Machine Learning Techniques for Developing Remotely Monitored Central Nervous System Biomarkers Using Wearable Sensors: A Narrative Literature Review
Abstract
1. Introduction
1.1. Motivation
1.2. What Is Machine Learning
1.3. Objectives
2. Methods
2.1. Information Sources and Search Strategy
2.2. Inclusion Criteria
2.3. Data Extraction
3. Results
3.1. Study Selection
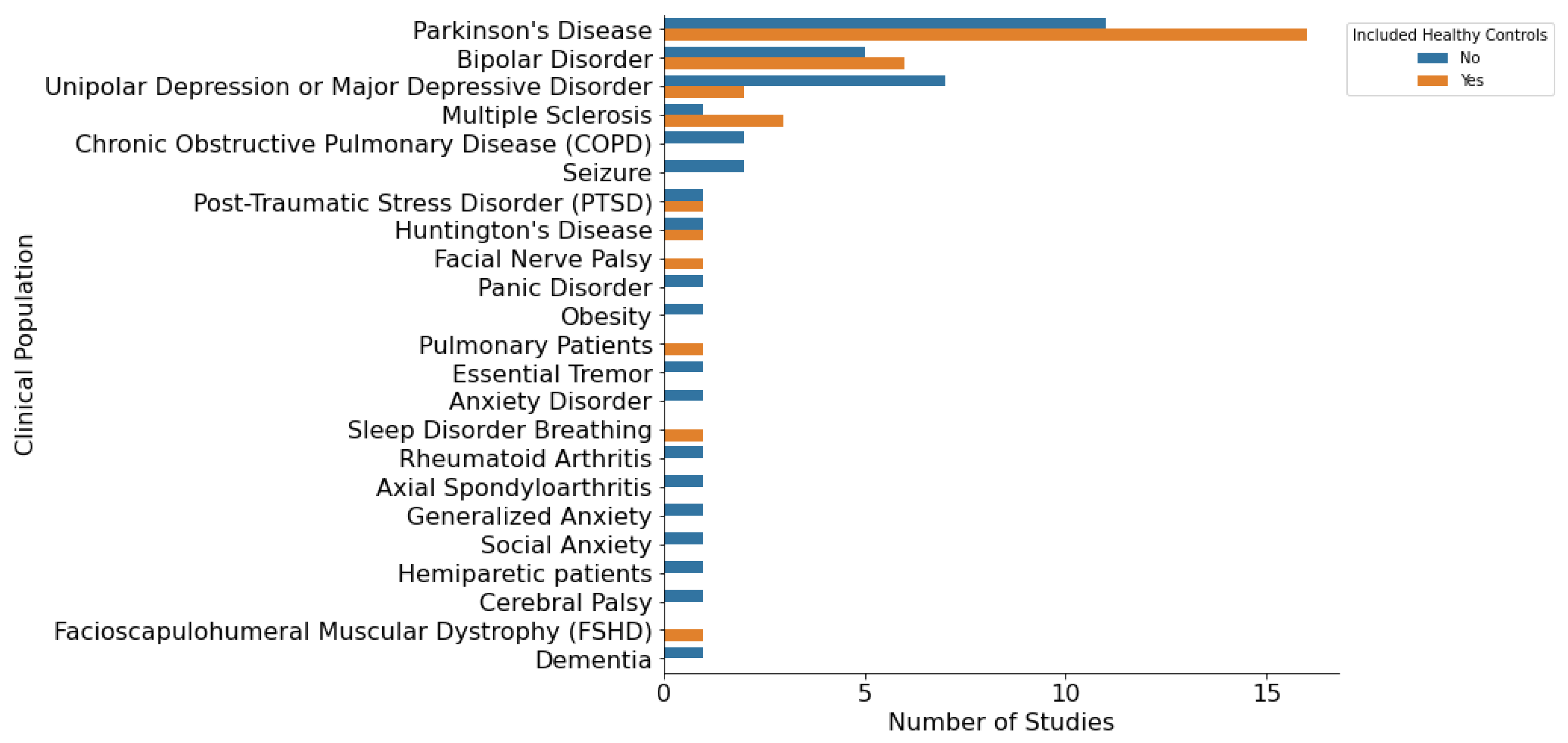
3.2. Study Characteristics
4. Missing and Outlier Data
4.1. Handling of Missing Data
4.2. Identification of Outliers
5. Feature Engineering
5.1. Feature Scaling
5.2. Expert Feature Engineering
5.3. Signal Processing
5.4. Principal Component Analysis
5.5. Clustering
5.6. Deep Learning
6. Feature Selection
6.1. Overfitting and Underfitting
6.2. Filter Methods
6.3. Embedded Methods
6.4. Wrapper Methods
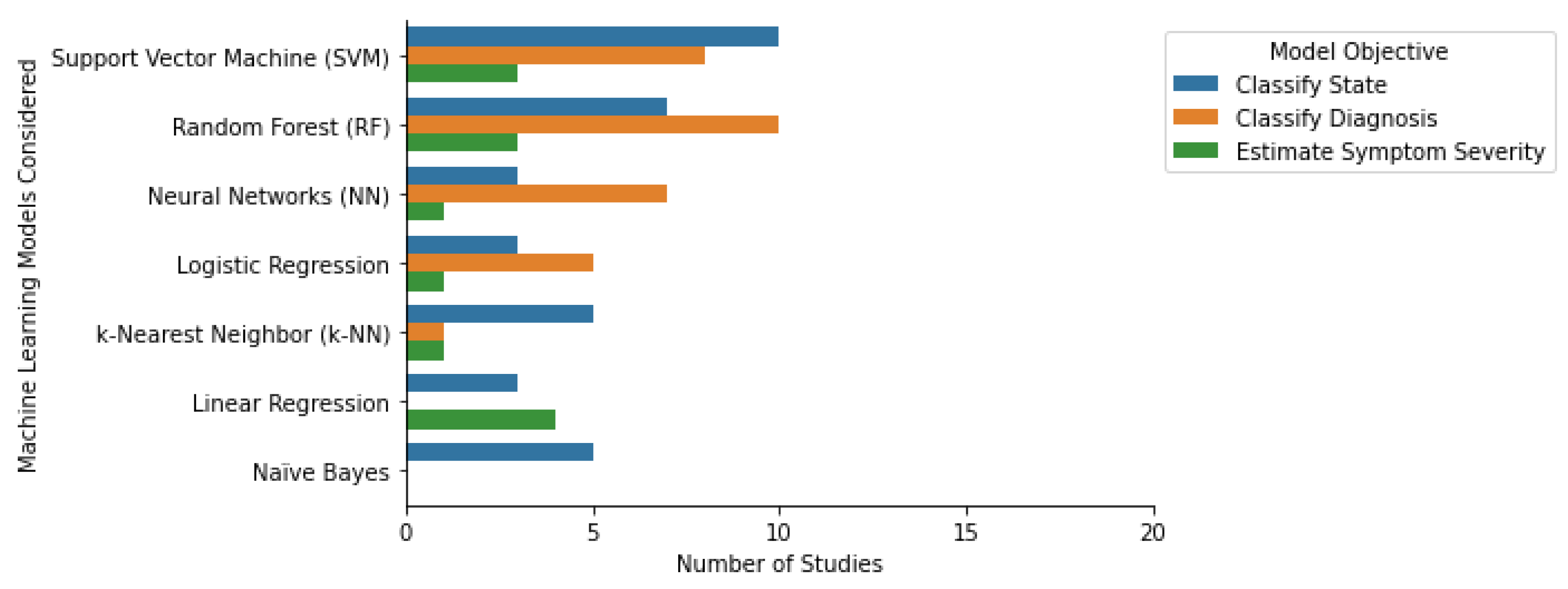
7. Machine Learning Algorithms
7.1. Tree-Based Models
7.2. Support Vector Machines
7.3. k-Nearest Neighbors
7.4. Naïve Bayes
7.5. Linear and Logistic Regression
7.6. Neural Networks
7.7. Transfer Learning
7.8. Multi-Task Learning
7.9. Generalized vs. Personalized
8. Model Hyperparameters
9. Model Evaluation
9.1. Classification Measures
9.2. Regression Measures
10. Model Validation
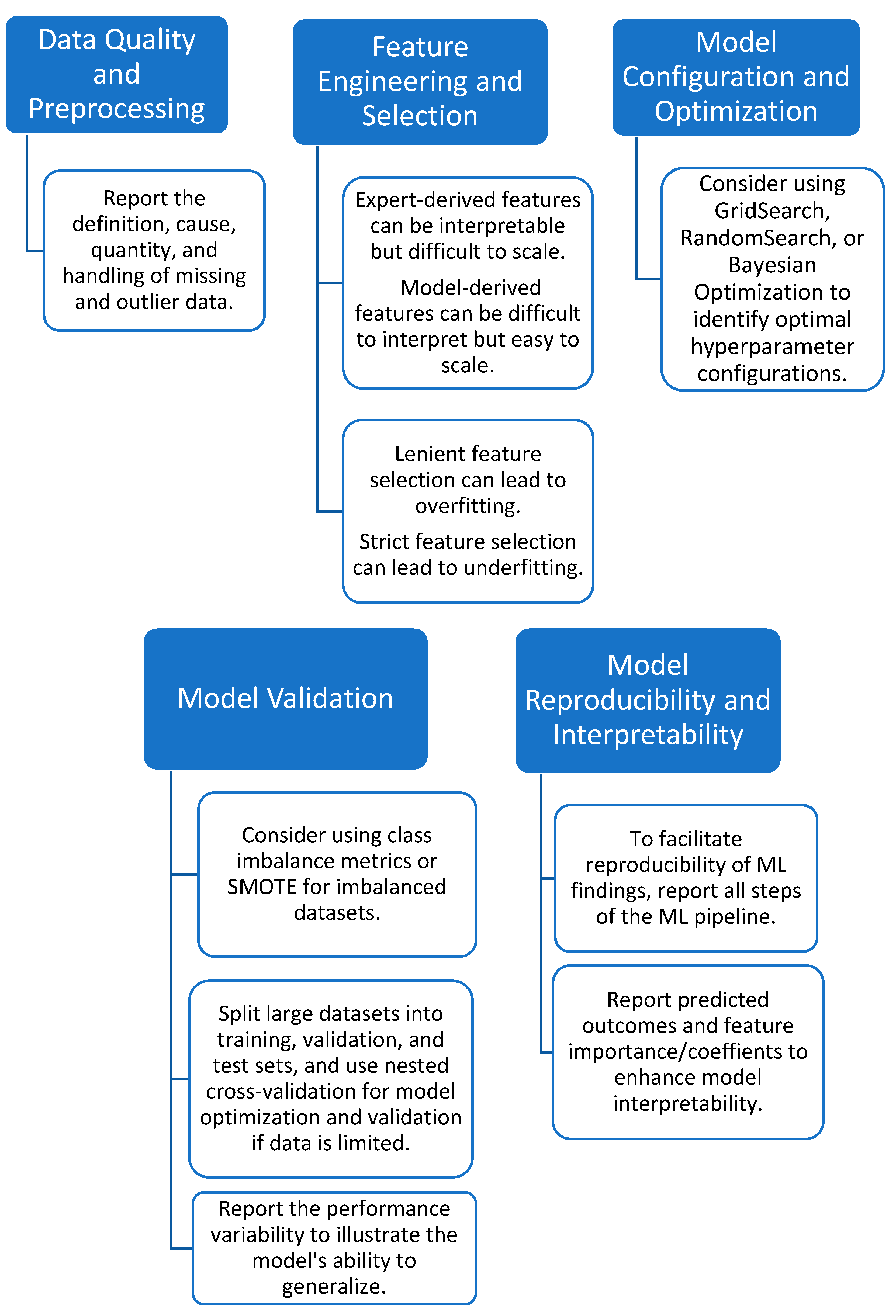
11. Recommendations
11.1. Inclusion of Healthy Controls
11.2. Data Quality and Preprocessing
11.3. Feature Engineering and Selection
11.4. Model Configuration and Optimization
11.5. Model Validation
11.6. Model Reproducibility and Interpretability
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Au, R.; Lin, H.; Kolachalama, V.B. Tele-Trials, Remote Monitoring, and Trial Technology for Alzheimer’s Disease Clinical Trials. In Alzheimer’s Disease Drug Development; Cambridge University Press: Cambridge, UK, 2022; pp. 292–300. [Google Scholar] [CrossRef]
- Inan, O.T.; Tenaerts, P.; Prindiville, S.A.; Reynolds, H.R.; Dizon, D.S.; Cooper-Arnold, K.; Turakhia, M.; Pletcher, M.J.; Preston, K.L.; Krumholz, H.M.; et al. Digitizing clinical trials. NPJ Digit. Med. 2020, 3, 101. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.X.; Davila, S.; Yang, C.; Hii, A.A.; Pua, C.J.; Yap, J.; Tan, S.Y.; Sahlén, A.; Chin, C.W.-L.; Teh, B.T.; et al. Digital phenotyping by consumer wearables identifies sleep-associated markers of cardiovascular disease risk and biological aging. bioRxiv 2019. [Google Scholar] [CrossRef]
- Brietzke, E.; Hawken, E.R.; Idzikowski, M.; Pong, J.; Kennedy, S.H.; Soares, C.N. Integrating digital phenotyping in clinical characterization of individuals with mood disorders. Neurosci. Biobehav. Rev. 2019, 104, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kourtis, L.C.; Regele, O.B.; Wright, J.M.; Jones, G.B. Digital biomarkers for Alzheimer’s disease: The mobile/wearable devices opportunity. NPJ Digit. Med. 2019, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Bhidayasiri, R.; Mari, Z. Digital phenotyping in Parkinson’s disease: Empowering neurologists for measurement-based care. Park. Relat. Disord. 2020, 80, 35–40. [Google Scholar] [CrossRef]
- Prosperi, M.; Min, J.S.; Bian, J.; Modave, F. Big data hurdles in precision medicine and precision public health. BMC Med. Inform. Decis. Mak. 2018, 18, 139. [Google Scholar] [CrossRef]
- Torres-Sospedra, J.; Ometov, A. Data from Smartphones and Wearables. Data 2021, 6, 45. [Google Scholar] [CrossRef]
- García-Santıllán, A.; del Flóres-Serrano, S.; López-Morales, J.S.; Rios-Alvarez, L.R. Factors Associated that Explain Anxiety toward Mathematics on Undergraduate Students. (An Empirical Study in Tierra Blanca Veracruz-México). Mediterr. J. Soc. Sci. 2014, 5. [Google Scholar] [CrossRef]
- Iniesta, R.; Stahl, D.; Mcguffin, P. Machine learning, statistical learning and the future of biological research in psychiatry. Psychol. Med. 2016, 46, 2455–2465. [Google Scholar] [CrossRef]
- Rajula, H.S.R.; Verlato, G.; Manchia, M.; Antonucci, N.; Fanos, V. Comparison of Conventional Statistical Methods with Machine Learning in Medicine: Diagnosis, Drug Development, and Treatment. Medicina 2020, 56, 455. [Google Scholar] [CrossRef]
- Getz, K.A.; Rafael, A.C. Trial watch: Trends in clinical trial design complexity. Nat. Rev. Drug. Discov. 2017, 16, 307. [Google Scholar] [CrossRef] [PubMed]
- Getz, K.A.; Stergiopoulos, S.; Marlborough, M.; Whitehill, J.; Curran, M.; Kaitin, K.I. Quantifying the Magnitude and Cost of Collecting Extraneous Protocol Data. Am. J. Ther. 2015, 22, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Getz, K.A.; Wenger, J.; Campo, R.A.; Seguine, E.S.; Kaitin, K.I. Assessing the Impact of Protocol Design Changes on Clinical Trial Performance. Am. J. Ther. 2008, 15, 450–457. [Google Scholar] [CrossRef]
- Globe Newswire. Rising Protocol Design Complexity Is Driving Rapid Growth in Clinical Trial Data Volume, According to Tufts Center for the Study of Drug Development. Available online: https://www.globenewswire.com/news-release/2021/01/12/2157143/0/en/Rising-Protocol-Design-Complexity-Is-Driving-Rapid-Growth-in-Clinical-Trial-Data-Volume-According-to-Tufts-Center-for-the-Study-of-Drug-Development.html (accessed on 12 January 2021).
- Santos, W.M.D.; Secoli, S.R.; de Araújo Püschel, V.A. The Joanna Briggs Institute approach for systematic reviews. Rev. Lat. Am. Enferm. 2018, 26, e3074. [Google Scholar] [CrossRef]
- Central Nervous System Diseases—MeSH—NCBI. 2023. Available online: https://www.ncbi.nlm.nih.gov/mesh?Db=mesh&Cmd=DetailsSearch&Term=%22Central+Nervous+System+Diseases%22%5BMeSH+Terms%5D (accessed on 5 January 2023).
- Martinez, G.J.; Mattingly, S.M.; Mirjafari, S.; Nepal, S.K.; Campbell, A.T.; Dey, A.K.; Striegel, A.D. On the Quality of Real-world Wearable Data in a Longitudinal Study of Information Workers. In Proceedings of the 2020 IEEE International Conference on Pervasive Computing and Communications Workshops, PerCom Workshops 2020, Austin, TX, USA, 23–27 March 2020. [Google Scholar] [CrossRef]
- Ruiz Blázquez, R.R.; Muñoz-Organero, M. Using Multivariate Outliers from Smartphone Sensor Data to Detect Physical Barriers While Walking in Urban Areas. Technologies 2020, 8, 58. [Google Scholar] [CrossRef]
- Poulos, J.; Valle, R. Missing Data Imputation for Supervised Learning. Appl. Artif. Intell. 2018, 32, 186–196. [Google Scholar] [CrossRef]
- Schafer, J.L.; Graham, J.W. Missing data: Our view of the state of the art. Psychol. Methods 2002, 7, 147–177. [Google Scholar] [CrossRef]
- Evers, L.J.; Raykov, Y.P.; Krijthe, J.H.; de Lima, A.L.S.; Badawy, R.; Claes, K.; Heskes, T.M.; Little, M.A.; Meinders, M.J.; Bloem, B.R. Real-life gait performance as a digital biomarker for motor fluctuations: The Parkinson@Home validation study. J. Med. Internet Res. 2020, 22, e19068. [Google Scholar] [CrossRef]
- Papadopoulos, A.; Kyritsis, K.; Klingelhoefer, L.; Bostanjopoulou, S.; Chaudhuri, K.R.; Delopoulos, A. Detecting Parkinsonian Tremor from IMU Data Collected In-The-Wild using Deep Multiple-Instance Learning. IEEE J. Biomed. Health Inform. 2019, 24, 2559–2569. [Google Scholar] [CrossRef]
- Tougui, I.; Jilbab, A.; El Mhamdi, J. Analysis of smartphone recordings in time, frequency, and cepstral domains to classify Parkinson’s disease. Healthc. Inform. Res. 2020, 26, 274–283. [Google Scholar] [CrossRef]
- Meyerhoff, J.; Liu, T.; Kording, K.P.; Ungar, L.H.; Kaiser, S.M.; Karr, C.J.; Mohr, D.C. Evaluation of Changes in Depression, Anxiety, and Social Anxiety Using Smartphone Sensor Features: Longitudinal Cohort Study. J. Med. Internet Res. 2021, 23, e22844. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, K.; Snyder, C.W.; Xiong, M.; Tarolli, C.G.; Sharma, S.; Dorsey, E.R.; Sharma, G.; Adams, J.L. A Longitudinal Wearable Sensor Study in Huntington’s Disease. J. Huntingt. Dis. 2020, 9, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.-H.; Lee, T.; Lee, H.-J. Mood Prediction of Patients with Mood Disorders by Machine Learning Using Passive Digital Phenotypes Based on the Circadian Rhythm: Prospective Observational Cohort Study. 2019. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6492069/ (accessed on 5 January 2023).
- Tanaka, T.; Kokubo, K.; Iwasa, K.; Sawa, K.; Yamada, N.; Komori, M. Intraday activity levels may better reflect the differences between major depressive disorder and bipolar disorder than average daily activity levels. Front. Psychol. 2018, 9, 2314. [Google Scholar] [CrossRef] [PubMed]
- Palmius, N.; Tsanas, A.; Saunders, K.E.A.; Bilderbeck, A.C.; Geddes, J.R.; Goodwin, G.M.; De Vos, M. Detecting bipolar depression from geographic location data. IEEE Trans. Biomed. Eng. 2017, 64, 1761–1771. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.; Matthews, M.; Frank, E.; Doherty, G.; Gay, G.; Choudhury, T. Automatic detection of social rhythms in bipolar disorder. J. Am. Med. Inform. Assoc. 2016, 23, 538–543. [Google Scholar] [CrossRef]
- Ramsperger, R.; Meckler, S.; Heger, T.; van Uem, J.; Hucker, S.; Braatz, U.; Graessner, H.; Berg, D.; Manoli, Y.; Serrano, J.A.; et al. Continuous leg dyskinesia assessment in Parkinson’s disease -clinical validity and ecological effect. Park. Relat. Disord. 2016, 26, 41–46. [Google Scholar] [CrossRef]
- Gossec, L.; Guyard, F.; Leroy, D.; Lafargue, T.; Seiler, M.; Jacquemin, C.; Molto, A.; Sellam, J.; Foltz, V.; Gandjbakhch, F.; et al. Detection of Flares by Decrease in Physical Activity, Collected Using Wearable Activity Trackers in Rheumatoid Arthritis or Axial Spondyloarthritis: An Application of Machine Learning Analyses in Rheumatology. Arthritis Care Res. 2019, 71, 1336–1343. [Google Scholar] [CrossRef]
- Bai, R.; Xiao, L.; Guo, Y.; Zhu, X.; Li, N.; Wang, Y.; Chen, Q.; Feng, L.; Wang, Y.; Yu, X.; et al. Tracking and monitoring mood stability of patients with major depressive disorder by machine learning models using passive digital data: Prospective naturalistic multicenter study. JMIR Mhealth Uhealth 2021, 9, e24365. [Google Scholar] [CrossRef]
- Schwab, P.; Karlen, W. A Deep Learning Approach to Diagnosing Multiple Sclerosis from Smartphone Data. IEEE J. Biomed. Health Inform. 2021, 25, 1284–1291. [Google Scholar] [CrossRef]
- Aghanavesi, S. Smartphone-Based Parkinson’s Disease Symptom Assessment. Licentiate Dissertation, Dalarna University, Falun, Sweden, 2017. [Google Scholar]
- Maleki, G.; Zhuparris, A.; Koopmans, I.; Doll, R.J.; Voet, N.; Cohen, A.; van Brummelen, E.; Groeneveld, G.J.; De Maeyer, J. Objective Monitoring of Facioscapulohumeral Dystrophy During Clinical Trials Using a Smartphone App and Wearables: Observational Study. JMIR Form. Res. 2022, 6, e31775. [Google Scholar] [CrossRef]
- Twose, J.; Licitra, G.; McConchie, H.; Lam, K.H.; Killestein, J. Early-warning signals for disease activity in patients diagnosed with multiple sclerosis based on keystroke dynamics. Chaos 2020, 30, 113133. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.H.; Lee, T.; Kim, M.G.; In, H.P.; Kim, L.; Lee, H.J. Mood prediction of patients with mood disorders by machine learning using passive digital phenotypes based on the circadian rhythm: Prospective observational cohort study. J. Med. Internet Res. 2019, 21, e11029. [Google Scholar] [CrossRef]
- Little, R.J.A.; Rubin, D.B. Complete-Case and Available-Case Analysis, Including Weighting Methods; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 41–58. [Google Scholar] [CrossRef]
- Demissie, S.; LaValley, M.P.; Horton, N.J.; Glynn, R.J.; Cupples, L.A. Bias due to missing exposure data using complete-case analysis in the proportional hazards regression model. Stat. Med. 2003, 22, 545–557. [Google Scholar] [CrossRef]
- Enders, C.K.; London, N.Y. Applied Missing Data Analysis; Guilford Press: New York, NY, USA, 2010. [Google Scholar]
- Zhang, Y.; Folarin, A.A. Predicting Depressive Symptom Severity Through Individuals’ Nearby Bluetooth Device Count Data Collected by Mobile Phones: Preliminary Longitudinal Study. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8367113/ (accessed on 5 January 2023).
- Creagh, A.P.; Dondelinger, F.; Lipsmeier, F.; Lindemann, M.; De Vos, M. Longitudinal Trend Monitoring of Multiple Sclerosis Ambulation using Smartphones. IEEE Open J. Eng. Med. Biol. 2022, 3, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-T.; Li, G.-H.; Huang, C.-T.; Cheng, Y.-C.; Chen, C.-H.; Chien, J.-Y.; Kuo, P.-H.; Kuo, L.-C.; Lai, F. Acute exacerbation of a chronic obstructive pulmonary disease prediction system using wearable device data, machine learning, and deep learning: Development and cohort study. JMIR Mhealth Uhealth 2021, 9, e22591. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, P.; Garcia-Ceja, E.; Riegler, M.; Stabell, L.A.; Nordgreen, T.; Torresen, J.; Fasmer, O.B.; Oedegaard, K.J. Applying machine learning in motor activity time series of depressed bipolar and unipolar patients compared to healthy controls. PLoS ONE 2020, 15, e0231995. [Google Scholar] [CrossRef]
- Lekkas, D.; Jacobson, N.C. Using artificial intelligence and longitudinal location data to differentiate persons who develop posttraumatic stress disorder following childhood trauma. Sci. Rep. 2021, 11, 10303. [Google Scholar] [CrossRef]
- Richman, M.B.; Trafalis, T.B.; Adrianto, I. Missing data imputation through machine learning algorithms. In Artificial Intelligence Methods in the Environmental Sciences; Springer: Dordrecht, The Netherlands, 2009; pp. 153–169. [Google Scholar] [CrossRef]
- Jerez, J.M.; Molina, I.; García-Laencina, P.J.; Alba, E.; Ribelles, N.; Martín, M.; Franco, L. Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artif. Intell. Med. 2010, 50, 105–115. [Google Scholar] [CrossRef]
- Lakshminarayan, K.; Harp, S.A.; Goldman, R.P.; Samad, T. Imputation of Missing Data Using Machine Learning Techniques. In KDD Proceedings 1996; AAAI Press: Palo Alto, CA, USA, 1996; Volume 96. [Google Scholar]
- Aggarwal, C.C. Data Mining; Springer International Publishing: Cham, Switzerland, 2015. [Google Scholar] [CrossRef]
- Ledolter, J.; Kardon, R.H. Does Testing More Frequently Shorten the Time to Detect Disease Progression? Transl. Vis. Sci. Technol. 2017, 6, 1. [Google Scholar] [CrossRef]
- Bazgir, O.; Habibi, S.A.H.; Palma, L.; Pierleoni, P.; Nafees, S. A classification system for assessment and home monitoring of tremor in patients with Parkinson’s disease. J. Med. Signals Sens. 2018, 8, 65–72. [Google Scholar] [CrossRef]
- Williamson, J.R.; Telfer, B.; Mullany, R.; Friedl, K.E. Detecting Parkinson’s Disease from Wrist-Worn Accelerometry in the U.K. Biobank. Sensors 2021, 21, 2047. [Google Scholar] [CrossRef] [PubMed]
- Buda, T.S.; Khwaja, M.; Matic, A. Outliers in Smartphone Sensor Data Reveal Outliers in Daily Happiness. Proc. ACM Interact. Mob. Wearable Ubiquitous Technol. 2021, 5, 1–19. [Google Scholar] [CrossRef]
- Buda, T.S.; Caglayan, B.; Assem, H. DeepAD: A generic framework based on deep learning for time series anomaly detection. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2018; pp. 577–588. [Google Scholar] [CrossRef]
- Arora, S.; Venkataraman, V.; Zhan, A.; Donohue, S.; Biglan, K.; Dorsey, E.; Little, M. Detecting and monitoring the symptoms of Parkinson’s disease using smartphones: A pilot study. Park. Relat. Disord. 2015, 21, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An Introduction to Feature Extraction. In Feature Extraction; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1–25. [Google Scholar] [CrossRef]
- Raju, V.N.G.; Lakshmi, K.P.; Jain, V.M.; Kalidindi, A.; Padma, V. Study the Influence of Normalization/Transformation process on the Accuracy of Supervised Classification. In Proceedings of the 2020 Third International Conference on Smart Systems and Inventive Technology (ICSSIT), Tirunelveli, India, 20–22 August 2022; pp. 729–735. [Google Scholar] [CrossRef]
- Dara, S.; Tumma, P. Feature Extraction by Using Deep Learning: A Survey. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; pp. 1795–1801. [Google Scholar] [CrossRef]
- Tizzano, G.R.; Spezialetti, M.; Rossi, S. A Deep Learning Approach for Mood Recognition from Wearable Data. In Proceedings of the IEEE Medical Measurements and Applications, MeMeA 2020—Conference Proceedings, Bari, Italy, 1 June–1 July 2020; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Garcia-Ceja, E.; Riegler, M.; Jakobsen, P.; Torresen, J.; Nordgreen, T.; Oedegaard, K.J.; Fasmer, O.B. Motor Activity Based Classification of Depression in Unipolar and Bipolar Patients. In Proceedings of the 2018 IEEE 31st International Symposium on Computer-Based Medical Systems (CBMS), Karlstad, Sweden, 18–21 June 2018; pp. 316–321. [Google Scholar] [CrossRef]
- Liu, H. Feature Engineering for Machine Learning and Data Analytics. In Feature Engineering for Machine Learning and Data Analytics; Taylor & Francis Group: Boca Raton, FL, USA, 2018. [Google Scholar]
- Nargesian, F.; Samulowitz, H.; Khurana, U.; Khalil, E.B.; Turaga, D. Learning feature engineering for classification. IJCAI Int. Jt. Conf. Artif. Intell. 2017, 2529–2535. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Feature Engineering and Selection: A Practical Approach for Predictive Models; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019. [Google Scholar] [CrossRef]
- Ronao, C.A.; Cho, S.-B. Human activity recognition with smartphone sensors using deep learning neural networks. Expert Syst. Appl. 2016, 59, 235–244. [Google Scholar] [CrossRef]
- Nweke, H.F.; Teh, Y.W.; Al-garadi, M.A.; Alo, U.R. Deep learning algorithms for human activity recognition using mobile and wearable sensor networks: State of the art and research challenges. Expert Syst. Appl. 2018, 105, 233–261. [Google Scholar] [CrossRef]
- Zdravevski, E.; Lameski, P.; Trajkovik, V.; Kulakov, A.; Chorbev, I.; Goleva, R.; Pombo, N.; Garcia, N. Improving Activity Recognition Accuracy in Ambient-Assisted Living Systems by Automated Feature Engineering. IEEE Access 2017, 5, 5262–5280. [Google Scholar] [CrossRef]
- McGinnis, R.S.; Mahadevan, N.; Moon, Y.; Seagers, K.; Sheth, N.; Wright, J.A., Jr.; Dicristofaro, S.; Silva, I.; Jortberg, E.; Ceruolo, M.; et al. A machine learning approach for gait speed estimation using skin-mounted wearable sensors: From healthy controls to individuals with multiple sclerosis. PLoS ONE 2017, 12, e0178366. [Google Scholar] [CrossRef]
- Maxhuni, A.; Muñoz-Meléndez, A.; Osmani, V.; Perez, H.; Mayora, O.; Morales, E.F. Classification of bipolar disorder episodes based on analysis of voice and motor activity of patients. Pervasive Mob. Comput. 2016, 31, 50–66. [Google Scholar] [CrossRef]
- Yamakawa, T.; Miyajima, M.; Fujiwara, K.; Kano, M.; Suzuki, Y.; Watanabe, Y.; Watanabe, S.; Hoshida, T.; Inaji, M.; Maehara, T. Wearable epileptic seizure prediction system with machine-learning-based anomaly detection of heart rate variability. Sensors 2020, 20, 3987. [Google Scholar] [CrossRef]
- Fuchs, C.; Nobile, M.S.; Zamora, G.; Degeneffe, A.; Kubben, P.; Kaymak, U. Tremor assessment using smartphone sensor data and fuzzy reasoning. BMC Bioinform. 2021, 22, 57. [Google Scholar] [CrossRef] [PubMed]
- Aich, S.; Pradhan, P.M.; Park, J.; Sethi, N.; Vathsa, V.S.S.; Kim, H.C. A validation study of freezing of gait (FoG) detection and machine-learning-based FoG prediction using estimated gait characteristics with a wearable accelerometer. Sensors 2018, 18, 3287. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martín, D.; Samà, A.; Pérez-López, C.; Català, A.; Arostegui, J.M.M.; Cabestany, J.; Bayés, À.; Alcaine, S.; Mestre, B.; Prats, A.; et al. Home detection of freezing of gait using Support Vector Machines through a single waist-worn triaxial accelerometer. PLoS ONE 2017, 12, e0171764. [Google Scholar] [CrossRef] [PubMed]
- Supratak, A.; Datta, G.; Gafson, A.R.; Nicholas, R.; Guo, Y.; Matthews, P.M. Remote monitoring in the home validates clinical gait measures for multiple sclerosis. Front. Neurol. 2018, 9, 561. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Kim, J.; Lim, J. A Deep Neural Network-Based Method for Prediction of Dementia Using Big Data. Int. J. Environ. Res. Public Health 2021, 18, 5386. [Google Scholar] [CrossRef]
- Clustering. In Principles of Data Mining; Springer: London, UK, 2007; pp. 221–238. [CrossRef]
- Arabie, P.; Hubert, L.J. An Overview of Combinatorial Data Analysis. In Clustering and Classification; World Scientific: Singapore, 1996; pp. 5–63. [Google Scholar] [CrossRef]
- Lu, J.; Shang, C.; Yue, C.; Morillo, R.; Ware, S.; Kamath, J.; Bamis, A.; Russell, A.; Wang, B.; Bi, J. Joint Modeling of Heterogeneous Sensing Data for Depression Assessment via Multi-task Learning. In Proceedings of the Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies; Association for Computing Machinery: New York, NY, USA, 2018; Volume 2, pp. 1–21. [Google Scholar] [CrossRef]
- Sabatelli, M.; Osmani, V.; Mayora, O.; Gruenerbl, A.; Lukowicz, P. Correlation of significant places with self-reported state of bipolar disorder patients. In Proceedings of the 2014 4th International Conference on Wireless Mobile Communication and Healthcare-Transforming Healthcare through Innovations in Mobile and Wireless Technologies (MOBIHEALTH), Athens, Greece, 3–5 November 2014; pp. 116–119. [Google Scholar] [CrossRef]
- Faurholt-Jepsen, M.; Busk, J.; Vinberg, M.; Christensen, E.M.; HelgaÞórarinsdóttir; Frost, M.; Bardram, J.E.; Kessing, L.V. Daily mobility patterns in patients with bipolar disorder and healthy individuals. J. Affect. Disord. 2021, 278, 413–422. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef]
- Marx, V. The big challenges of big data. Nature 2013, 498, 255–260. [Google Scholar] [CrossRef]
- Li, Y.; Ding, L.; Gao, X. On the decision boundary of deep neural networks. arXiv 2018, arXiv:1808.05385. [Google Scholar]
- Juen, J.; Cheng, Q.; Schatz, B. A Natural Walking Monitor for Pulmonary Patients Using Mobile Phones. IEEE J. Biomed. Health Inform. 2015, 19, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Cole, B.T.; Roy, S.H.; De Luca, C.J.; Nawab, S.H. Dynamical learning and tracking of tremor and dyskinesia from wearable sensors. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Peraza, L.R.; Kinnunen, K.M.; McNaney, R.; Craddock, I.J.; Whone, A.L.; Morgan, C.; Joules, R.; Wolz, R. An automatic gait analysis pipeline for wearable sensors: A pilot study in parkinson’s disease. Sensors 2021, 21, 8286. [Google Scholar] [CrossRef] [PubMed]
- Saeys, Y.; Abeel, T.; Van De Peer, Y. Robust feature selection using ensemble feature selection techniques. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2008; pp. 313–325. [Google Scholar] [CrossRef]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature selection in machine learning: A new perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Jabar, H.; Khan, R.Z. Methods to avoid over-fitting and under-fitting in supervised machine learning (comparative study). Comput. Sci. Commun. Instrum. Devices 2015, 70, 163–172. [Google Scholar]
- Hall, M.A. Correlation-based Feature Selection for Machine Learning. Ph.D. Thesis, The University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- Hall, M.A.; Smith, L.A. Feature selection for machine learning: Comparing a correlation-based filter approach to the wrapper. In Proceedings of the FLAIRS Conference 1999, Orlando, FL, USA, 1–5 May 1999; Volume 1999, pp. 235–239. [Google Scholar]
- Galperin, I.; Hillel, I.; Del Din, S.; Bekkers, E.M.; Nieuwboer, A.; Abbruzzese, G.; Avanzino, L.; Nieuwhof, F.; Bloem, B.R.; Rochester, L.; et al. Associations between daily-living physical activity and laboratory-based assessments of motor severity in patients with falls and Parkinson’s disease. Park. Relat. Disord. 2019, 62, 85–90. [Google Scholar] [CrossRef]
- Dong, C.; Ye, T.; Long, X.; Aarts, R.M.; van Dijk, J.P.; Shang, C.; Liao, X.; Chen, W.; Lai, W.; Chen, L.; et al. A Two-Layer Ensemble Method for Detecting Epileptic Seizures Using a Self-Annotation Bracelet with Motor Sensors. IEEE Trans. Instrum. Meas. 2022, 71, 4005013. [Google Scholar] [CrossRef]
- Creagh, A.P.; Simillion, C.; Bourke, A.K.; Scotland, A.; Lipsmeier, F.; Bernasconi, C.; van Beek, J.; Baker, M.; Gossens, C.; Lindemann, M.; et al. Smartphone-and Smartwatch-Based Remote Characterisation of Ambulation in Multiple Sclerosis during the Two-Minute Walk Test. IEEE J. Biomed. Health Inform. 2021, 25, 838–849. [Google Scholar] [CrossRef]
- Chen, O.Y.; Lipsmeier, F.; Phan, H.; Prince, J.; Taylor, K.I.; Gossens, C.; Lindemann, M.; de Vos, M. Building a Machine-Learning Framework to Remotely Assess Parkinson’s Disease Using Smartphones. IEEE Trans. Biomed. Eng. 2020, 67, 3491–3500. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Eijkemans, M.J.C.; Habbema, J.D.F. Stepwise selection in small data sets: A simulation study of bias in logistic regression analysis. J. Clin. Epidemiol. 1999, 52, 935–942. [Google Scholar] [CrossRef]
- Austin, P.C.; Tu, J.V. Bootstrap Methods for Developing Predictive Models. Am. Stat. 2004, 58, 131–137. [Google Scholar] [CrossRef]
- Zimmerman, D.W. Power Functions of the Test and Mann-Whitney Test Under Violation of Parametric Assumptions. Percept. Mot. Skills 1985, 61, 467–470. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Meeker, M.; la Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef]
- Kira, K.; Rendell, L.A. A Practical Approach to Feature Selection; Elsevier: Amsterdam, The Netherlands, 1992; pp. 249–256. [Google Scholar] [CrossRef]
- Verma, N.K.; Salour, A. Feature selection. Stud. Syst. Decis. Control 2020, 256, 175–200. [Google Scholar] [CrossRef]
- Yaman, O.; Ertam, F.; Tuncer, T. Automated Parkinson’s disease recognition based on statistical pooling method using acoustic features. Med. Hypotheses 2020, 135, 109483. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Molinero, A.; Samà, A.; Pérez-Martínez, D.A.; López, C.P.; Romagosa, J.; Bayes, A.; Sanz, P.; Calopa, M.; Gálvez-Barrón, C.; De Mingo, E.; et al. Validation of a portable device for mapping motor and gait disturbances in Parkinson’s disease. JMIR Mhealth Uhealth 2015, 3, e9. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Goldsmith, J.; Bobb, J.; Crainiceanu, C.M.; Caffo, B.; Reich, D. Penalized functional regression. J. Comput. Graph. Stat. 2011, 20, 830–851. [Google Scholar] [CrossRef]
- Prince, J.; Andreotti, F.; De Vos, M. Multi-Source Ensemble Learning for the Remote Prediction of Parkinson’s Disease in the Presence of Source-Wise Missing Data. IEEE Trans. Biomed. Eng. 2019, 66, 1402–1411. [Google Scholar] [CrossRef]
- Motin, M.A.; Pah, N.D.; Raghav, S.; Kumar, D.K. Parkinson’s Disease Detection Using Smartphone Recorded Phonemes in Real World Conditions. IEEE Access 2022, 10, 97600–97609. [Google Scholar] [CrossRef]
- Cakmak, A.S.; Alday, E.A.P.; Da Poian, G.; Rad, A.B.; Metzler, T.J.; Neylan, T.C.; House, S.L.; Beaudoin, F.L.; An, X.; Stevens, J.S.; et al. Classification and Prediction of Post-Trauma Outcomes Related to PTSD Using Circadian Rhythm Changes Measured via Wrist-Worn Research Watch in a Large Longitudinal Cohort. IEEE J. Biomed. Health Inform. 2021, 25, 2866–2876. [Google Scholar] [CrossRef] [PubMed]
- Tracy, J.M.; Özkanca, Y.; Atkins, D.C.; Ghomi, R.H. Investigating voice as a biomarker: Deep phenotyping methods for early detection of Parkinson’s disease. J. Biomed. Inform. 2020, 104, 103362. [Google Scholar] [CrossRef] [PubMed]
- Abdulhafedh, A. Comparison between Common Statistical Modeling Techniques Used in Research, Including: Discriminant Analysis vs Logistic Regression, Ridge Regression vs LASSO, and Decision Tree vs Random Forest. OAlib 2022, 9, 1–19. [Google Scholar] [CrossRef]
- Sánchez-Maroño, N.; Alonso-Betanzos, A.; Tombilla-Sanromán, M. Filter methods for feature selection—A comparative study. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2007; pp. 178–187. [Google Scholar] [CrossRef]
- Porter, B.W.; Bareiss, R.; Holte, R.C. Concept learning and heuristic classification in weak-theory domains. Artif. Intell. 1990, 45, 229–263. [Google Scholar] [CrossRef]
- Wu, C.-T.; Wang, S.-M.; Su, Y.-E.; Hsieh, T.-T.; Chen, P.-C.; Cheng, Y.-C.; Tseng, T.-W.; Chang, W.-S.; Su, C.-S.; Kuo, L.-C.; et al. A Precision Health Service for Chronic Diseases: Development and Cohort Study Using Wearable Device, Machine Learning, and Deep Learning. IEEE J. Transl. Eng. Health Med. 2022, 10, 2700414. [Google Scholar] [CrossRef] [PubMed]
- de Lima, A.L.S.; Evers, L.J.; Hahn, T.; de Vries, N.M.; Daeschler, M.; Boroojerdi, B.; Terricabras, D.; Little, M.A.; Bloem, B.R.; Faber, M.J. Impact of motor fluctuations on real-life gait in Parkinson’s patients. Gait Posture 2018, 62, 388–394. [Google Scholar] [CrossRef]
- Pulliam, C.; Eichenseer, S.; Goetz, C.; Waln, O.; Hunter, C.; Jankovic, J.; Vaillancourt, D.; Giuffrida, J.; Heldman, D. Continuous in-home monitoring of essential tremor. Park. Relat. Disord. 2014, 20, 37–40. [Google Scholar] [CrossRef]
- Goni, M.; Eickhoff, S.B.; Far, M.S.; Patil, K.R.; Dukart, J. Smartphone-Based Digital Biomarkers for Parkinson’s Disease in a Remotely-Administered Setting. IEEE Access 2022, 10, 28361–28384. [Google Scholar] [CrossRef]
- Livingston, E.; Cao, J.; Dimick, J.B. Tread carefully with stepwise regression. Arch. Surg. 2010, 145, 1039–1040. [Google Scholar] [CrossRef]
- Li, F.; Yang, Y. Analysis of recursive feature elimination methods. In Proceedings of the the 28th ACM/SIGIR International Symposium on Information Retrieval 2005, Salvador, Brazil, 15–19 August 2005. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K.; Kuhn, M.; Johnson, K. An Introduction to Feature Selection. In Applied Predictive Modeling; Springer: New York, NY, USA, 2013; pp. 487–519. [Google Scholar] [CrossRef]
- Senturk, Z.K. Early diagnosis of Parkinson’s disease using machine learning algorithms. Med. Hypotheses 2020, 138, 109603. [Google Scholar] [CrossRef]
- Zhang, X.D. Machine Learning. In A Matrix Algebra Approach to Artificial Intelligence; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Russell, S.J.; Norvig, P. Artificial Intelligence: A Modern Approach, 4th ed.; Prentice Hall: Hoboken, NJ, USA, 2020. [Google Scholar]
- Tinschert, P.; Rassouli, F.; Barata, F.; Steurer-Stey, C.; Fleisch, E.; Puhan, M.; Kowatsch, T.; Brutsche, M.H. Smartphone-Based Cough Detection Predicts Asthma Control—Description of a Novel, Scalable Digital Biomarker; European Respiratory Society (ERS): Lausanne, Switzerland, 2020; p. 4569. [Google Scholar] [CrossRef]
- ZhuParris, A.; Kruizinga, M.D.; van Gent, M.; Dessing, E.; Exadaktylos, V.; Doll, R.J.; Stuurman, F.E.; Driessen, G.A.; Cohen, A.F. Development and Technical Validation of a Smartphone-Based Cry Detection Algorithm. Front. Pediatr. 2021, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Fatima, M.; Pasha, M. Survey of Machine Learning Algorithms for Disease Diagnostic. J. Intell. Learn. Syst. Appl. 2017, 9, 1–16. [Google Scholar] [CrossRef]
- Ensari, I.; Caceres, B.A.; Jackman, K.B.; Suero-Tejeda, N.; Shechter, A.; Odlum, M.L.; Bakken, S. Digital phenotyping of sleep patterns among heterogenous samples of Latinx adults using unsupervised learning. Sleep. Med. 2021, 85, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.-F.; Kuo, P.-H.; Wang, C.-F.; Chen, Y.-J.; Chuang, P.-C.; Li, S.-Z.; Chen, B.-W.; Yang, F.-C.; Lo, Y.-C.; Yang, Y.; et al. Quantification Analysis of Sleep Based on Smartwatch Sensors for Parkinson’s Disease. Biosensors 2022, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Farhan, A.A.; Yue, C.; Morillo, R.; Ware, S.; Lu, J.; Bi, J.; Kamath, J.; Russell, A.; Bamis, A.; Wang, B. Behavior vs. introspection: Refining prediction of clinical depression via smartphone sensing data. In Proceedings of the 2016 IEEE Wireless Health (WH), Bethesda, MD, USA, 25–27 October 2016. [Google Scholar] [CrossRef]
- Derungs, A.; Schuster-Amft, C.; Amft, O. Longitudinal walking analysis in hemiparetic patients using wearable motion sensors: Is there convergence between body sides? Front. Bioeng. Biotechnol. 2018, 6, 57. [Google Scholar] [CrossRef]
- Freedman, D.A. Statistical Models. In Statistical Models: THeory and Practice; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Ahmed, S.T.; Basha, S.M.; Arumugam, S.R.; Kodabagi, M.M. Pattern Recognition: An Introduction, 1st ed.; MileStone Research Publications: Bengaluru, India, 2021. [Google Scholar]
- Ruppert, D. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. J. Am. Stat. Assoc. 2004, 99, 567. [Google Scholar] [CrossRef]
- Opitz, D.; Maclin, R. Popular Ensemble Methods: An Empirical Study. J. Artif. Intell. Res. 1999, 11, 169–198. [Google Scholar] [CrossRef]
- Kosasi, S. Perancangan Prototipe Sistem Pemesanan Makanan dan Minuman Menggunakan Mobile Device. Indones. J. Netw. Secur. 2015, 1, 1–10. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: New York, NY, USA, 2013. [Google Scholar]
- San-Segundo, R.; Zhang, A.; Cebulla, A.; Panev, S.; Tabor, G.; Stebbins, K.; Massa, R.E.; Whitford, A.; de la Torre, F.; Hodgins, J. Parkinson’s disease tremor detection in the wild using wearable accelerometers. Sensors 2020, 20, 5817. [Google Scholar] [CrossRef]
- Ahmadi, M.N.; O’neil, M.E.; Baque, E.; Boyd, R.N.; Trost, S.G. Machine learning to quantify physical activity in children with cerebral palsy: Comparison of group, group-personalized, and fully-personalized activity classification models. Sensors 2020, 20, 3976. [Google Scholar] [CrossRef]
- Faurholt-Jepsen, M.; Busk, J.; Frost, M.; Vinberg, M.; Christensen, E.M.; Winther, O.; Bardram, J.E.; Kessing, L.V. Voice analysis as an objective state marker in bipolar disorder. Transl. Psychiatry 2016, 6, e856. [Google Scholar] [CrossRef]
- Jacobson, N.C.; Lekkas, D.; Huang, R.; Thomas, N. Deep learning paired with wearable passive sensing data predicts deterioration in anxiety disorder symptoms across 17–18 years. J. Affect. Disord. 2021, 282, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J. Statistics the Elements of Statistical Learning. Math. Intell. 2009, 27, 83–85. [Google Scholar]
- Patle, A.; Chouhan, D.S. SVM kernel functions for classification. In Proceedings of the 2013 International Conference on Advances in Technology and Engineering, ICATE 2013, Mumbai, India, 23–25 January 2013. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, S.Y.; Kim, Y.H.; Park, K.S. A smartphone-based automatic diagnosis system for facial nerve palsy. Sensors 2015, 15, 26756–26768. [Google Scholar] [CrossRef] [PubMed]
- Luca, S.; Karsmakers, P.; Cuppens, K.; Croonenborghs, T.; Van de Vel, A.; Ceulemans, B.; Lagae, L.; Van Huffel, S.; Vanrumste, B. Detecting rare events using extreme value statistics applied to epileptic convulsions in children. Artif. Intell. Med. 2014, 60, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Ghoraani, B.; Hssayeni, M.D.; Bruack, M.M.; Jimenez-Shahed, J. Multilevel Features for Sensor-Based Assessment of Motor Fluctuation in Parkinson’s Disease Subjects. IEEE J. Biomed. Health Inform. 2020, 24, 1284–1295. [Google Scholar] [CrossRef]
- Kramer, O. K-Nearest Neighbors. In Dimensionality Reduction with Unsupervised Nearest Neighbors. Intelligent Systems Reference Library; Springer: Berlin/Heidelberg, Germany, 2013; Volume 51. [Google Scholar] [CrossRef]
- Jeon, H.; Lee, W.; Park, H.; Lee, H.J.; Kim, S.K.; Kim, H.B.; Jeon, B.; Park, K.S. Automatic classification of tremor severity in Parkinson’s disease using awearable device. Sensors 2017, 17, 2067. [Google Scholar] [CrossRef]
- Grunerbl, A.; Muaremi, A.; Osmani, V.; Bahle, G.; Ohler, S.; Troster, G.; Mayora, O.; Haring, C.; Lukowicz, P. Smartphone-based recognition of states and state changes in bipolar disorder patients. IEEE J. Biomed. Health Inform. 2015, 19, 140–148. [Google Scholar] [CrossRef]
- Pranckevičius, T.; Marcinkevičius, V. Comparison of Naive Bayes, Random Forest, Decision Tree, Support Vector Machines, and Logistic Regression Classifiers for Text Reviews Classification. Balt. J. Mod. Comput. 2017, 5, 221–232. [Google Scholar] [CrossRef]
- Worster, A.; Fan, J.; Ismaila, A. Understanding linear and logistic regression analyses. Can. J. Emerg. Med. 2007, 9, 111–113. [Google Scholar] [CrossRef]
- Morrow-Howell, N. The M word: Multicollinearity in multiple regression. Soc. Work. Res. 1994, 18, 247–251. [Google Scholar] [CrossRef]
- Schwenk, M.; Hauer, K.; Zieschang, T.; Englert, S.; Mohler, J.; Najafi, B. Sensor-derived physical activity parameters can predict future falls in people with dementia. Gerontology 2014, 60, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.V. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 1996, 49, 1225–1231. [Google Scholar] [CrossRef]
- Mudiyanselage, T.K.B.; Xiao, X.; Zhang, Y.; Pan, Y. Deep Fuzzy Neural Networks for Biomarker Selection for Accurate Cancer Detection. IEEE Trans. Fuzzy Syst. 2020, 28, 3219–3228. [Google Scholar] [CrossRef]
- Yagin, F.H.; Cicek, I.B.; Alkhateeb, A.; Yagin, B.; Colak, C.; Azzeh, M.; Akbulut, S. Explainable artificial intelligence model for identifying COVID-19 gene biomarkers. Comput. Biol. Med. 2023, 154, 106619. [Google Scholar] [CrossRef]
- Wang, Y.; Lucas, M.; Furst, J.; Fawzi, A.A.; Raicu, D. Explainable Deep Learning for Biomarker Classification of OCT Images. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 204–210. [Google Scholar] [CrossRef]
- Fisher, J.M.; Hammerla, N.Y.; Ploetz, T.; Andras, P.; Rochester, L.; Walker, R.W. Unsupervised home monitoring of Parkinson’s disease motor symptoms using body-worn accelerometers. Park. Relat. Disord. 2016, 33, 44–50. [Google Scholar] [CrossRef]
- Frogner, J.I.; Noori, F.M.; Halvorsen, P.; Hicks, S.A.; Garcia-Ceja, E.; Torresen, J.; Riegler, M.A. One-dimensional convolutional neural networks on motor activity measurements in detection of depression. In Proceedings of the HealthMedia 2019—Proceedings of the 4th International Workshop on Multimedia for Personal Health and Health Care, Co-Located with MM 2019, Nice, France, 21–25 October 2019; pp. 9–15. [Google Scholar] [CrossRef]
- Meisel, C.; el Atrache, R.; Jackson, M.; Schubach, S.; Ufongene, C.; Loddenkemper, T. Machine learning from wristband sensor data for wearable, noninvasive seizure forecasting. Epilepsia 2020, 61, 2653–2666. [Google Scholar] [CrossRef]
- Matarazzo, M.; Arroyo-Gallego, T.; Montero, P.; Puertas-Martín, V.; Butterworth, I.; Mendoza, C.S.; Ledesma-Carbayo, M.J.; Catalán, M.J.; Molina, J.A.; Bermejo-Pareja, F.; et al. Remote Monitoring of Treatment Response in Parkinson’s Disease: The Habit of Typing on a Computer. Mov. Disord. 2019, 34, 1488–1495. [Google Scholar] [CrossRef]
- Weiss, K.; Khoshgoftaar, T.M.; Background, D.W. A survey of transfer learning. J. Big Data 2016, 3, 1345–1459. [Google Scholar] [CrossRef]
- Kamishima, T.; Hamasaki, M.; Akaho, S. TrBagg: A Simple Transfer Learning Method and its Application to Personalization in Collaborative Tagging. In Proceedings of the 2009 Ninth IEEE International Conference on Data Mining, Miami, FL, USA, 6–9 December 2009; pp. 219–228. [Google Scholar] [CrossRef]
- Fu, Z.; He, X.; Wang, E.; Huo, J.; Huang, J.; Wu, D. Personalized Human Activity Recognition Based on Integrated Wearable Sensor and Transfer Learning. Sensors 2021, 21, 885. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qin, X.; Wang, J.; Yu, C.; Gao, W. FedHealth: A Federated Transfer Learning Framework for Wearable Healthcare. IEEE Intell. Syst. 2020, 35, 83–93. [Google Scholar] [CrossRef]
- Goschenhofer, J.; Pfister, F.M.J.; Yuksel, K.A.; Bischl, B.; Fietzek, U.; Thomas, J. Wearable-Based Parkinson’s Disease Severity Monitoring Using Deep Learning. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2020; Volume 11908 LNAI, pp. 400–415. [Google Scholar] [CrossRef]
- Hssayeni, M.D.; Jimenez-Shahed, J.; Burack, M.A.; Ghoraani, B. Ensemble deep model for continuous estimation of Unified Parkinson’s Disease Rating Scale III. Biomed. Eng. Online 2021, 20, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Q. Special Topic: Machine Learning An overview of multi-task learning. Natl. Sci. Rev. 2018, 5, 30–43. [Google Scholar] [CrossRef]
- Lee, G.; Yang, E.; Hwang, S. Asymmetric multi-task learning based on task relatedness and loss. In Proceedings of the International Conference on Machine Learning 2016, New York, NY, USA, 19–24 June 2016; pp. 230–238. [Google Scholar]
- Xin, W.; Bi, J.; Yu, S.; Sun, J.; Song, M. Multiplicative Multitask Feature Learning. J. Mach. Learn. Res. JMLR 2016, 17, 1–33. [Google Scholar]
- Zhang, Z.; Jung, T.P.; Makeig, S.; Pi, Z.; Rao, B.D. Spatiotemporal sparse Bayesian learning with applications to compressed sensing of multichannel physiological signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 1186–1197. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Vlachos, M. Personalization of deep learning. In Data Science–Analytics and Applications: Proceedings of the 3rd International Data Science Conference–iDSC2020; Springer: Wiesbaden, Geramny, 2021; pp. 89–96. [Google Scholar]
- Khademi, A.; El-Manzalawy, Y.; Buxton, O.M.; Honavar, V. Toward personalized sleep-wake prediction from actigraphy. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical and Health Informatics, BHI 2018, Vegas, NV, USA, 4–7 March 2018; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2018; pp. 414–417. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Putin, E.; Mamoshina, P.; Aliper, A.; Korzinkin, M.; Moskalev, A.; Kolosov, A.; Ostrovskiy, A.; Cantor, C.; Vijg, J.; Zhavoronkov, A. Deep biomarkers of human aging: Application of deep neural networks to biomarker development. Aging 2016, 8, 1021–1033. [Google Scholar] [CrossRef]
- Yang, L.; Shami, A. On hyperparameter optimization of machine learning algorithms: Theory and practice. Neurocomputing 2020, 415, 295–316. [Google Scholar] [CrossRef]
- Waring, J.; Lindvall, C.; Umeton, R. Automated machine learning: Review of the state-of-the-art and opportunities for healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef]
- Bergstra, J.; Ca, J.B.; Ca, Y.B. Random Search for Hyper-Parameter Optimization Yoshua Bengio. 2012. Available online: http://scikit-learn.sourceforge.net (accessed on 5 January 2023).
- Beam, A.L.; Manrai, A.K.; Ghassemi, M. Challenges to the Reproducibility of Machine Learning Models in Health Care. JAMA 2020, 323, 305. [Google Scholar] [CrossRef] [PubMed]
- Ahlrichs, C.; Samà, A.; Lawo, M.; Cabestany, J.; Rodríguez-Martín, D.; Pérez-López, C.; Sweeney, D.; Quinlan, L.R.; Laighin, G.Ò.; Counihan, T.; et al. Detecting freezing of gait with a tri-axial accelerometer in Parkinson’s disease patients. Med. Biol. Eng. Comput. 2016, 54, 223–233. [Google Scholar] [CrossRef]
- Rosenwein, T.; Dafna, E.; Tarasiuk, A.; Zigel, Y. Detection of Breathing Sounds during Sleep Using Non-Contact Audio Recordings; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Pérez-López, C.; Samà, A.; Rodríguez-Martín, D.; Moreno-Aróstegui, J.M.; Cabestany, J.; Bayes, A.; Mestre, B.; Alcaine, S.; Quispe, P.; Laighin, G.; et al. Dopaminergic-induced dyskinesia assessment based on a single belt-worn accelerometer. Artif. Intell. Med. 2016, 67, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Bernad-Elazari, H.; Herman, T.; Mirelman, A.; Gazit, E.; Giladi, N.; Hausdorff, J.M. Objective characterization of daily living transitions in patients with Parkinson’s disease using a single body-fixed sensor. J. Neurol. 2016, 263, 1544–1551. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.V.; Pereira, E.M.; Cardoso, J.S. Machine Learning Interpretability: A Survey on Methods and Metrics. Electronics 2019, 8, 832. [Google Scholar] [CrossRef]
- Zhou, J.; Gandomi, A.H.; Chen, F.; Holzinger, A. Evaluating the Quality of Machine Learning Explanations: A Survey on Methods and Metrics. Electronics 2021, 10, 593. [Google Scholar] [CrossRef]
- Hossin, M.; Sulaiman, M.N. A Review on Evaluation Metrics for Data Classification Evaluations. Int. J. Data Min. Knowl. Manag. Process 2015, 5, 1–11. [Google Scholar] [CrossRef]
- He, H.; Ma, Y. Imbalanced Learning; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Wan, S.; Liang, Y.; Zhang, Y.; Guizani, M. Deep Multi-Layer perceptron classifier for behavior analysis to estimate Parkinson’s disease severity using smartphones. IEEE Access 2018, 6, 36825–36833. [Google Scholar] [CrossRef]
- Chicco, D.; Tötsch, N.; Jurman, G. The matthews correlation coefficient (Mcc) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation. BioData Min. 2021, 14, 1–22. [Google Scholar] [CrossRef]
- Jurman, G.; Riccadonna, S.; Furlanello, C. A Comparison of MCC and CEN Error Measures in Multi-Class Prediction. PLoS ONE 2012, 7, e41882. [Google Scholar] [CrossRef]
- Faurholt-Jepsen, M.; Busk, J.; HelgaÞórarinsdóttir; Frost, M.; Bardram, J.E.; Vinberg, M.; Kessing, L.V. Objective smartphone data as a potential diagnostic marker of bipolar disorder. Aust. N. Z. J. Psychiatry 2019, 53, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shen, D.; Nie, T.; Kou, Y. A hybrid sampling algorithm combining M-SMOTE and ENN based on Random forest for medical imbalanced data. J. Biomed. Inform. 2020, 107, 103465. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Zou, B.; Wei, F.; Liu, X.; Wang, L. Effective prediction of three common diseases by combining SMOTE with Tomek links technique for imbalanced medical data. In Proceedings of the 2016 IEEE International Conference of Online Analysis and Computing Science, ICOACS 2016, Chongqing, China, 28–29 May 2016; pp. 225–228. [Google Scholar] [CrossRef]
- Botchkarev, A. Performance Metrics (Error Measures) in Machine Learning Regression, Forecasting and Prognostics: Properties and Typology. Interdiscip. J. Inf. Knowl. Manag. 2018, 14, 45–76. [Google Scholar] [CrossRef]
- di Bucchianico, A. Coefficient of Determination. In Encyclopedia of Statistics in Quality and Reliability; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Piepho, H. A coefficient of determination (R2) for generalized linear mixed models. Biom. J. 2019, 61, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Gelman, A.; Pardoe, I. Bayesian Measures of Explained Variance and Pooling in Multilevel (Hierarchical) Models. Technometrics 2006, 48, 241–251. [Google Scholar] [CrossRef]
- Hodson, T.O. Root-mean-square error (RMSE) or mean absolute error (MAE): When to use them or not. Geosci. Model. Dev. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]
- Mezzadri, G.; Laloë, T.; Mathy, F.; Reynaud-Bouret, P. Hold-out strategy for selecting learning models: Application to categorization subjected to presentation orders. J. Math. Psychol. 2022, 109, 102691. [Google Scholar] [CrossRef]
- Gholamiangonabadi, D.; Kiselov, N.; Grolinger, K. Deep Neural Networks for Human Activity Recognition with Wearable Sensors: Leave-One-Subject-Out Cross-Validation for Model Selection. IEEE Access 2020, 8, 133982–133994. [Google Scholar] [CrossRef]
- Little, M.A.; Varoquaux, G.; Saeb, S.; Lonini, L.; Jayaraman, A.; Mohr, D.C.; Kording, K.P. Using and understanding cross-validation strategies. Perspectives on Saeb et al. Gigascience 2017, 6, 1–6. [Google Scholar] [CrossRef]
- Peterson, R.A.; Cavanaugh, J.E. Ordered quantile normalization: A semiparametric transformation built for the cross-validation era. J. Appl. Stat. 2020, 47, 2312–2327. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y. Cross-validation for selecting a model selection procedure. J. Econom. 2015, 187, 95–112. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-Validation. In Encyclopedia of Database Systems; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–7. [Google Scholar] [CrossRef]
- Browne, M.W. Cross-validation methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Wainer, J.; Cawley, G. Nested cross-validation when selecting classifiers is overzealous for most practical applications. Expert Syst. Appl. 2021, 182, 115222. [Google Scholar] [CrossRef]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. 1995. Available online: http://robotics.stanford.edu/~ronnyk (accessed on 5 January 2023).
- Vanwinckelen, G.; Blockeel, H. On estimating model accuracy with repeated cross-validation. In BeneLearn 2012: Proceedings of the 21st Belgian-Dutch conference on machine learning; Benelearn 2012 Organization Committee: Ghent, Belgium, 2012; pp. 39–44. [Google Scholar]
- Parvandeh, S.; Yeh, H.-W.; Paulus, M.P.; McKinney, B.A. Consensus Features Nested Cross-Validation. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Goble, C.; Cohen-Boulakia, S.; Soiland-Reyes, S.; Garijo, D.; Gil, Y.; Crusoe, M.; Peters, K.; Schober, D. Fair computational workflows. Data Intell. 2020, 2, 108–121. [Google Scholar] [CrossRef]
- Muehlematter, U.J.; Daniore, P.; Vokinger, K.N. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): A comparative analysis. Lancet Digit. Health 2021, 3, e195–e203. [Google Scholar] [CrossRef]
- Beckers, R.; Kwade, Z.; Zanca, F. The EU medical device regulation: Implications for artificial intelligence-based medical device software in medical physics. Phys. Med. 2021, 83, 1–8. [Google Scholar] [CrossRef]
- van Oirschot, J.; Ooms, G. Interpreting the EU Artificial Intelligence Act for the Health Sector; Health Action International: Amsterdam, The Netherlands, February 2022. [Google Scholar]
- Agarwal, S.; LeFevre, A.; Lee, J.; L’engle, K.; Mehl, G.; Sinha, C.; Labrique, A. Guidelines for reporting of health interventions using mobile phones: Mobile health (mHealth) evidence reporting and assessment (mERA) checklist. BMJ 2016, 352, i1174. [Google Scholar] [CrossRef]
- Yang, J.; Soltan, A.A.S.; Clifton, D.A. Machine learning generalizability across healthcare settings: Insights from multi-site COVID-19 screening. NPJ Digit. Med. 2022, 5, 69. [Google Scholar] [CrossRef]
- Petersen, E.; Potdevin, Y.; Mohammadi, E.; Zidowitz, S.; Breyer, S.; Nowotka, D.; Henn, S.; Pechmann, L.; Leucker, M.; Rostalski, P.; et al. Responsible and Regulatory Conform Machine Learning for Medicine: A Survey of Challenges and Solutions. IEEE Access 2022, 10, 58375–58418. [Google Scholar] [CrossRef]
- FAIR Principles—GO FAIR. Available online: https://www.go-fair.org/fair-principles/ (accessed on 16 December 2021).
- Fletcher, R.R.; Nakeshimana, A.; Olubeko, O. Addressing Fairness, Bias, and Appropriate Use of Artificial Intelligence and Machine Learning in Global Health. Front. Artif. Intell. 2021, 3, 116. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Desrosiers, C.; Frasnelli, J. Machine Learning for the Diagnosis of Parkinson’s Disease: A Review of Literature. Front. Aging Neurosci. 2021, 13, 633752. [Google Scholar] [CrossRef] [PubMed]
- Chekroud, A.M.; Bondar, J.; Delgadillo, J.; Doherty, G.; Wasil, A.; Fokkema, M.; Cohen, Z.; Belgrave, D.; DeRubeis, R.; Iniesta, R.; et al. The promise of machine learning in predicting treatment outcomes in psychiatry. World Psychiatry 2021, 20, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Kruizinga, M.D.; Zhuparris, A.; Dessing, E.; Krol, F.J.; Sprij, A.J.; Doll, R.; Stuurman, F.E.; Exadaktylos, V.; Driessen, G.J.A.; Cohen, A.F. Development and technical validation of a smartphone-based pediatric cough detection algorithm. Pediatr. Pulmonol. 2022, 57, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Doshi-Velez, F.; Kim, B. Towards A Rigorous Science of Interpretable Machine Learning. arXiv 2017, arXiv:1702.08608. [Google Scholar]
- Ignatiev, A. Towards Trustable Explainable AI. 2020. Available online: https://www.kaggle.com/uciml/zoo-animal-classification (accessed on 5 January 2023).
- Walsh, I.; Fishman, D.; Garcia-Gasulla, D.; Titma, T.; Pollastri, G.; Capriotti, E.; Casadio, R.; Capella-Gutierrez, S.; Cirillo, D.; Del Conte, A.; et al. DOME: Recommendations for supervised machine learning validation in biology. Nat. Methods 2021, 18, 1122–1127. [Google Scholar] [CrossRef]
- Zippel, C.; Bohnet-Joschko, S. Rise of Clinical Studies in the Field of Machine Learning: A Review of Data Registered in ClinicalTrials.gov. Int. J. Environ. Res. Public Health 2021, 18, 5072. [Google Scholar] [CrossRef] [PubMed]


| Stage | Objective | Example |
|---|---|---|
| The ML pipeline is provided with a study objective in which the features and corresponding outputs are defined. The ML model aims to identify the associations between the features and outputs. | The study objective is to classify Parkinson’s Disease patients and control groups using smartphone-based features. |
| Data preprocessing filters and transforms raw data to guarantee or enhance the ML training process. | To improve the model performance, one may identify and exclude any missing or outlier data. |
| Feature engineering uses raw data to create new features that are not readily available in the dataset. Feature selection selects the most relevant features for the model objective by removing redundant or noisy features. Together, the goal is to simplify and accelerate the computational process while also improving the model process. For deep learning methods, the concept of “feature engineering” is typically embedded within the model architecture and training process, although substantial preprocessing steps may occur prior to that. | An interaction of two or more predictors (such as a ratio or product) or re-representation of a predictor are examples of feature engineering. Removing highly correlated or non-informative features are examples of feature selection. Note: The feature selection step can occur during model training |
| During training, the ML model(s) iterates through all the examples in the training dataset and optimizes the parameters of the mathematical function to minimize the prediction error. To evaluate the performance of the trained ML model, the predictions of an unseen test set are compared with a known ground truth label. | Cross-validation can be used to optimize and evaluate model performance. Classification models may be evaluated based on their prediction accuracy, sensitivity, and specificity, while regression models may be evaluated using variance explained (R2) and Mean Absolute Error. |
| Domain | Search String |
|---|---|
| Technology | ((“smartphone”[tiab] OR “wearable”[tiab] OR “remote + monitoring”[tiab] OR “home + monitoring”[tiab] OR “mobile + sensors”[tiab] OR “mobile + montoring”[tiab] OR “behavioral + sensing”[tiab] OR “geolocation”[tiab] OR “mHealth”[tiab] OR “passive + monitoring”[tiab] OR “digital + phenotype”[tiab] OR “digital + phenotyping”[tiab] OR “digital + biomarker”[tiab]) |
| Analysis | AND (“machine + learning”[tiab] OR “deep + learning”[tiab] OR “random + forest”[tiab] OR “neural + network”[tiab] OR “time + series”[tiab] OR “regression”[tiab] OR “SVM”[tiab] OR “knn”[tiab] OR “dynamics + model”[tiab] OR “decision + tree”[tiab] OR “discriminant + analysis”[tiab] OR “feature + engineering”[tiab] OR “feature + selection”[tiab] OR “data + mining”[tiab] OR “model”[tiab] OR “classification”[tiab] OR “diagnostic”[tiab] OR “prognostic”[tiab] OR “symptom + severity”[tiab] OR “prediction”[tiab] OR “monitoring”[tiab]) |
| Population | AND (“disease”[tiab] OR “disorder”[tiab] OR “diagnosis”[tiab] OR “prognosis” OR “alzheimer”[tiab] OR “parkinson”[tiab] OR “Huntington”[tiab] OR “neurodegenerative”[tiab] OR “degenerative” OR “tremor”[tiab] OR “bipolar”[tiab] OR “depression”[tiab] OR “manic”[tiab] OR “anxiety”[tiab] OR “vocal + biomarker”[tiab] OR “amyotrophic + lateral + sclerosis”[tiab] OR “central + nervous + system”[tiab] OR “symptom”[tiab] OR “psychosis”[tiab] OR “stroke”[tiab] OR “muscular dystrophy”[tiab] OR “Facioscapulohumeral Dystrophy”[tiab] OR “autoimmune”[tiab] OR “seizure”[tiab] OR “multiple + sclerosis”[tiab]) |
| Date | AND (“2012/01/01”[PDAT]:”2022/12/31”[PDAT]) |
| Language | AND (English[lang]) |
| Exclusion Criteria | NOT(“animals”[tiab] OR “implant”[tiab] OR “hospital”[tiab] OR “caregiver”[tiab] OR “telemedicine”[tiab] OR “telerehabilitation”[tiab] OR “smartphone + addiction”[tiab] OR “nursing”[tiab] OR”screening”[tiab] OR “recruitment”[tiab] OR “diabetes”[tiab] OR “malaria”[tiab] OR “self-care”[tiab] OR “self-management”[tiab] OR “self-help”[tiab]) |
| Article Type | AND (clinicalstudy[Filter] OR clinicaltrial[Filter] OR clinicaltrialphasei[Filter] OR clinicaltrialphaseii[Filter] OR clinicaltrialphaseiii[Filter] OR clinicaltrialphaseiv[Filter] OR controlledclinicaltrial[Filter] OR meta-analysis[Filter] observationalstudy[Filter] OR randomizedcontrolledtrial[Filter] OR systematicreview[Filter]) |
| Category | Criteria |
|---|---|
| Population | The study must be initiated by a research organization and not by the participants. The participants must have a clinical diagnosis that is affected by the CNS. Hence, studies that collected data from participants with no clinically confirmed diagnosis were not considered. |
| Intervention | The study must include the use of smartphone or non-invasive wearables to remotely monitor and quantify passive biomarkers under free-living conditions. |
| Comparator | A ground truth comparator for digital phenotyping such as clinical assessment, medical records, or self-reported outcomes. |
| Outcomes | A ML model that is used to classify a clinical label (such as a diagnosis, or clinical event), estimate symptom severity, or to detect treatment effects. |
| Study Type | The paper must be about a human-centered observational study (cohort or longitudinal) where the data were collected outside the clinic, lab, or hospital (free-living conditions). Hence, studies that use smartphones or wearables as a form of intervention or as screening tools are not of interest. The study must show if the ML models had ecological validity by validating the models using free-living data. The study has to have been written or translated into English and published within the last 10 years (2012 onwards). |
| Term | Title 2 | Title 3 |
|---|---|---|
| Accuracy | Out of all the predictions, how many predictions were correctly identified as positive or negative? | |
| Precision | How many predictions were correctly labeled as patients out of all correctly classified patients and misclassified healthy controls? | |
| Specificity | How many predictions were correctly labeled as healthy controls out of all healthy controls? In other words, of all healthy controls, who were correctly identified as such? | |
| Recall/Sensitivity | Of all the patients, who were correctly classified/identified as such? | |
| F1-score | How many predictions were correctly labeled as patients (recall) and what was the accuracy with regards to correctly predicted patients (precision)? | |
| Mean Square Error | What is the absolute difference between the true scores and the predicted scores? | |
| Root Mean Square Error | What is the average difference between the true and the predicted scores (in the same unit of the true scores)? | |
| R2 | What fraction of the variance in the data is captured by the model? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
ZhuParris, A.; de Goede, A.A.; Yocarini, I.E.; Kraaij, W.; Groeneveld, G.J.; Doll, R.J. Machine Learning Techniques for Developing Remotely Monitored Central Nervous System Biomarkers Using Wearable Sensors: A Narrative Literature Review. Sensors 2023, 23, 5243. https://doi.org/10.3390/s23115243
ZhuParris A, de Goede AA, Yocarini IE, Kraaij W, Groeneveld GJ, Doll RJ. Machine Learning Techniques for Developing Remotely Monitored Central Nervous System Biomarkers Using Wearable Sensors: A Narrative Literature Review. Sensors. 2023; 23(11):5243. https://doi.org/10.3390/s23115243
Chicago/Turabian StyleZhuParris, Ahnjili, Annika A. de Goede, Iris E. Yocarini, Wessel Kraaij, Geert Jan Groeneveld, and Robert Jan Doll. 2023. "Machine Learning Techniques for Developing Remotely Monitored Central Nervous System Biomarkers Using Wearable Sensors: A Narrative Literature Review" Sensors 23, no. 11: 5243. https://doi.org/10.3390/s23115243
APA StyleZhuParris, A., de Goede, A. A., Yocarini, I. E., Kraaij, W., Groeneveld, G. J., & Doll, R. J. (2023). Machine Learning Techniques for Developing Remotely Monitored Central Nervous System Biomarkers Using Wearable Sensors: A Narrative Literature Review. Sensors, 23(11), 5243. https://doi.org/10.3390/s23115243








