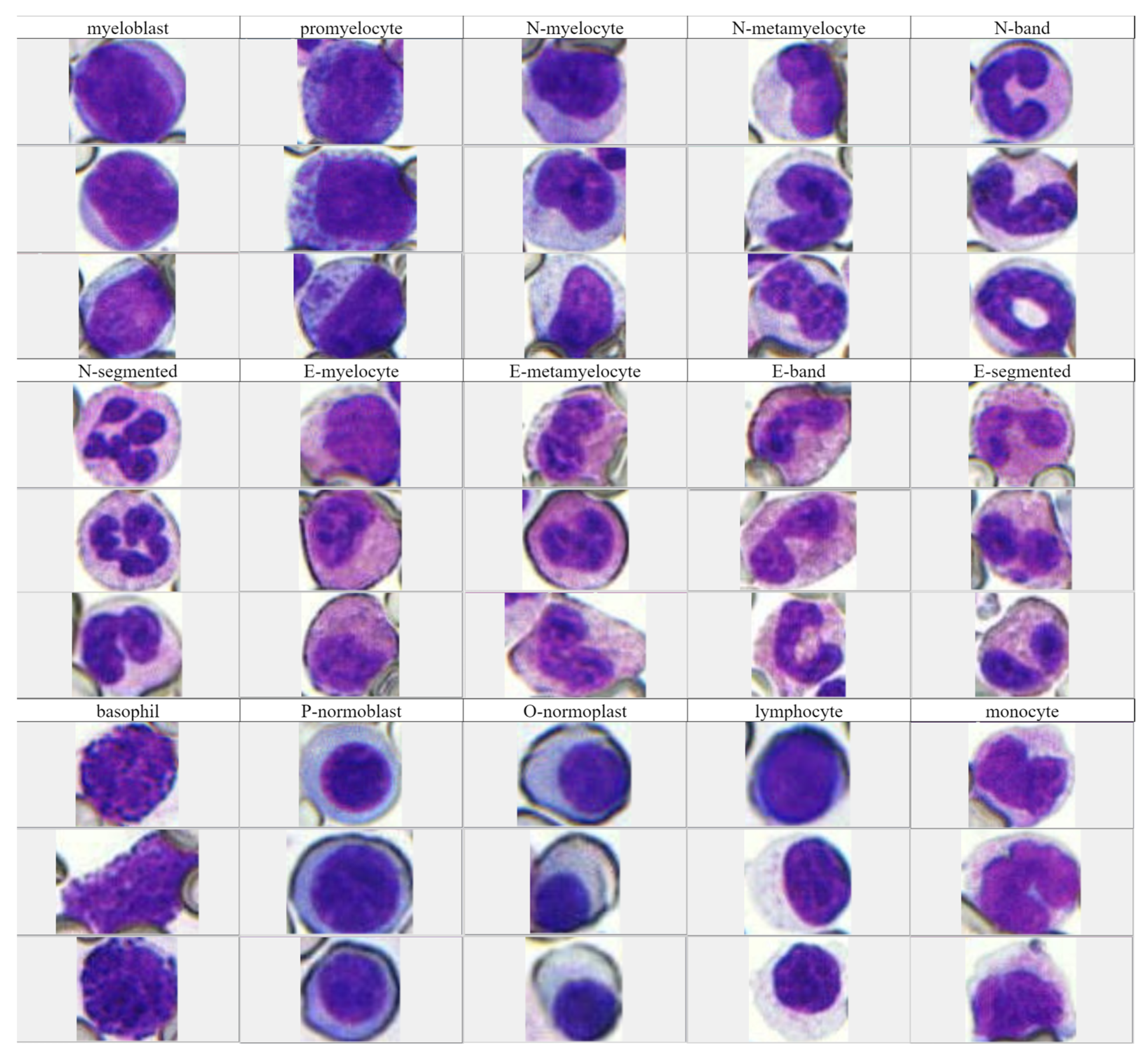
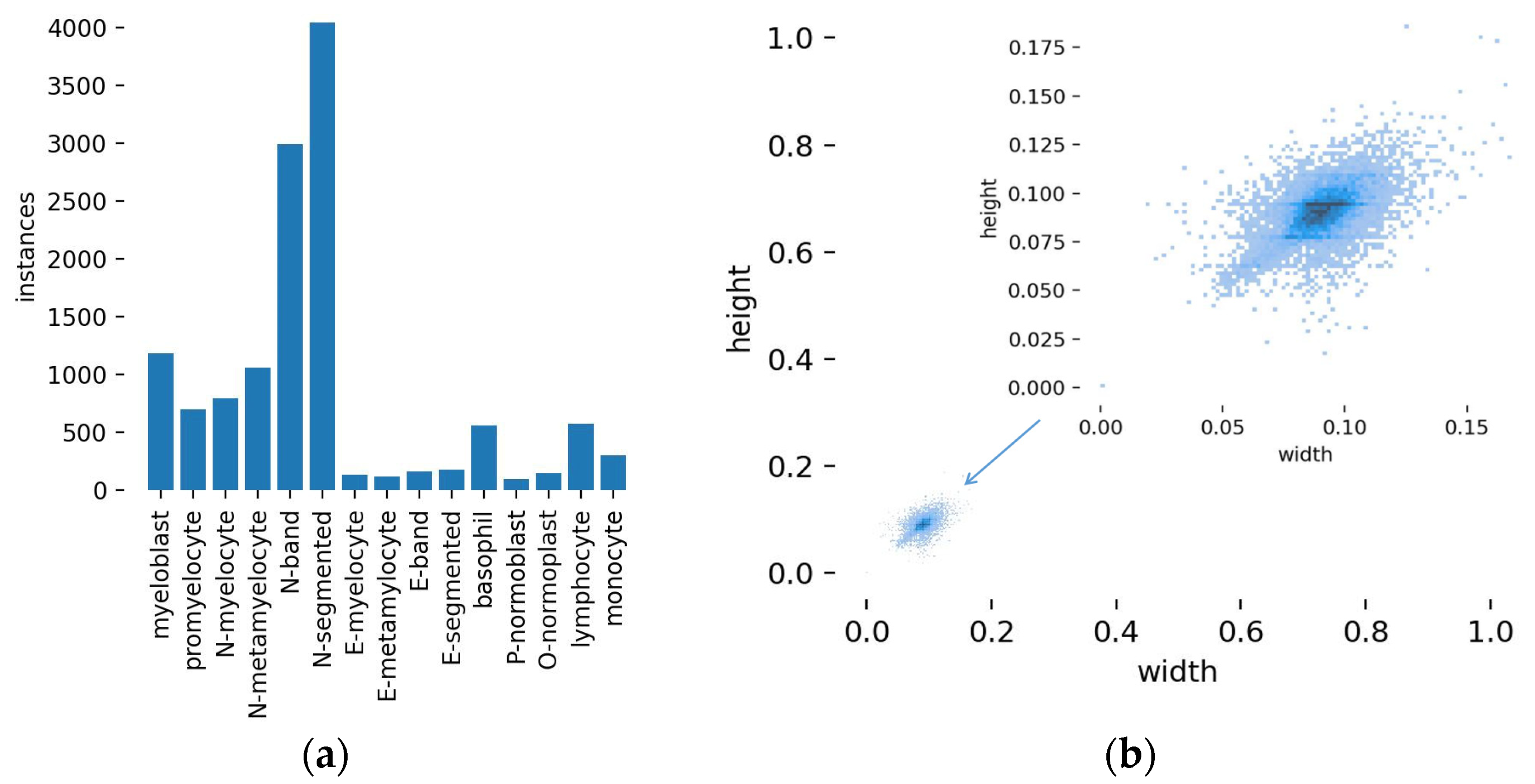
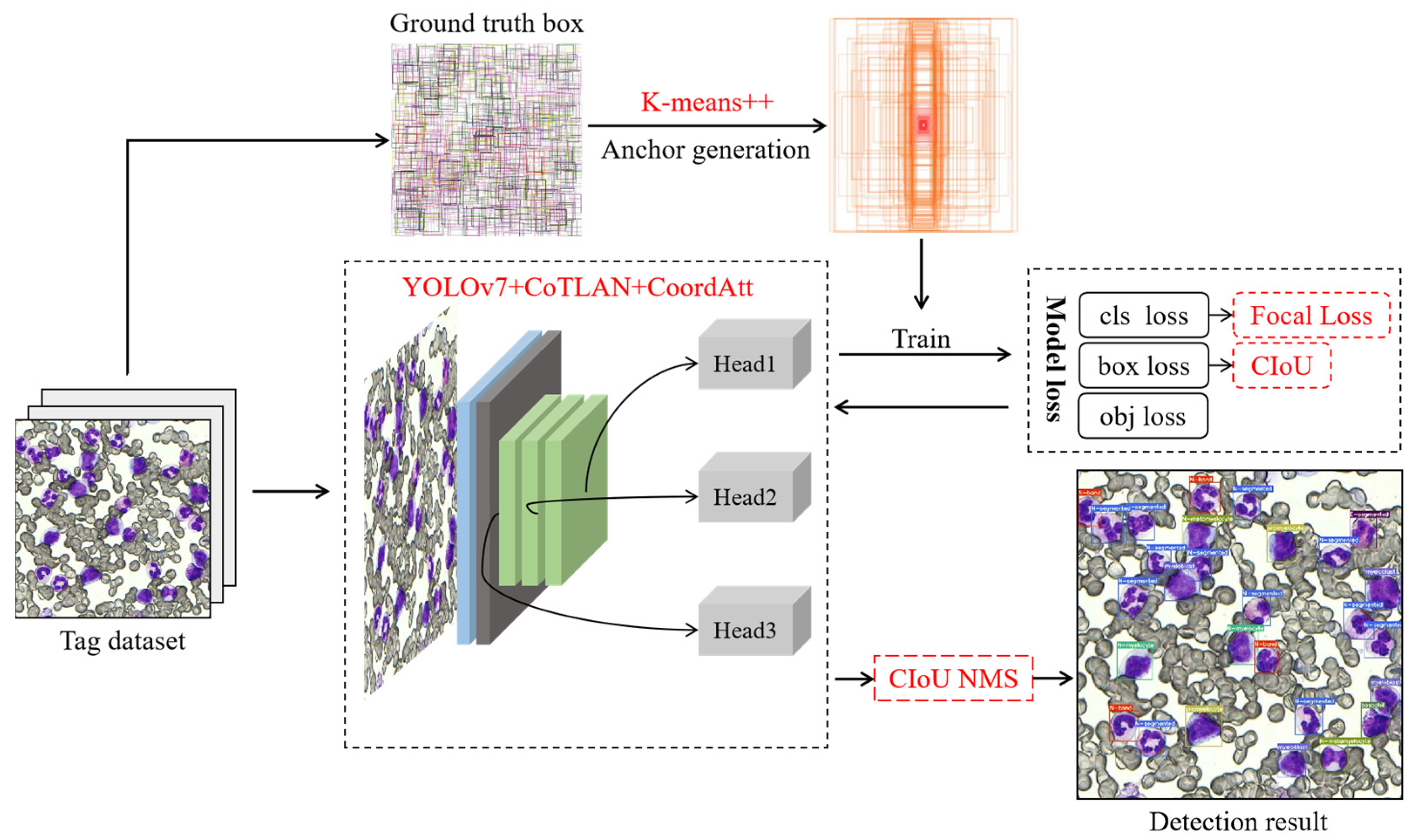
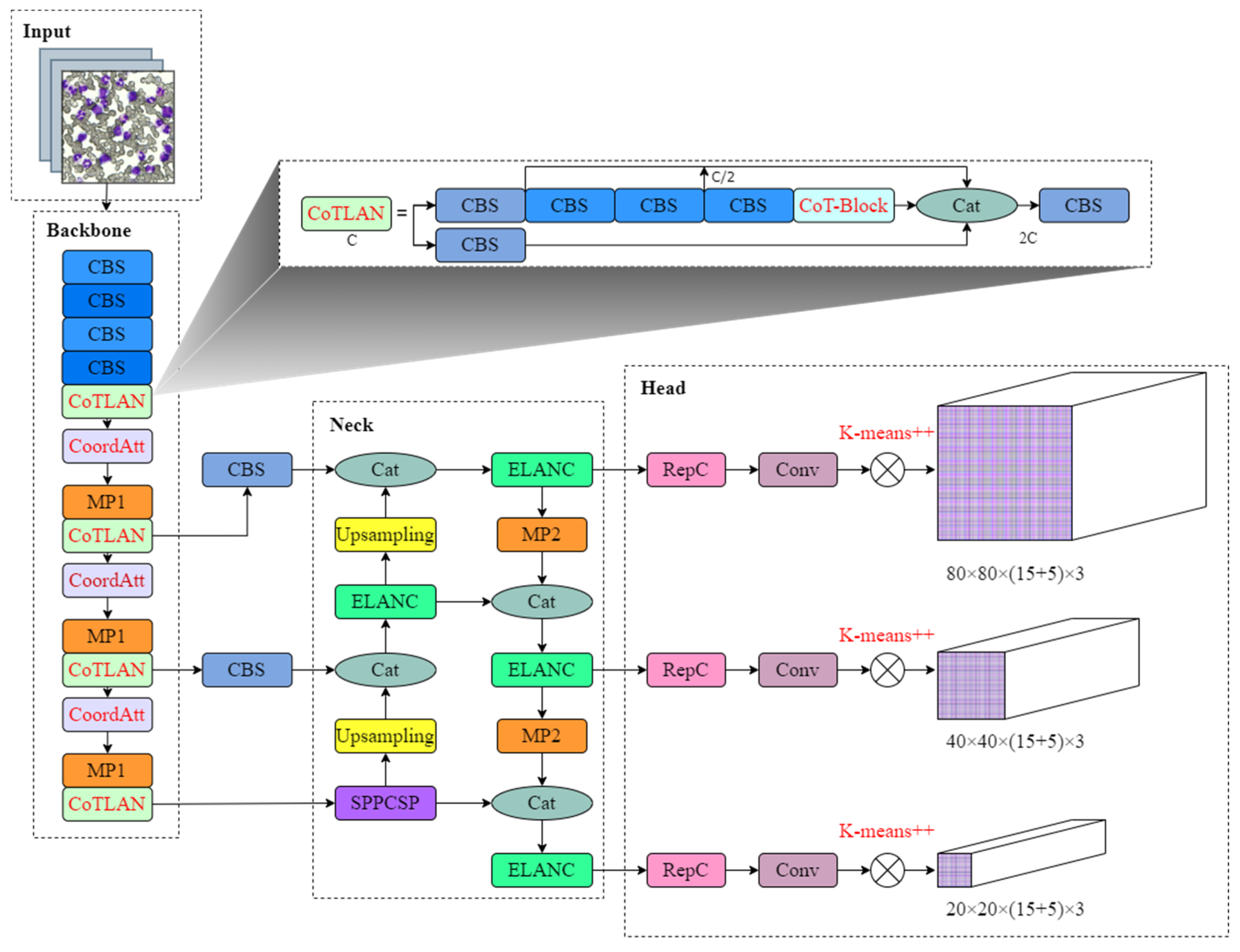
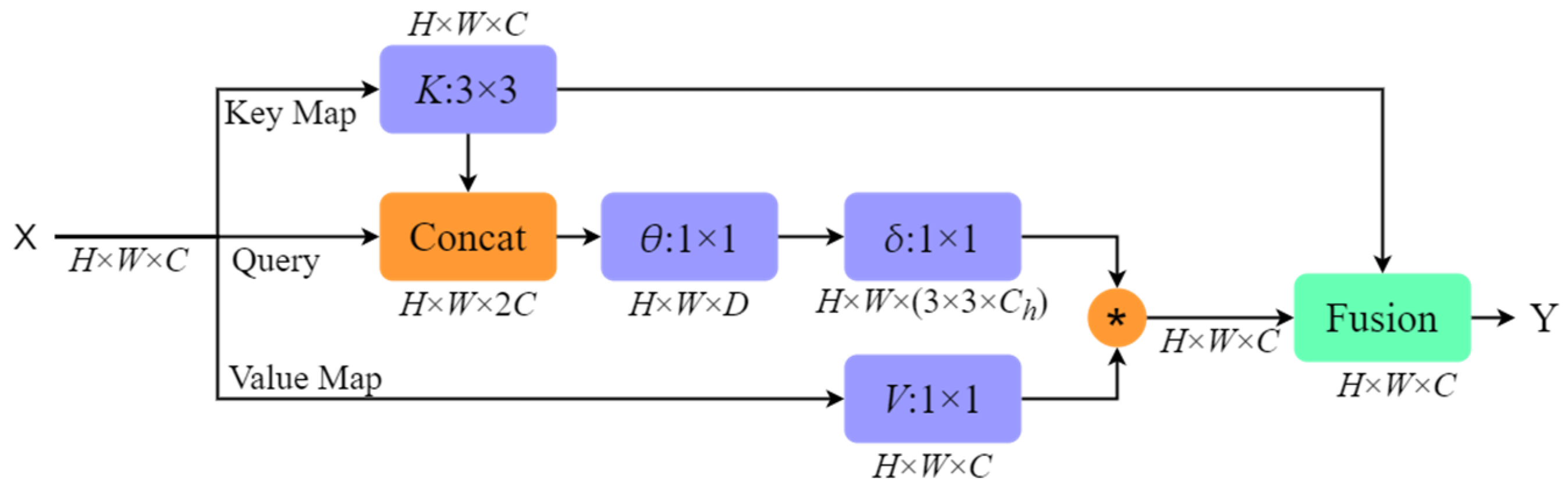
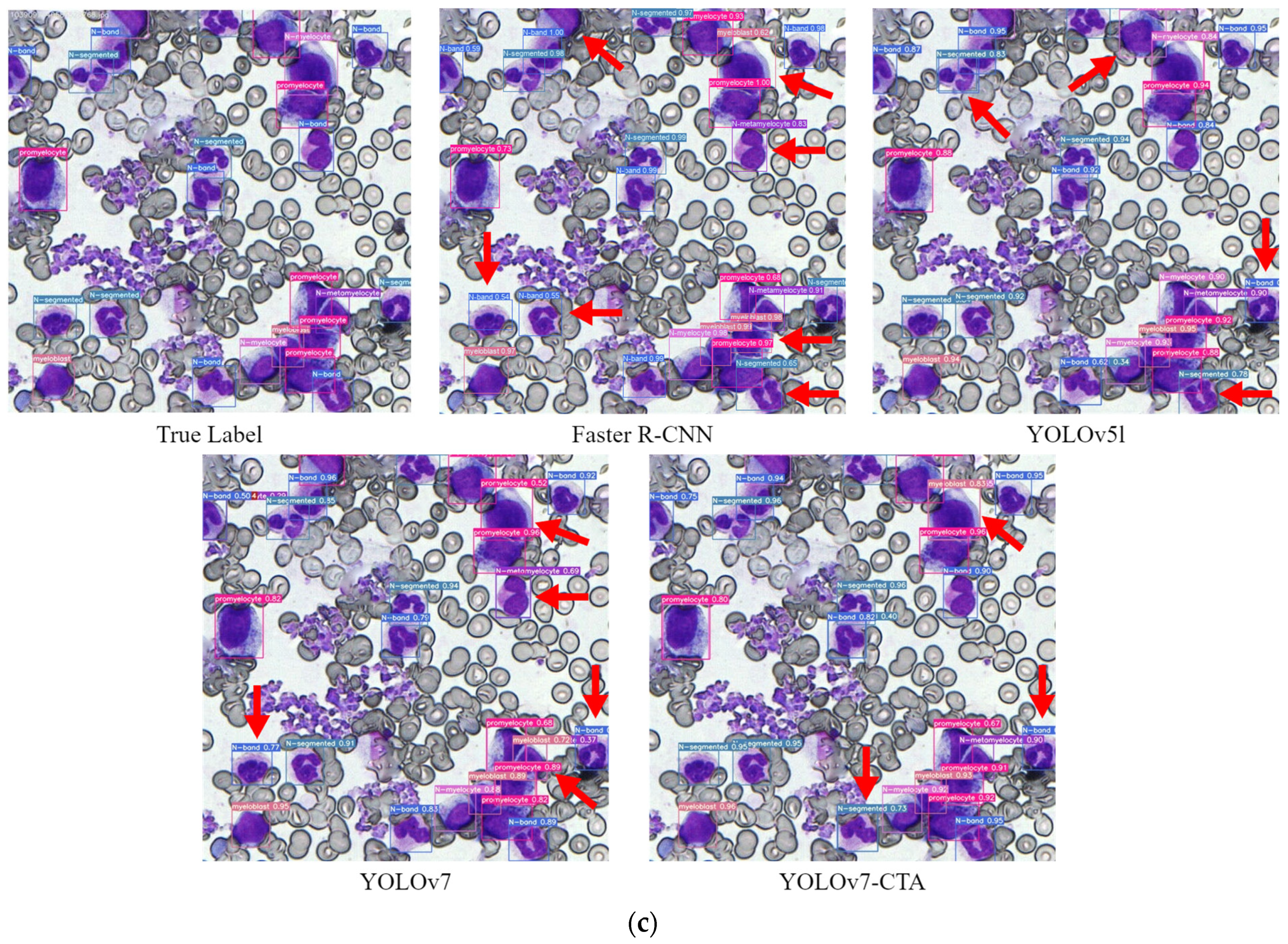
3.2. Experimental Results and Analysis
Firstly, we conducted comparative experiments and ablation experiments to improve the model network structure using the YOLOv7 model as the baseline. The comparative experiment analyzed three of the most popular attention mechanisms to identify the optimal one, while the ablation experiment aimed to validate the effectiveness of the proposed model-structure improvement method. Secondly, we used the improved YOLOv7-CTA model as a baseline to determine a suitable localization loss function, and conducted ablation experiments on the setting of anchor box hyperparameters and the use of the improved loss function to further optimize the model. Finally, we compared the improved model’s performance with the original model and other classic models and verified its superiority. Since the main purpose of this paper is to improve the detection performance of the model, we used metrics such as precision, recall, mAP, and FPS (Frames Per Second) to evaluate the results.
To incorporate suitable attention mechanisms into the network, this paper added SE, CBAM, and CoordAtt attention mechanisms between ELAN modules for training and comparison. The results of the experiments are presented in
Table 3.
The impact of adding different attention mechanisms to the backbone of model detection varies. While the SE attention mechanism ignores positional information, it considers channel attention. The result shows that adding SE attention to the backbone improves the mAP, but it improves the least. The CBAM attention aims to leverage positional information by reducing the input tensor’s channel dimension and computing spatial attention through convolutional operations. However, convolution can only capture local relationships and cannot model the long-term dependencies required for visual tasks. The incorporation of CBAM attention into the backbone improves the mAP, but it significantly increases the number of parameter counts and computational costs, and decreases by 4 FPS. In contrast, the CoordAtt attention mechanism embeds positional information into channel attention, capturing long-range dependencies in one spatial direction while preserving accurate positional information in the other. The results show that after adding CoordAtt to the backbone network, although the recall is reduced by 1.2%, the precision and the most important evaluation index, mAP, increase by 8.0% and 3.6%, respectively, which are significantly higher than the previous two methods, and the increase in parameters and calculation is not significant. This indicates that using the CoordAtt attention mechanism can enable the network model to detect targets more extensively and improve the network’s detection ability.
Ablation experiments were conducted in this study to validate the beneficial effects of the improved strategy proposed on the network. The results are presented in
Table 4, where the adoption of the corresponding enhancement method is denoted by “√” and the absence of the enhancement method is denoted by “×”.
The detection outcomes of the original YOLOv7 network are presented in the first row of the table. As can be seen from the above table, when the ELAN module of the original model is replaced with the new designed structure, CoTLAN, compared with the original YOLOv7 without any improvement strategy, precision, recall, and mAP are improved by 6.4%, 0.4%, and 3.6%, respectively. The network model integrates the two modules of CoTLAN and CoordAtt. The results show that the recall rate and FPS of the combined method have decreased, but the accuracy and mAP have been greatly improved. Among them, the detection accuracy and mAP increased by 14.9% and 5.2%, respectively, which effectively improved the detection ability of the model.
After the network model was determined, we used the improved YOLOv7-CTA model as the baseline and conducted comparative experiments on different localization loss functions. We compared the performances of three new methods: SIoU [
37], EIoU [
38], and CIoU (ours). The results are shown in
Table 5.
As seen from
Table 5, when the CIoU loss function is used, the accuracy value and mAP value of the improved model are the highest. From this, we conclude that using the CIoU loss function can help the improved model to have a better detection effect on BM cells.
This paper finally uses CIoU loss as the localization loss function of the YOLOv7-CTA model, and conducts ablation experiments on the setting of anchor box hyperparameters and the use of the improved loss function, as shown in
Table 6.
After replacing the original initial anchor box with the new anchor box generated by the K-means++ algorithm, the mAP of the improved model increased by 0.4%. This proves the effectiveness of the method. When the cross-entropy loss function in the improved model is replaced by the Focal loss function, the improved model improves mAP by 0.6%. For the combination of these two improvements, the mAP is significantly improved to 88.6%, which surpasses the previous two methods. For the above improvement schemes, this paper selects the improvement scheme with the highest mAP to participate in the subsequent experimental comparison.
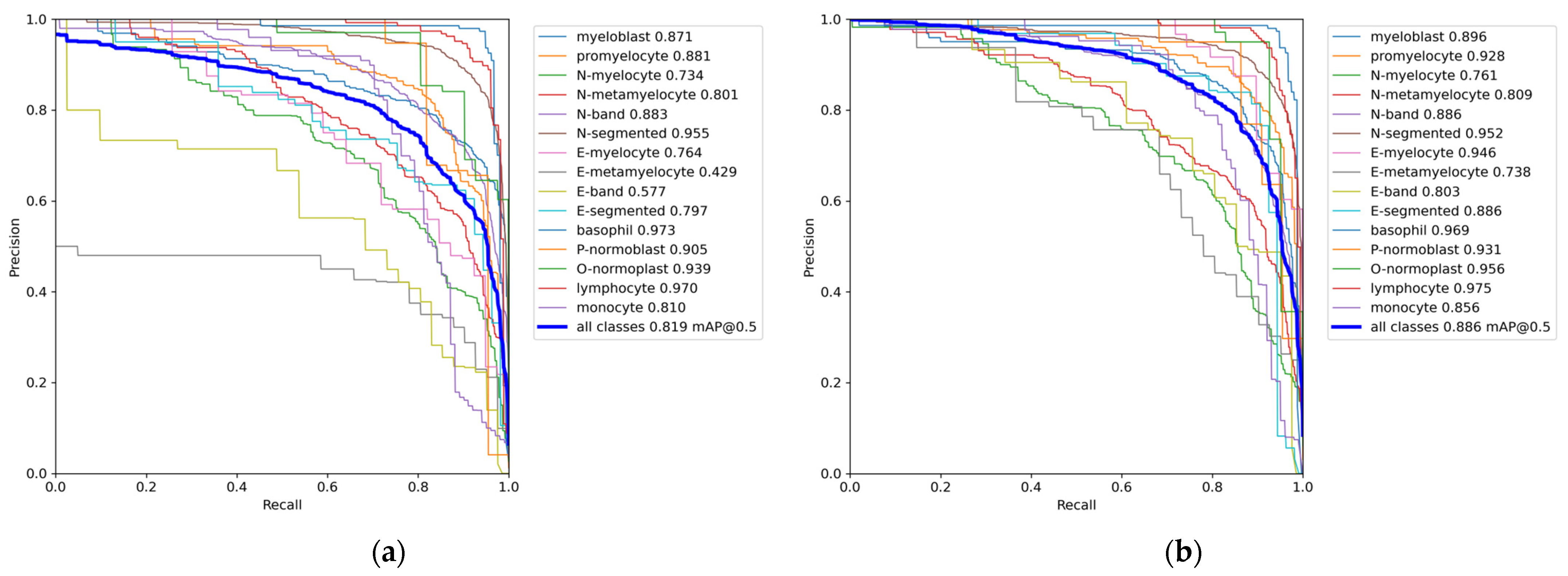
Since the PR curve comprehensively considers the accuracy and recall of each category detected by the model, in order to better measure the performance of the model, we further used the PR curve to measure the performance of the model.
Figure 8 depicts the PR curve of the present model, where the horizontal axis indicates the recall rate, and the vertical axis represents the precision. The PR curve provides a visualization of how precision changes as the recall rate increases. If the area enclosed by the curve and the coordinate axis in the figure is larger, it means the overall performance of the model is better. Compared with the original model, the performance improvement of the model is very obvious. It can be seen from the figure that the mAP of N-segmented and basophil decreased slightly, and the mAP of other cells increased to varying degrees. Among them, the mAP of seven types of cells reached more than 92.5%, indicating the effectiveness of the YOLOv7-CTA model.
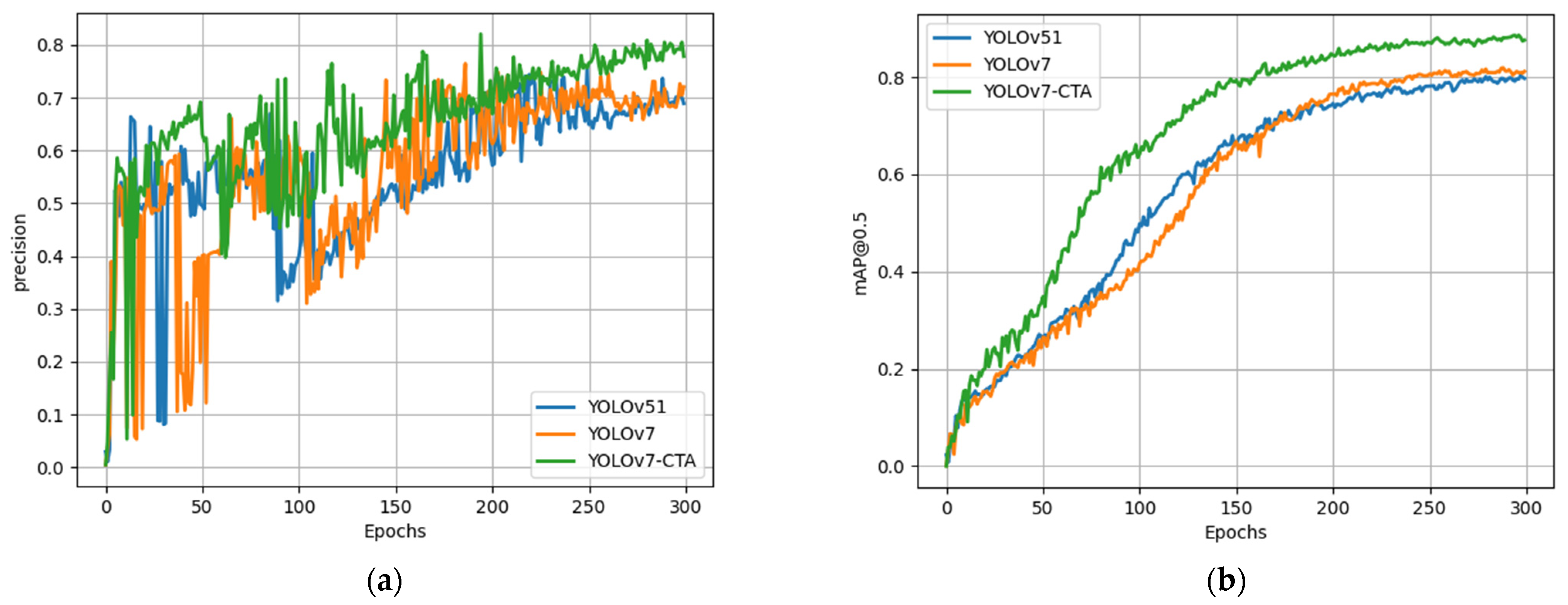
While the ablation experiments confirmed the effectiveness of the improved strategy proposed in this study relative to the original algorithm, it remains to be established whether it can achieve an advanced level. As such, a series of comparative experiments were conducted on BM-cell datasets, under identical experimental conditions, to compare the proposed method with the current mainstream object-detection approach.
Figure 9 illustrates the comparison of training outcomes among multiple models. The results demonstrate that the proposed algorithm in this paper achieved notably higher precision and mAP than the other two models.
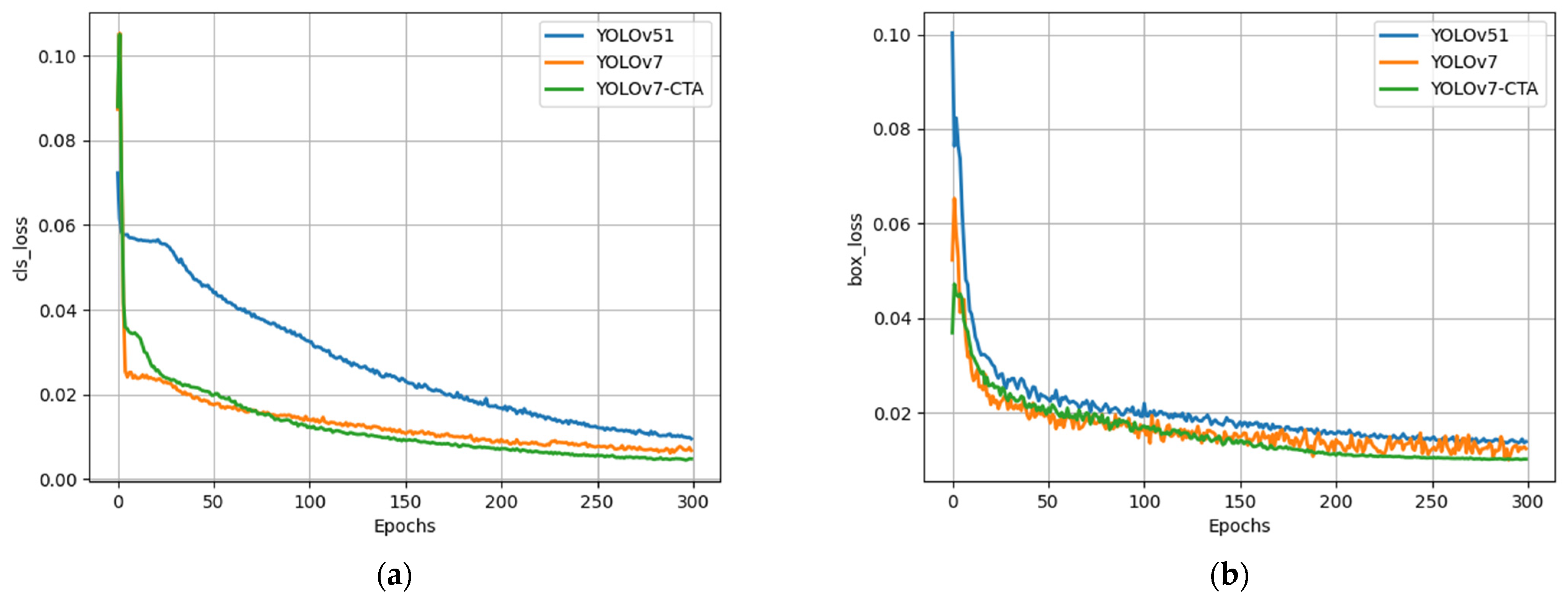
The training loss curves of various models are compared in
Figure 10. The curves of all models stabilized and converged after 250 iterations. The results indicate that YOLOv5l exhibited a significantly inferior performance to YOLOv7 concerning both regression and classification losses. From the figure, the improved network structure in this paper resulted in a slightly slower convergence rate of the model compared with that of YOLOv7. Nonetheless, after approximately 70 iterations, the proposed model exhibited a superior decline rate and convergence ability to YOLOv7, suggesting the efficacy of the loss function adjustment in enhancing the network’s convergence performance.
Finally, this paper lists the comparison results of the performance metrics of different models, as shown in
Table 7. The table shows that, compared with other models, the recall, precision, and mAP of YOLOv7-CTA are significantly higher. Among them, compared with YOLOv7, the precision, recall, and mAP of the improved model have increased by 10.1%, 0.6%, and 6.7%, respectively. Although the processing speed of YOLOv7-CTA is slightly lower than that of YOLOv7, it is still much faster than the two-stage model Faster R-CNN. The FPS indicator shows that YOLOv7-CTA can recognize 22 images of BM cells with a resolution of 600 × 600 per second, so the expected running time for the detection and recognition of 500 BM cells is <2.5 s, which can help pathologists greatly reduce their workload. In conclusion, the improved model has achieved high speed and high efficiency in predicting the task of BM cells.
In addition, we calculated the parameter count and computational cost of these models. Although the parameters of the two-stage model are small, the model structure is very complicated because the detection and classification are performed separately, resulting in a particularly large amount of calculations and high training costs. However, our proposed YOLOv7-CTA has the lowest computational cost, which reduces 2.7GFLOPs on the basis of YOLOv7, and its parameter count is also reduced by 0.6 M. Therefore, the YOLOV7-CTA does not have high requirements for hardware equipment, and has lower requirements than other models.