composition and Structure by Direct Gradient Analysis and Nearest Neighbor Imputation in Coastal Oregon, USA. Can. J. For. Res. 2002, 32, 725–741. [Google Scholar] [CrossRef]
- Guo, Z.; Peng, S.; Wang, B. Using TM data to extract forest biomass in western Guangdong. Ecol. J. 2002, 22, 1832–1839+2022. [Google Scholar]





| Factor | B2 | B3 | B4 | B5 | B6 | B7 | |||||
| Correlation coefficient | −0.342 ** | −0.290 ** | −0.302 ** | −0.157 * | −0.337 ** | −0.307 ** | |||||
| Factor | ARVI | DVI | EVI | NDPI | NDVI | RVI | |||||
| Correlation coefficient | 0.250 ** | 0.249 ** | −0.228 ** | 0.323 ** | 0.313 ** | 0.322 ** | |||||
| Factor | SVAI | VH | VV | altitude | slope | canopy closure | |||||
| Correlation coefficient | 0.310 ** | −0.070 | −0.004 | 0.404 ** | −0.015 | 0.328 ** | |||||
| Mean | Variance | Homogeneity | Contrast | Dissimilarity | Entropy | Second Moment | Correlation | ASM | MAX | Energy | |
| B2 | −0.310 ** | −0.056 | 0.069 | −0.036 | −0.063 | −0.087 | 0.083 | 0.107 | |||
| B3 | −0.271 ** | −0.092 | 0.162 * | −0.084 | −0.137 | −0.185 * | 0.184 * | −0.040 | |||
| B4 | −0.286 ** | −0.052 | 0.171 * | −0.068 | −0.163 * | −0.186 * | 0.169 * | −0.007 | |||
| B5 | −0.158 * | −0.046 | 0.004 | 0.001 | −0.006 | 0.023 | −0.063 | 0.004 | |||
| B6 | −0.322 ** | −0.168 * | 0.145 | −0.152 | −0.159 * | −0.211 ** | 0.194 * | 0.002 | |||
| B7 | −0.285 ** | −0.132 | 0.175 * | −0.156 * | −0.174 * | −0.201 * | 0.184 * | 0.037 | |||
| VH | −0.174 * | −0.191 * | −0.197 * | 0.119 | 0.148 | 0.196 * | −0.053 | −0.139 | −0.136 | −0.168 * | |
| VV | −0.126 | −0.148 | −0.143 | 0.065 | 0.093 | 0.241 ** | 0.004 | −0.136 | −0.099 | −0.176 * | |
| Model | Unstandardized Coefficient | Standardized Coefficient | t | Sig. | |
|---|---|---|---|---|---|
| B | Standard Error | ||||
| Constant | −243.422 | 126.015 | −1.932 | 0.055 | |
| Altitude | 0.090 | 0.025 | 0.276 | 3.533 | 0.001 |
| Canopy closure | 125.943 | 33.185 | 0.267 | 3.795 | 0.000 |
| ARVI | −156.917 | 64.266 | −0.472 | −2.442 | 0.016 |
| EVI | 73.454 | 35.773 | 0.236 | 2.053 | 0.042 |
| RVI | 148.340 | 43.052 | 0.676 | 3.446 | 0.001 |
| VVEntropy | 34.447 | 18.391 | 0.127 | 1.873 | 0.063 |
| B6Mean | 24.306 | 13.932 | 0.417 | 1.745 | 0.083 |
| B6 | −245.049 | 93.419 | −0.656 | −2.623 | 0.010 |
| Model | R2 | Time |
|---|---|---|
| Stepwise regression model | 0.5468 | 0.0862 s |
| BP neural network model | 0.78226 | 0.13 s |
| POS improved neural network model | 0.83204 | 235.16 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Ni, H. Inversion of Forest Biomass Based on Multi-Source Remote Sensing Images. Sensors 2023, 23, 9313. https://doi.org/10.3390/s23239313
Zhang D, Ni H. Inversion of Forest Biomass Based on Multi-Source Remote Sensing Images. Sensors. 2023; 23(23):9313. https://doi.org/10.3390/s23239313
Chicago/Turabian StyleZhang, Danhua, and Hui Ni. 2023. "Inversion of Forest Biomass Based on Multi-Source Remote Sensing Images" Sensors 23, no. 23: 9313. https://doi.org/10.3390/s23239313
APA StyleZhang, D., & Ni, H. (2023). Inversion of Forest Biomass Based on Multi-Source Remote Sensing Images. Sensors, 23(23), 9313. https://doi.org/10.3390/s23239313




