Deep-Learning-Aided Evaluation of Spondylolysis Imaged with Ultrashort Echo Time Magnetic Resonance Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Imaging Data
2.2. Deep Learning: Image Regression Model
Matlab Code for Image Regression
- % data directories
- imageDir = “directory for MRI images”
- labelDir = “directory for CT images”
- % create image and label datastores
- imds = imageDatastore(imageDir)
- pxds = imageDatastore(labelDir)
- % combined training datastore
- dsTrain = combine(imds,pxds)
- % create the baseline U-net
- imageSize = [384 224]
- encoderDepth = 5;
- network1 = unetLayers(imageSize,2,‘EncoderDepth’,encoderDepth)
- % create 2D convolution and regression layers to replace base Unet
- layer_conv = “2D convolution layer with 1 channel output”
- layer_reg = “regression layer with 1 channel output”
- % remove softmax and segmentation layers and replace with regression layer
- network2 = “command to remove segmentation layer from network1”
- network2 = “command to replace final 2D convolution layer with layer_conv”
- network2 = “command to replace softmax layer with layer_reg”
- % change options to match your PC hardware
- train_options = trainingOptions(‘adam’,…
- ‘LearnRateDropFactor’,0.05, …
- ‘LearnRateDropPeriod’,5, …
- ‘Shuffle’,’every-epoch’,…
- ‘MaxEpochs’,100, …
- ‘MiniBatchSize’,8); %, …
- % start training
- Regression_Network = trainNetwork (dsTrain, network2, train_options)
- %%%%%%%% inference post training %%%%%%%%
- CT_like_image = predict(Trained_Network, input_MRI)
2.3. Deep Learning: Saliency Mapping Model
Matlab Code for Saliency Mapping
- % data directories
- imageDir = “directory for MRI images”
- labelDir = “directory for annotations for pars defects”
- % create image and label datastores
- imds = imageDatastore(imageDir)
- pxds = imageDatastore(labelDir)
- % combined training datastore
- dsTrain = combine(imds,pxds)
- % create the baseline U-net
- imageSize = [384 224]
- encoderDepth = 5;
- network1 = unetLayers(imageSize,2,‘EncoderDepth’,encoderDepth)
- % add class weights since the volume of defect is very small
- tbl = countEachLabel(pxds);
- numberPixels = tbl.ImagePixelCount;
- frequency = tbl.PixelCount ./ numberPixels;
- classWeights = 1 ./ frequency;
- % replace the last layer with a weighted classification layer
- layer1 = “segmentation layer with weighted class weights”
- network2 = “command to replace the last segmentation layer with layer1”
- % change options to match your PC hardware
- train_options = trainingOptions(‘adam’,…
- ‘InitialLearnRate’,1e-5,…
- ‘Shuffle’,’every-epoch’,…
- ‘Verbose’,true,…
- ‘MaxEpochs’,100, …
- ‘MiniBatchSize’,8);
- % start training
- Saliency_Network = trainNetwork (dsTrain, network2, train_options)
- %%%%%%%% saliency mapping post training %%%%%%%%
- testimg = imread(“test image”);
- act = activations(net1,testimg,’Softmax-Layer’); % “see” the activation layer
- figure
- imagesc(act(:,:,2))
2.4. Outcome Measures
2.4.1. Similarity Measures between CT and CT-like Images
2.4.2. Contrast-to-Noise Ratio (CNR)
2.4.3. Width Measurement of Pars Defects
2.5. Statistics
3. Results
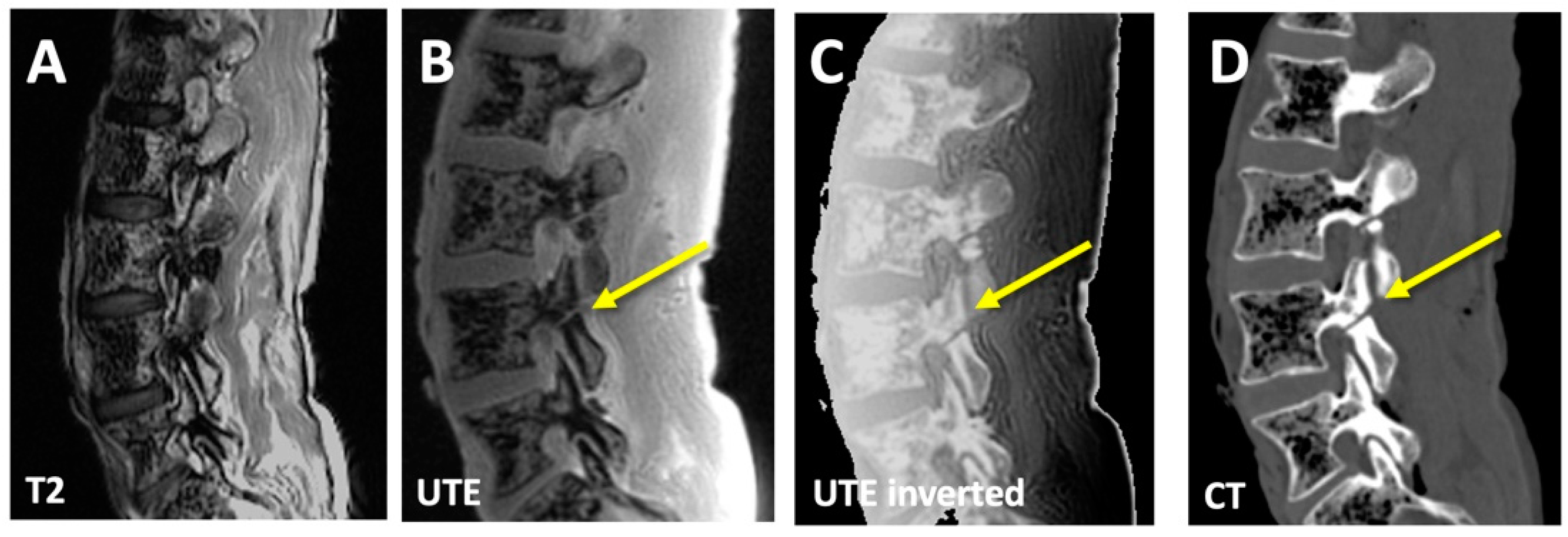
3.1. CT-like Images
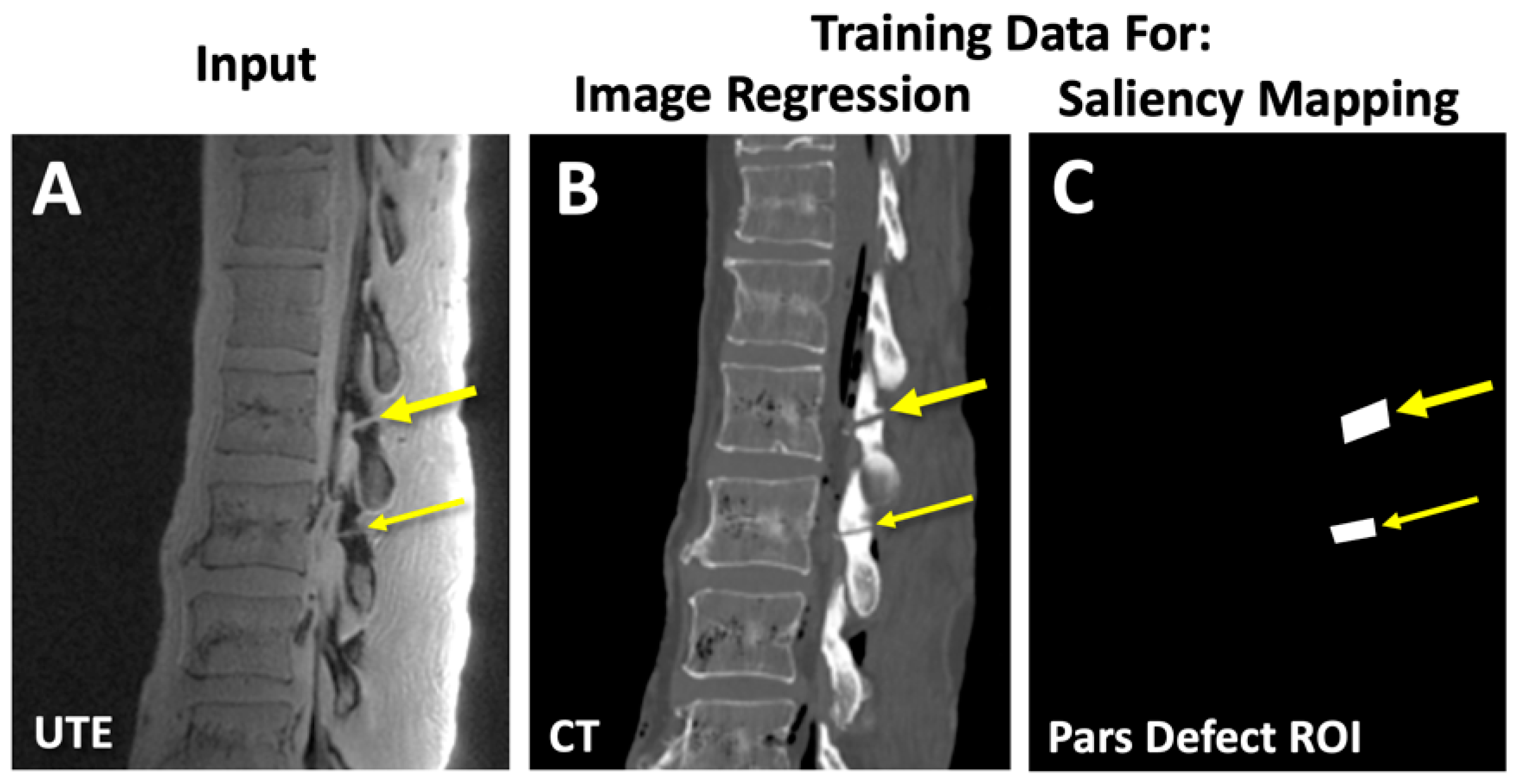
3.2. Saliency Mapping
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wiltse, L.L.; Newman, P.H.; Macnab, I. Classification of spondylolisis and spondylolisthesis. Clin. Orthop. Relat. Res. 1976, 117, 23–29. [Google Scholar] [CrossRef]
- Fredrickson, B.E.; Baker, D.; McHolick, W.J.; Yuan, H.A.; Lubicky, J.P. The natural history of spondylolysis and spondylolisthesis. J. Bone Jt. Surg. Am. 1984, 66, 699–707. [Google Scholar] [CrossRef]
- Bechtel, W.; Griffiths, H.; Eisenstadt, R. The Pathogenesis of Spondylolysis. Investig. Radiol. 1982, 17, S29. [Google Scholar] [CrossRef]
- Micheli, L.J.; Wood, R. Back pain in young athletes: Significant differences from adults in causes and patterns. Arch. Pediatr. Adolesc. Med. 1995, 149, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.L.; Anderson, R.L.; Dearwater, S.R.; Kriska, A.M.; Cauley, J.A.; Aaron, D.J.; LaPorte, R.E. The epidemiology of low back pain in an adolescent population. Am. J. Public Health 1992, 82, 606–608. [Google Scholar] [CrossRef]
- Selhorst, M.; MacDonald, J.; Martin, L.C.; Rodenberg, R.; Krishnamurthy, R.; Ravindran, R.; Fischer, A. Immediate functional progression program in adolescent athletes with a spondylolysis. Phys. Ther. Sport 2021, 52, 140–146. [Google Scholar] [CrossRef]
- Soler, T.; Calderon, C. The prevalence of spondylolysis in the Spanish elite athlete. Am. J. Sports Med. 2000, 28, 57–62. [Google Scholar] [CrossRef]
- Reitman, C.A.; Gertzbein, S.D.; Francis, W.R., Jr. Lumbar isthmic defects in teenagers resulting from stress fractures. Spine J. 2002, 2, 303–306. [Google Scholar] [CrossRef]
- Deyo, R.A.; Weinstein, J.N. Low back pain. N. Engl. J. Med. 2001, 344, 363–370. [Google Scholar] [CrossRef]
- Lim, M.R.; Yoon, S.C.; Green, D.W. Symptomatic spondylolysis: Diagnosis and treatment. Curr. Opin. Pediatr. 2004, 16, 37–46. [Google Scholar] [CrossRef]
- McCleary, M.D.; Congeni, J.A. Current concepts in the diagnosis and treatment of spondylolysis in young athletes. Curr. Sports Med. Rep. 2007, 6, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.W.; Wiltse, L.L.; Dingeman, R.D.; Hayes, M. Stress reactions involving the pars interarticularis in young athletes. Am. J. Sports Med. 1981, 9, 304–312. [Google Scholar] [CrossRef]
- Masci, L.; Pike, J.; Malara, F.; Phillips, B.; Bennell, K.; Brukner, P. Use of the one-legged hyperextension test and magnetic resonance imaging in the diagnosis of active spondylolysis. Br. J. Sports Med. 2006, 40, 940–946; discussion 946. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Kobayashi, T.; Kato, K.; Higuchi, H.; Takagishi, K. Diagnosis of radiographically occult lumbar spondylolysis in young athletes by magnetic resonance imaging. Am. J. Sports Med. 2013, 41, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Beck, N.A.; Sampson, N.R.; Zhu, X.; Flynn, J.M.; Drummond, D. Imaging modalities for low back pain in children: A review of spondyloysis and undiagnosed mechanical back pain. J. Pediatr. Orthop. 2013, 33, 282–288. [Google Scholar] [CrossRef]
- West, A.M.; d’Hemecourt, P.A.; Bono, O.J.; Micheli, L.J.; Sugimoto, D. Diagnostic Accuracy of Magnetic Resonance Imaging and Computed Tomography Scan in Young Athletes With Spondylolysis. Clin Pediatr. 2019, 58, 671–676. [Google Scholar] [CrossRef]
- Yamane, T.; Yoshida, T.; Mimatsu, K. Early diagnosis of lumbar spondylolysis by MRI. J. Bone Jt. Surg. Br. 1993, 75, 764–768. [Google Scholar] [CrossRef]
- Ganiyusufoglu, A.K.; Onat, L.; Karatoprak, O.; Enercan, M.; Hamzaoglu, A. Diagnostic accuracy of magnetic resonance imaging versus computed tomography in stress fractures of the lumbar spine. Clin. Radiol. 2010, 65, 902–907. [Google Scholar] [CrossRef]
- Little, C.B.; Mittaz, L.; Belluoccio, D.; Rogerson, F.M.; Campbell, I.K.; Meeker, C.T.; Bateman, J.F.; Pritchard, M.A.; Fosang, A.J. ADAMTS-1-knockout mice do not exhibit abnormalities in aggrecan turnover in vitro or in vivo. Arthritis Rheum. 2005, 52, 1461–1472. [Google Scholar] [CrossRef]
- Dhouib, A.; Tabard-Fougere, A.; Hanquinet, S.; Dayer, R. Diagnostic accuracy of MR imaging for direct visualization of lumbar pars defect in children and young adults: A systematic review and meta-analysis. Eur. Spine J. 2018, 27, 1058–1066. [Google Scholar] [CrossRef]
- Yamaguchi, K.T., Jr.; Skaggs, D.L.; Acevedo, D.C.; Myung, K.S.; Choi, P.; Andras, L. Spondylolysis is frequently missed by MRI in adolescents with back pain. J. Child. Orthop. 2012, 6, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, J.L.; Mathews, V.P.; Elster, A.D.; Mark, L.P.; Daniels, D.L.; Mueller, W. MR imaging of lumbar spondylolysis: The importance of ancillary observations. AJR Am. J. Roentgenol. 1997, 169, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Dunn, A.J.; Campbell, R.S.; Mayor, P.E.; Rees, D. Radiological findings and healing patterns of incomplete stress fractures of the pars interarticularis. Skelet. Radiol. 2008, 37, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Rush, J.K.; Astur, N.; Scott, S.; Kelly, D.M.; Sawyer, J.R.; Warner, W.C., Jr. Use of magnetic resonance imaging in the evaluation of spondylolysis. J. Pediatr. Orthop. 2015, 35, 271–275. [Google Scholar] [CrossRef]
- Williams, A.; Qian, Y.; Golla, S.; Chu, C.R. UTE-T2 * mapping detects sub-clinical meniscus injury after anterior cruciate ligament tear. Osteoarthr. Cartil. 2012, 20, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Springer, F.; Steidle, G.; Martirosian, P.; Syha, R.; Claussen, C.D.; Schick, F. Rapid assessment of longitudinal relaxation time in materials and tissues with extremely fast signal decay using UTE sequences and the variable flip angle method. Investig. Radiol. 2011, 46, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Finkenstaedt, T.; Siriwanarangsun, P.; Achar, S.; Carl, M.; Finkenstaedt, S.; Abeydeera, N.; Chung, C.B.; Bae, W.C. Ultrashort Time-to-Echo Magnetic Resonance Imaging at 3 T for the Detection of Spondylolysis in Cadaveric Spines: Comparison With CT. Investig. Radiol. 2019, 54, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.D.; Gatehouse, P.D.; Bydder, M.; Bydder, G.M. Magnetic resonance: An introduction to ultrashort TE (UTE) imaging. J. Comput. Assist. Tomogr. 2003, 27, 825–846. [Google Scholar] [CrossRef]
- Techawiboonwong, A.; Song, H.K.; Wehrli, F.W. In vivo MRI of submillisecond T(2) species with two-dimensional and three-dimensional radial sequences and applications to the measurement of cortical bone water. NMR Biomed. 2008, 21, 59–70. [Google Scholar] [CrossRef]
- Wu, Y.; Dai, G.; Ackerman, J.L.; Hrovat, M.I.; Glimcher, M.J.; Snyder, B.D.; Nazarian, A.; Chesler, D.A. Water- and fat-suppressed proton projection MRI (WASPI) of rat femur bone. Magn. Reson. Med. 2007, 57, 554–567. [Google Scholar] [CrossRef]
- Rahmer, J.; Bornert, P.; Groen, J.; Bos, C. Three-dimensional radial ultrashort echo-time imaging with T2 adapted sampling. Magn. Reson. Med. 2006, 55, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Weiger, M.; Pruessmann, K.P.; Hennel, F. MRI with zero echo time: Hard versus sweep pulse excitation. Magn. Reson. Med. 2011, 66, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Boada, F.E. Acquisition-weighted stack of spirals for fast high-resolution three-dimensional ultra-short echo time MR imaging. Magn. Reson. Med. 2008, 60, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Idiyatullin, D.; Corum, C.; Park, J.Y.; Garwood, M. Fast and quiet MRI using a swept radiofrequency. J. Magn. Reson. 2006, 181, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Carl, M.; Bae, W.C.; Statum, S.; Chang, E.Y.; Bydder, G.M.; Chung, C.B. Dual inversion recovery ultrashort echo time (DIR-UTE) imaging and quantification of the zone of calcified cartilage (ZCC). Osteoarthr. Cartil. 2012, 21, 77–85. [Google Scholar] [CrossRef]
- Bae, W.C.; Biswas, R.; Chen, K.; Chang, E.Y.; Chung, C.B. UTE MRI of the Osteochondral Junction. Curr. Radiol. Rep. 2014, 2, 35. [Google Scholar] [CrossRef]
- Bae, W.C.; Chen, P.C.; Chung, C.B.; Masuda, K.; D’Lima, D.; Du, J. Quantitative ultrashort echo time (UTE) MRI of human cortical bone: Correlation with porosity and biomechanical properties. J. Bone Min. Res. 2012, 27, 848–857. [Google Scholar] [CrossRef]
- Bharadwaj, U.U.; Coy, A.; Motamedi, D.; Sun, D.; Joseph, G.B.; Krug, R.; Link, T.M. CT-like MRI: A qualitative assessment of ZTE sequences for knee osseous abnormalities. Skelet. Radiol. 2022, 51, 1585–1594. [Google Scholar] [CrossRef]
- Cheng, K.Y.; Moazamian, D.; Ma, Y.; Jang, H.; Jerban, S.; Du, J.; Chung, C.B. Clinical application of ultrashort echo time (UTE) and zero echo time (ZTE) magnetic resonance (MR) imaging in the evaluation of osteoarthritis. Skelet. Radiol. 2024. online ahead of print. [Google Scholar] [CrossRef]
- Geiger, D.; Bae, W.C.; Statum, S.; Du, J.; Chung, C.B. Quantitative 3D ultrashort time-to-echo (UTE) MRI and micro-CT (muCT) evaluation of the temporomandibular joint (TMJ) condylar morphology. Skelet. Radiol. 2014, 43, 19–25. [Google Scholar] [CrossRef]
- Sun, H.; Xi, Q.; Fan, R.; Sun, J.; Xie, K.; Ni, X.; Yang, J. Synthesis of pseudo-CT images from pelvic MRI images based on an MD-CycleGAN model for radiotherapy. Phys. Med. Biol. 2022, 67, 035006. [Google Scholar] [CrossRef] [PubMed]
- Dovletov, G.; Pham, D.D.; Lorcks, S.; Pauli, J.; Gratz, M.; Quick, H.H. Grad-CAM Guided U-Net for MRI-based Pseudo-CT Synthesis. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2022, 2022, 2071–2075. [Google Scholar] [CrossRef]
- Wiesinger, F.; Bylund, M.; Yang, J.; Kaushik, S.; Shanbhag, D.; Ahn, S.; Jonsson, J.H.; Lundman, J.A.; Hope, T.; Nyholm, T.; et al. Zero TE-based pseudo-CT image conversion in the head and its application in PET/MR attenuation correction and MR-guided radiation therapy planning. Magn. Reson. Med. 2018, 80, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Burgos, N.; Guerreiro, F.; McClelland, J.; Presles, B.; Modat, M.; Nill, S.; Dearnaley, D.; deSouza, N.; Oelfke, U.; Knopf, A.C.; et al. Iterative framework for the joint segmentation and CT synthesis of MR images: Application to MRI-only radiotherapy treatment planning. Phys. Med. Biol. 2017, 62, 4237–4253. [Google Scholar] [CrossRef] [PubMed]
- Eshraghi Boroojeni, P.; Chen, Y.; Commean, P.K.; Eldeniz, C.; Skolnick, G.B.; Merrill, C.; Patel, K.B.; An, H. Deep-learning synthesized pseudo-CT for MR high-resolution pediatric cranial bone imaging (MR-HiPCB). Magn. Reson. Med. 2022, 88, 2285–2297. [Google Scholar] [CrossRef]
- Bourbonne, V.; Jaouen, V.; Hognon, C.; Boussion, N.; Lucia, F.; Pradier, O.; Bert, J.; Visvikis, D.; Schick, U. Dosimetric Validation of a GAN-Based Pseudo-CT Generation for MRI-Only Stereotactic Brain Radiotherapy. Cancers 2021, 13, 1082. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, D.; Van Leemput, K.; Hansen, R.H.; Andersen, J.A.; Edmund, J.M. Patch-based generation of a pseudo CT from conventional MRI sequences for MRI-only radiotherapy of the brain. Med. Phys. 2015, 42, 1596–1605. [Google Scholar] [CrossRef]
- Johnstone, E.; Wyatt, J.J.; Henry, A.M.; Short, S.C.; Sebag-Montefiore, D.; Murray, L.; Kelly, C.G.; McCallum, H.M.; Speight, R. Systematic Review of Synthetic Computed Tomography Generation Methodologies for Use in Magnetic Resonance Imaging-Only Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 199–217. [Google Scholar] [CrossRef]
- Dowling, J.A.; Sun, J.; Pichler, P.; Rivest-Henault, D.; Ghose, S.; Richardson, H.; Wratten, C.; Martin, J.; Arm, J.; Best, L.; et al. Automatic Substitute Computed Tomography Generation and Contouring for Magnetic Resonance Imaging (MRI)-Alone External Beam Radiation Therapy From Standard MRI Sequences. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 1144–1153. [Google Scholar] [CrossRef]
- Sjolund, J.; Forsberg, D.; Andersson, M.; Knutsson, H. Generating patient specific pseudo-CT of the head from MR using atlas-based regression. Phys. Med. Biol. 2015, 60, 825–839. [Google Scholar] [CrossRef]
- Korhonen, J.; Kapanen, M.; Keyrilainen, J.; Seppala, T.; Tenhunen, M. A dual model HU conversion from MRI intensity values within and outside of bone segment for MRI-based radiotherapy treatment planning of prostate cancer. Med. Phys. 2014, 41, 011704. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Karlsson, M.; Nyholm, T. CT substitute derived from MRI sequences with ultrashort echo time. Med. Phys. 2011, 38, 2708–2714. [Google Scholar] [CrossRef] [PubMed]
- Largent, A.; Barateau, A.; Nunes, J.C.; Mylona, E.; Castelli, J.; Lafond, C.; Greer, P.B.; Dowling, J.A.; Baxter, J.; Saint-Jalmes, H.; et al. Comparison of Deep Learning-Based and Patch-Based Methods for Pseudo-CT Generation in MRI-Based Prostate Dose Planning. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Kalmet, P.H.S.; Sanduleanu, S.; Primakov, S.; Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Hulst, L.V.; Lambin, P.; Poeze, M. Deep learning in fracture detection: A narrative review. Acta Orthop. 2020, 91, 215–220. [Google Scholar] [CrossRef]
- Jenkinson, M.; Smith, S.M. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001, 5, 143–156. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Spriinger: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Eckert, M.P.; Bradley, A.P. Perceptual quality metrics applied to still image compression. Signal Process. 1998, 70, 177–200. [Google Scholar] [CrossRef]
- Zhou, W.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar]
- Edelstein, W.A.; Bottomley, P.A.; Hart, H.R.; Leue, W.M.; Schenck, J.F.; Redington, R.W. NMR imaging at 5.1 MHz: Work in progress. In Proceedings of the International Symposium on NMR Imaging, Society of Nuclear Medicine and Molecular Imaging, Winston-Salem, NC, USA, 1 November 1982; Witcofski, R., Karstaedt, N., Partain, C.L., Eds.; Bowman Gray School of Medicine: Winston-Salem, NC, USA, 1982; pp. 139–145. [Google Scholar]
- Li, J.; Wu, L.H.; Xu, M.Y.; Ren, J.L.; Li, Z.; Liu, J.R.; Wang, A.J.; Chen, B. Improving Image Quality and Reducing Scan Time for Synthetic MRI of Breast by Using Deep Learning Reconstruction. Biomed. Res. Int. 2022, 2022, 3125426. [Google Scholar] [CrossRef]
- Massa, H.A.; Johnson, J.M.; McMillan, A.B. Comparison of deep learning synthesis of synthetic CTs using clinical MRI inputs. Phys. Med. Biol. 2020, 65, 23NT03. [Google Scholar] [CrossRef]
- Li, Y.; Li, W.; Xiong, J.; Xia, J.; Xie, Y. Comparison of Supervised and Unsupervised Deep Learning Methods for Medical Image Synthesis between Computed Tomography and Magnetic Resonance Images. Biomed. Res. Int. 2020, 2020, 5193707. [Google Scholar] [CrossRef]
- Xiang, L.; Wang, Q.; Nie, D.; Zhang, L.; Jin, X.; Qiao, Y.; Shen, D. Deep embedding convolutional neural network for synthesizing CT image from T1-Weighted MR image. Med. Image Anal. 2018, 47, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Emami, H.; Dong, M.; Nejad-Davarani, S.P.; Glide-Hurst, C.K. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med. Phys. 2018, 45, 3627–3636. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.B.; Kim, H.; Liu, M.; Jung, W.; Joo, S.; Park, E.; Ahn, Y.S.; Han, I.H.; Lee, J.I.; Cui, X. Deep CT to MR Synthesis Using Paired and Unpaired Data. Sensors 2019, 19, 2361. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Harms, J.; Wang, T.; Liu, Y.; Shu, H.K.; Jani, A.B.; Curran, W.J.; Mao, H.; Liu, T.; Yang, X. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med. Phys. 2019, 46, 3565–3581. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Sun, J.; Carass, A.; Zhao, C.; Lee, J.; Prince, J.L.; Xu, Z. Unsupervised MR-to-CT Synthesis Using Structure-Constrained CycleGAN. IEEE Trans. Med. Imaging 2020, 39, 4249–4261. [Google Scholar] [CrossRef]
- Lakshmanaprabu, S.K.; Mohanty, S.N.; Shankar, K.; Arunkumar, N.; Ramirez, G. Optimal deep learning model for classification of lung cancer on CT images. In Future Generation Computer Systems; Elsevier: Amsterdam, The Netherlands, 2019; Volume 92, pp. 374–382. ISSN 0167-739X. [Google Scholar] [CrossRef]





| Inverted UTE vs. CT | CT-like vs. CT | p-Value | |
|---|---|---|---|
| MSE | 0.0528 ± 0.0340 | 0.0038 ± 0.0054 | 2.60 × 10−133 |
| PSNR (dB) | 16.5 ± 14.2 | 28.6 ± 6.1 | 6.60 × 10−53 |
| SSIM | 0.68 ± 0.19 | 0.73 ± 0.28 | 0.0012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Achar, S.; Hwang, D.; Finkenstaedt, T.; Malis, V.; Bae, W.C. Deep-Learning-Aided Evaluation of Spondylolysis Imaged with Ultrashort Echo Time Magnetic Resonance Imaging. Sensors 2023, 23, 8001. https://doi.org/10.3390/s23188001
Achar S, Hwang D, Finkenstaedt T, Malis V, Bae WC. Deep-Learning-Aided Evaluation of Spondylolysis Imaged with Ultrashort Echo Time Magnetic Resonance Imaging. Sensors. 2023; 23(18):8001. https://doi.org/10.3390/s23188001
Chicago/Turabian StyleAchar, Suraj, Dosik Hwang, Tim Finkenstaedt, Vadim Malis, and Won C. Bae. 2023. "Deep-Learning-Aided Evaluation of Spondylolysis Imaged with Ultrashort Echo Time Magnetic Resonance Imaging" Sensors 23, no. 18: 8001. https://doi.org/10.3390/s23188001
APA StyleAchar, S., Hwang, D., Finkenstaedt, T., Malis, V., & Bae, W. C. (2023). Deep-Learning-Aided Evaluation of Spondylolysis Imaged with Ultrashort Echo Time Magnetic Resonance Imaging. Sensors, 23(18), 8001. https://doi.org/10.3390/s23188001





