Improved Yield Prediction of Winter Wheat Using a Novel Two-Dimensional Deep Regression Neural Network Trained via Remote Sensing †
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
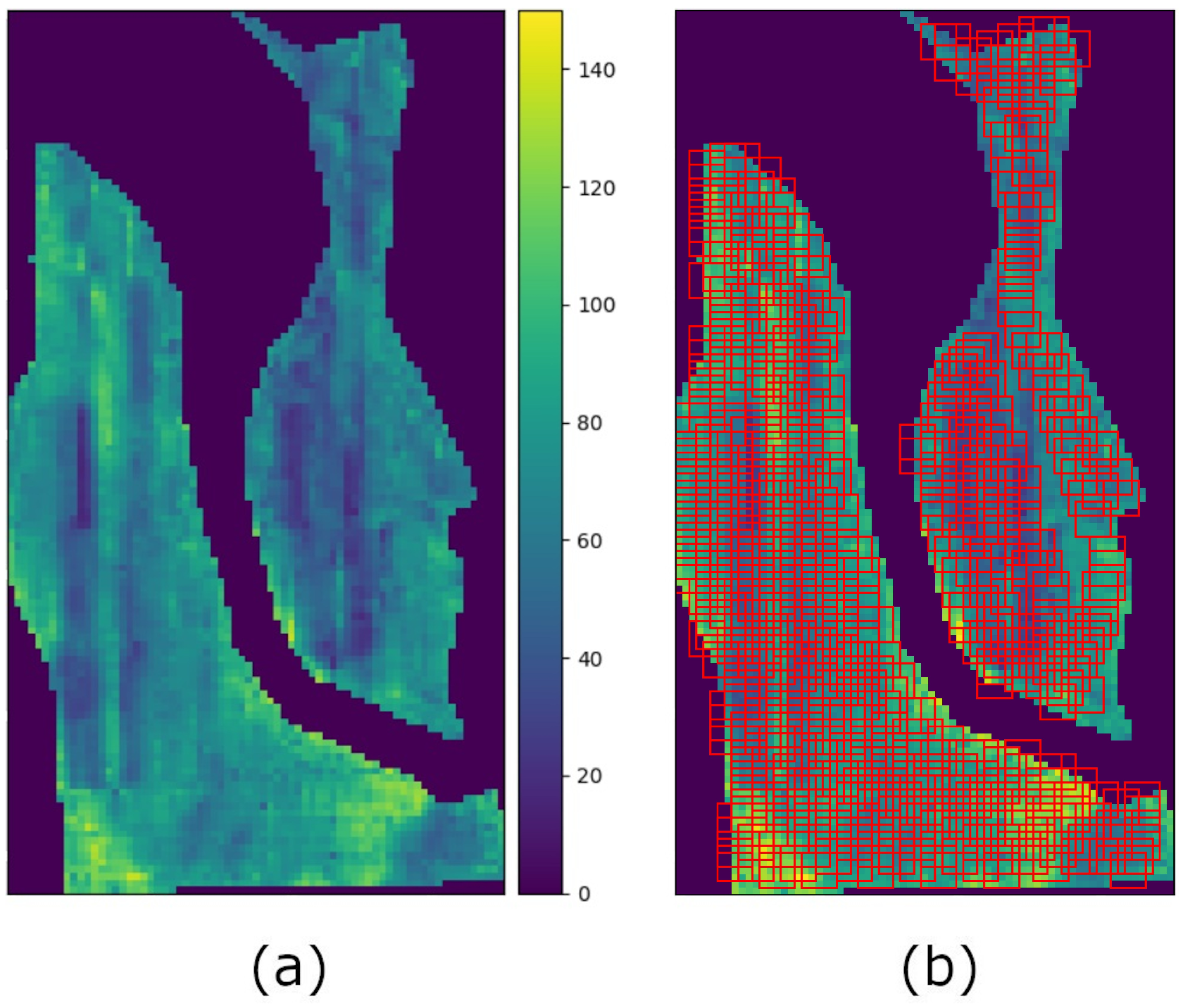
3.1. Datasets
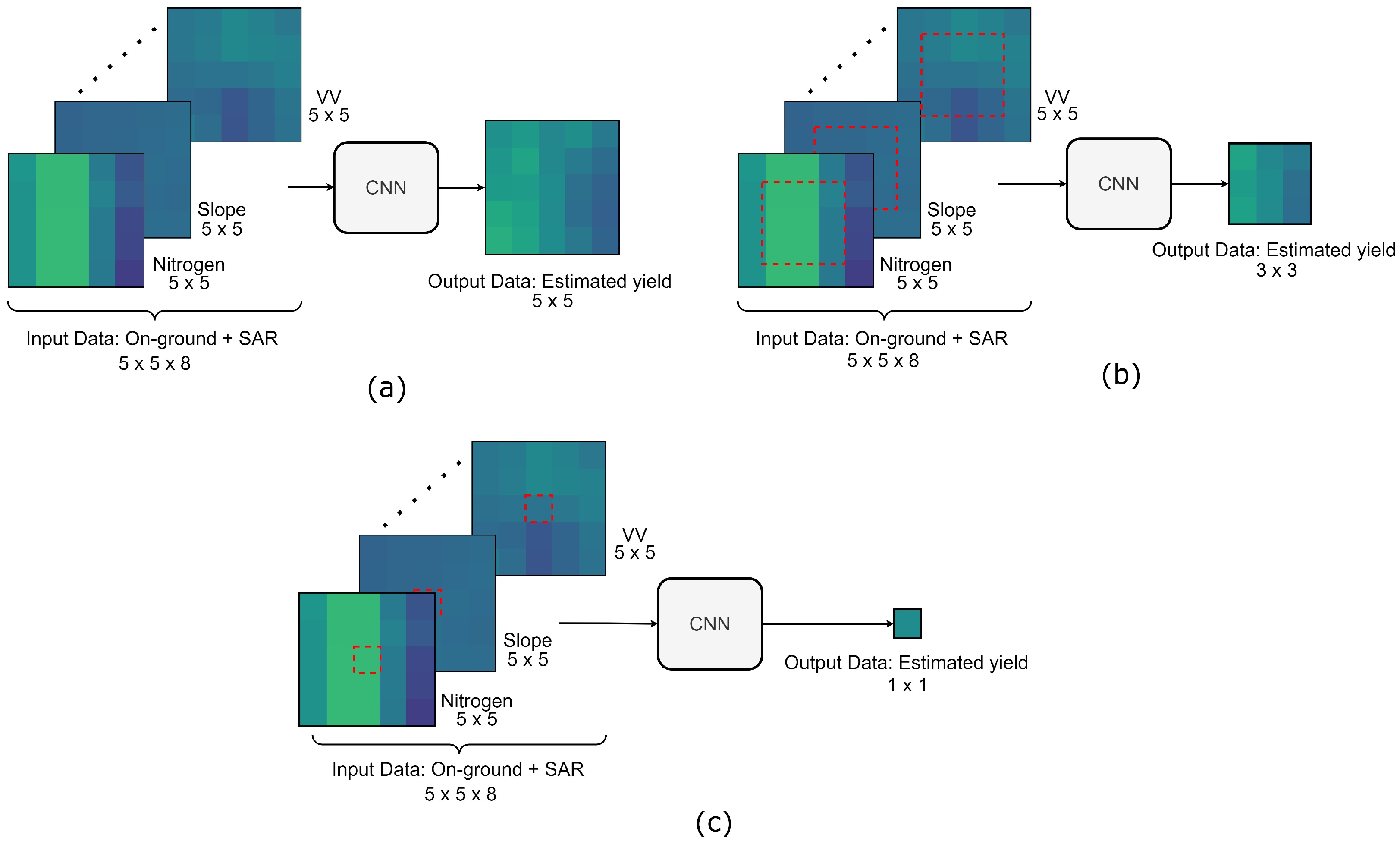
3.2. Yield Prediction Model
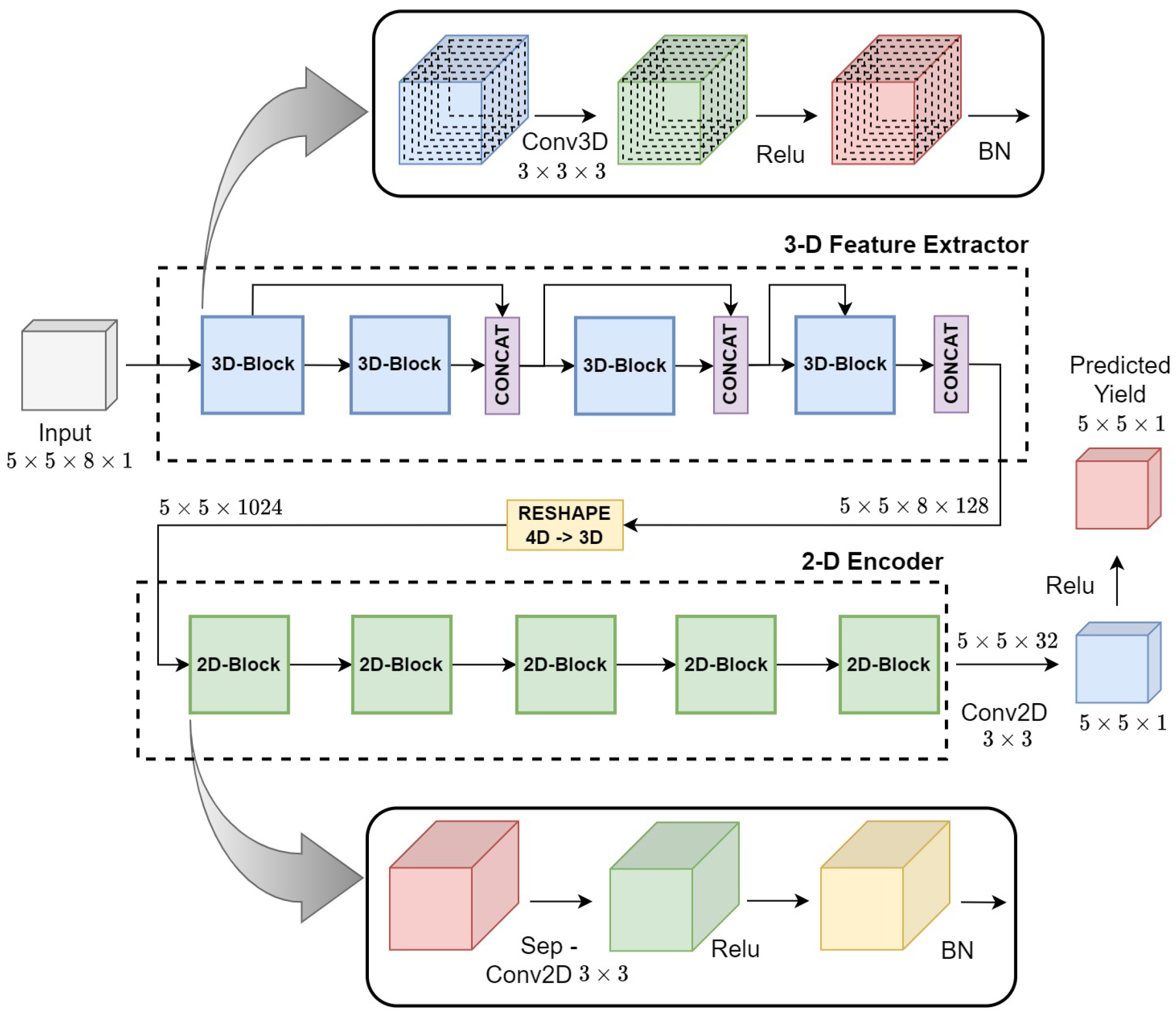
3.3. Hyper3DNetReg Architecture
3.4. Predicted Yield Map Generation
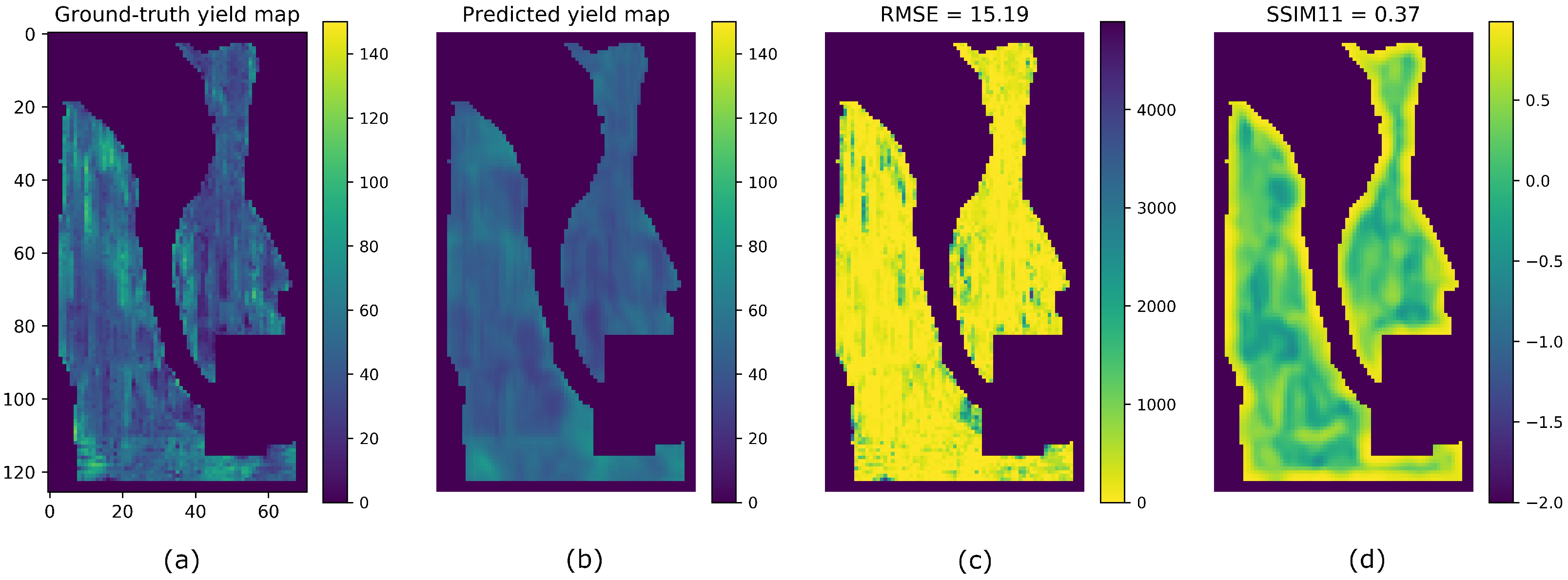
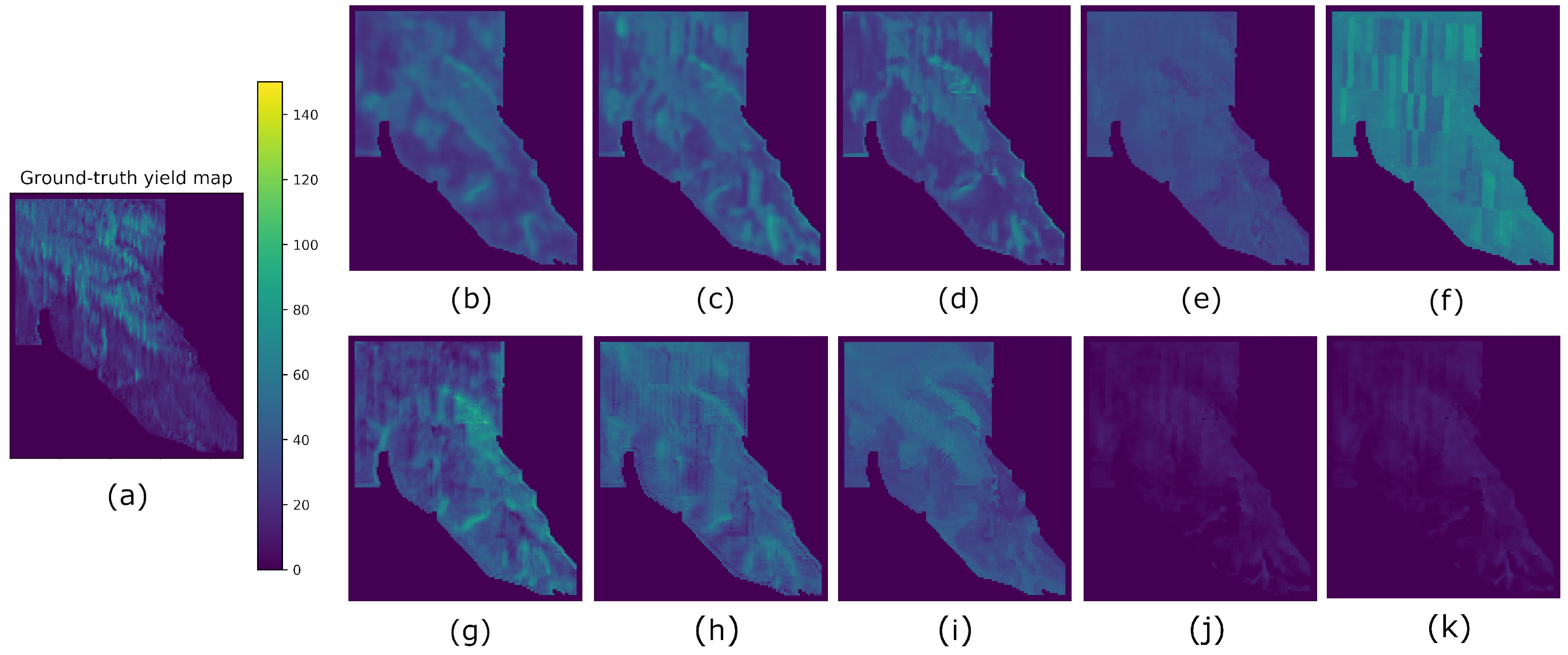
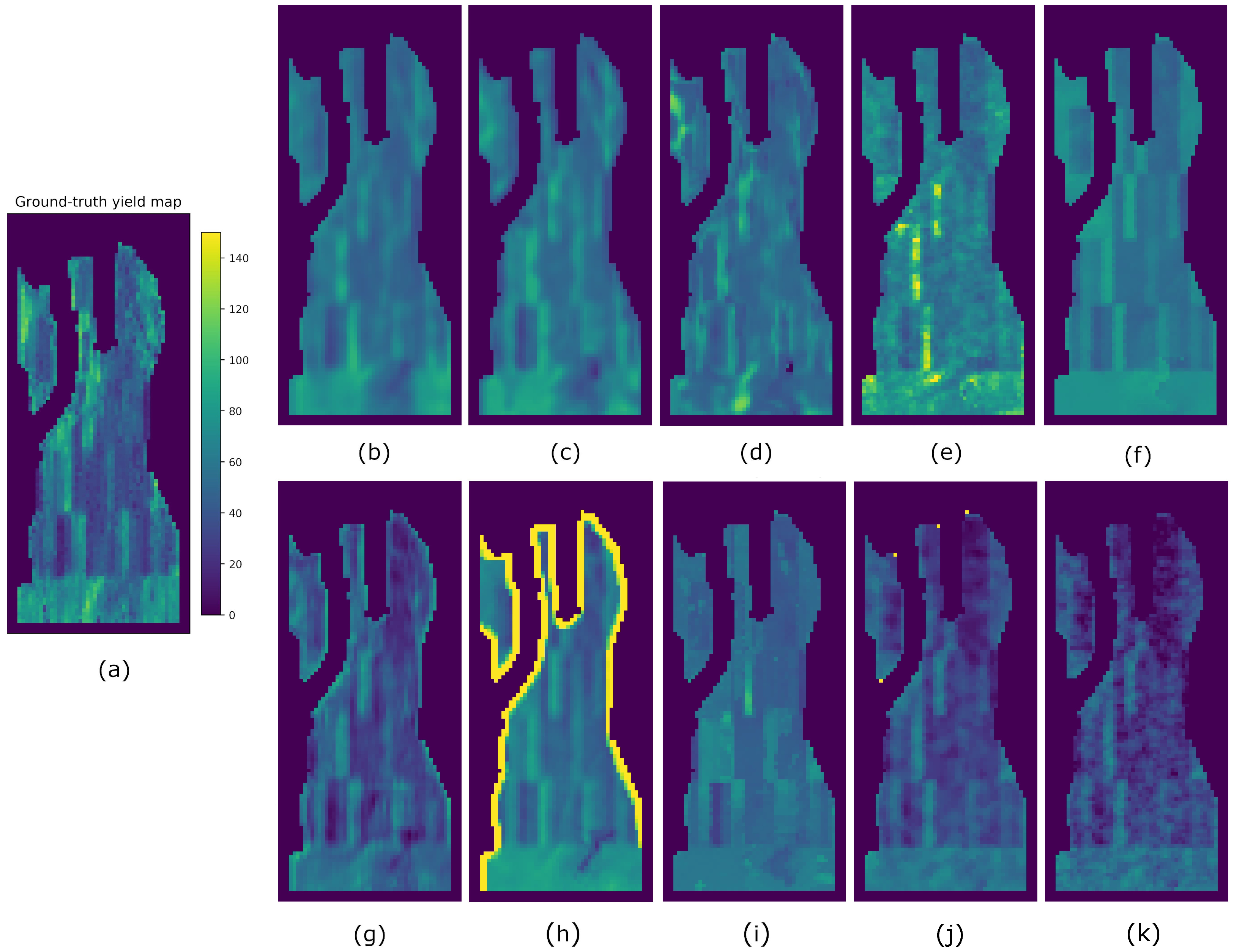
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Society for Precision Agriculture. Precision Agriculture Definition. Available online: https://www.ispag.org/about/definition (accessed on 1 November 2022).
- Gebbers, R.; Adamchuk, V.I. Precision Agriculture and Food Security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef] [PubMed]
- McBratney, A.; Whelan, B.; Ancev, T.; Bouma, J. Future Directions of Precision Agriculture. Precis. Agric. 2005, 6, 7–23. [Google Scholar] [CrossRef]
- Shafi, U.; Mumtaz, R.; García-Nieto, J.; Hassan, S.; Zaidi, S.; Iqbal, N. Precision Agriculture Techniques and Practices: From Considerations to Applications. Sensors 2019, 19, 3796. [Google Scholar] [CrossRef] [PubMed]
- Cisternas, I.; Velásquez, I.; Caro, A.; Rodríguez, A. Systematic literature review of implementations of precision agriculture. Comput. Electron. Agric. 2020, 176, 105626. [Google Scholar] [CrossRef]
- Cook, S.; Lacoste, M.; Evans, F.; Ridout, M.; Gibberd, M.; Oberthür, T. An On-Farm experimental philosophy for farmer-centric digital innovation. In Proceedings of the 14th International Conference on Precision Agriculture, Montreal, QC, Canada, 24–27 June 2018. [Google Scholar]
- Bullock, D.S.; Boerngen, M.; Tao, H.; Maxwell, B.; Luck, J.D.; Shiratsuchi, L.; Puntel, L.; Martin, N.F. The Data-Intensive Farm Management Project: Changing Agronomic Research Through On-Farm Precision Experimentation. Agron. J. 2019, 111, 2736–2746. [Google Scholar] [CrossRef]
- Vuran, M.; Salam, A.; Wong, R.; Irmak, S. Internet of underground things in precision agriculture: Architecture and technology aspects. Ad Hoc Netw. 2018, 81, 160–173. [Google Scholar] [CrossRef]
- Hunt, E.; Daughtry, C.S.T. What good are unmanned aircraft systems for agricultural remote sensing and precision agriculture? Int. J. Remote Sens. 2018, 39, 5345–5376. [Google Scholar] [CrossRef]
- Van Klompenburg, T.; Kassahun, A.; Catal, C. Crop yield prediction using machine learning: A systematic literature review. Comput. Electron. Agric. 2020, 177, 105709. [Google Scholar] [CrossRef]
- Patrício, D.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Horie, T.; Yajima, M.; Nakagawa, H. Yield forecasting. Agric. Syst. 1992, 40, 211–236. [Google Scholar] [CrossRef]
- Maxwell, B.; Hegedus, P.; Davis, P.; Bekkerman, A.; Payn, R.; Sheppard, J.; Silverman, N.; Izurieta, C. Can optimization associated with on-farm experimentation using site-specific technologies improve producer management decisions? In Proceedings of the 14th International Conference on Precision Agriculture, Montreal, QC, Canada, 24–27 June 2018. [Google Scholar]
- Schimmelpfennig, D.; Lowenberg-DeBoer, J. Farm Types and Precision Agriculture Adoption: Crops, Regions, Soil Variability, and Farm Size Farm Types and Precision Agriculture Adoption: Crops, Regions, Soil Variability, and Farm Size. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Russello, H. Convolutional Neural Networks for Crop Yield Prediction Using Satellite Images. Master’s Thesis, University of Amsterdam, Amsterdam, The Netherlands, 2018. [Google Scholar]
- Pandey, P.C.; Mandal, V.P.; Katiyar, S.; Kumar, P.; Tomar, V.; Patairiya, S.; Ravisankar, N.; Gangwar, B. Geospatial Approach to Assess the Impact of Nutrients on Rice Equivalent Yield Using MODIS Sensors’-Based MOD13Q1-NDVI Data. IEEE Sens. J. 2015, 15, 6108–6115. [Google Scholar] [CrossRef]
- Gómez, D.; Salvador, P.; Sanz, J.; Casanova, J. Potato Yield Prediction Using Machine Learning Techniques and Sentinel 2 Data. Remote Sens. 2019, 11, 1745. [Google Scholar] [CrossRef]
- Peerlinck, A.; Sheppard, J.; Senecal, J. Adaboost with neural networks for yield and protein prediction in precision agriculture. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019. [Google Scholar]
- Barbosa, A.; Trevisan, R.; Hovakimyan, N.; Martin, N. Modeling yield response to crop management using convolutional neural networks. Comput. Electron. Agric. 2020, 170, 105197. [Google Scholar] [CrossRef]
- Peerlinck, A.; Sheppard, J.; Maxwell, B. Using Deep Learning in Yield and Protein Prediction of Winter Wheat Based on Fertilization Prescriptions in Precision Agriculture. In Proceedings of the 14th International Conference on Precision Agriculture, Montreal, QC, Canada, 24–27 June 2018. [Google Scholar]
- Morales, G.; Sheppard, J. Two-dimensional deep regression for early yield prediction of winter wheat. In Proceedings of the SPIE Future Sensing Technologies 2021, Online, 15–19 November 2021; Kimata, M., Shaw, J.A., Valenta, C.R., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2021; Volume 11914, p. 119140H. [Google Scholar]
- Bullock, D.G.; Bullock, D.S. Quadratic and Quadratic-Plus-Plateau Models for Predicting Optimal Nitrogen Rate of Corn: A Comparison. Agron. J. 1994, 86, 191–195. [Google Scholar] [CrossRef]
- Roberts, R.K.; Mahajanashetti, S.B.; English, B.C.; Larson, J.A.; Tyler, D.D. Variable Rate Nitrogen Application on Corn Fields: The Role of Spatial Variability and Weather. J. Agric. Appl. Econ. 2002, 34, 111–129. [Google Scholar] [CrossRef]
- Ackello-Ogutu, C.; Paris, Q.; Williams, W.A. Testing a von Liebig Crop Response Function against Polynomial Specifications. Am. J. Agric. Econ. 1985, 67, 873–880. [Google Scholar] [CrossRef]
- Boyer, C.N.; Larson, J.A.; Roberts, R.K.; McClure, A.T.; Tyler, D.D.; Zhou, V. Stochastic Corn Yield Response Functions to Nitrogen for Corn after Corn, Corn after Cotton, and Corn after Soybeans. J. Agric. Appl. Econ. 2013, 45, 669–681. [Google Scholar] [CrossRef]
- Anselin, L.; Bongiovanni, R.; Lowenberg-DeBoer, J. A Spatial Econometric Approach to the Economics of Site-Specific Nitrogen Management in Corn Production. Am. J. Agric. Econ. 2004, 86, 675–687. [Google Scholar] [CrossRef]
- Bolton, D.; Friedl, M. Forecasting crop yield using remotely sensed vegetation indices and crop phenology metrics. Agric. For. Meteorol. 2013, 173, 74–84. [Google Scholar] [CrossRef]
- Johnson, D. An assessment of pre- and within-season remotely sensed variables for forecasting corn and soybean yields in the United States. Remote Sens. Environ. 2014, 141, 116–128. [Google Scholar] [CrossRef]
- Paul, M.; Vishwakarma, S.K.; Verma, A. Analysis of Soil Behaviour and Prediction of Crop Yield Using Data Mining Approach. In Proceedings of the 2015 International Conference on Computational Intelligence and Communication Networks (CICN), Jabalpur, India, 12–14 December 2015; pp. 766–771. [Google Scholar]
- Gonzalez-Sanchez, A.; Frausto-Solis, J.; Ojeda-Bustamante, W. Predictive ability of machine learning methods for massive crop yield prediction. Span. J. Agric. Res. 2014, 12, 313–328. [Google Scholar] [CrossRef]
- Kim, N.; Lee, Y. Machine Learning Approaches to Corn Yield Estimation Using Satellite Images and Climate Data: A Case of Iowa State. J. Korean Soc. Surv. Geod. Photogramm. Cartogr. 2016, 34, 383–390. [Google Scholar] [CrossRef]
- Wei, M.; Maldaner, L.; Ottoni, P.; Molin, J. Carrot Yield Mapping: A Precision Agriculture Approach Based on Machine Learning. AI 2020, 1, 229–241. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S. Deep Gaussian Process for Crop Yield Prediction Based on Remote Sensing Data. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Ulaby, F.T. Microwave Remote Sensing Active and Passive. In Rader Remote Sensing and Surface Scattering and Emission Theory; Longman Higher Education: Harlow, UK, 1982; pp. 848–902. [Google Scholar]
- Álvarez Mozos, J.; Verhoest, N.E.; Larrañaga, A.; Casalí, J.; González-Audícana, M. Influence of Surface Roughness Spatial Variability and Temporal Dynamics on the Retrieval of Soil Moisture from SAR Observations. Sensors 2009, 9, 463–489. [Google Scholar] [CrossRef]
- Zhang, L.; Lv, X.; Chen, Q.; Sun, G.; Yao, J. Estimation of Surface Soil Moisture during Corn Growth Stage from SAR and Optical Data Using a Combined Scattering Model. Remote Sens. 2020, 12, 1844. [Google Scholar] [CrossRef]
- Betbeder, J.; Fieuzal, R.; Philippets, Y.; Ferro-Famil, L.; Baup, F. Contribution of multitemporal polarimetric synthetic aperture radar data for monitoring winter wheat and rapeseed crops. J. Appl. Remote Sens. 2016, 10, 026020. [Google Scholar] [CrossRef]
- Clauss, K.; Ottinger, M.; Leinenkugel, P.; Kuenzer, C. Estimating rice production in the Mekong Delta, Vietnam, utilizing time series of Sentinel-1 SAR data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 574–585. [Google Scholar] [CrossRef]
- Zhuo, W.; Huang, J.; Li, L.; Zhang, X.; Ma, H.; Gao, X.; Huang, H.; Xu, B.; Xiao, X. Assimilating Soil Moisture Retrieved from Sentinel-1 and Sentinel-2 Data into WOFOST Model to Improve Winter Wheat Yield Estimation. Remote Sens. 2019, 11, 1618. [Google Scholar] [CrossRef]
- Attema, E.; Ulaby, F.T. Vegetation modeled as a water cloud. Radio Sci. 1978, 13, 357–364. [Google Scholar] [CrossRef]
- Van Diepen, C.; Wolf, J.; Van Keulen, H.; Rappoldt, C. WOFOST: A simulation model of crop production. Soil Use Manag. 1989, 5, 16–24. [Google Scholar] [CrossRef]
- Filipponi, F. Sentinel-1 GRD Preprocessing Workflow. Proceedings 2019, 18, 11. [Google Scholar]
- Morales, G.; Sheppard, J.; Scherrer, B.; Shaw, J. Reduced-Cost Hyperspectral Convolutional Neural Networks. J. Appl. Remote Sens. 2020, 14, 036519. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Sifre, L. Rigid-Motion Scattering For Image Classification. Ph.D. Thesis, Ecole Polytechnique, Palaiseau, France, 2014. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Zeiler, M.D. ADADELTA: An Adaptive Learning Rate Method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Wang, Z.; Bovik, A.; Sheikh, H.; Simoncelli, E. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Armstrong, R.A. Should Pearson’s correlation coefficient be avoided? Ophthalmic Physiol. Opt. 2019, 39, 316–327. [Google Scholar] [CrossRef]






| Field | # Samples 1st Year | # Samples 2nd Year | # Samples 3rd Year | Observed Years |
|---|---|---|---|---|
| F1 | 408 | 316 | 317 | 2016, 2018, 2020 |
| G1 | 484 | 497 | 614 | 2016, 2018, 2020 |
| G2 | 1014 | 920 | 1014 | 2016, 2019, 2021 |
| G3 | 290 | 414 | — | 2017, 2020 |
| Field | Split | Training + Validation | Test |
|---|---|---|---|
| F1 | A | 2016, 2018 | 2020 |
| B | 2016, 2020 | 2018 | |
| C | 2018, 2020 | 2016 | |
| G1 | A | 2016, 2018 | 2020 |
| B | 2016, 2020 | 2018 | |
| C | 2018, 2020 | 2016 | |
| G2 | A | 2016, 2019 | 2021 |
| B | 2016, 2021 | 2019 | |
| C | 2019, 2021 | 2016 | |
| G3 | A | 2017 | 2020 |
| B | 2020 | 2017 |
| Layer Name | Kernel Size | Padding Size | Output Size |
|---|---|---|---|
| Input | — | — | (5, 5, n, 1) |
| Conv3D + ReLU + BN | (3, 3, 3) | (1, 1, 1) | (5, 5, n, 32) |
| Conv3D + ReLU + BN | (3, 3, 3) | (1, 1, 1) | (5, 5, n, 32) |
| CONCAT | — | — | (5, 5, n, 64) |
| Conv3D + ReLU + BN | (3, 3, 3) | (1, 1, 1) | (5, 5, n, 32) |
| CONCAT | — | — | (5, 5, n, 96) |
| Conv3D + ReLU + BN | (3, 3, 3) | (1, 1, 1) | (5, 5, n, 32) |
| CONCAT | — | — | (5, 5, n, 128) |
| Reshape | — | — | (5, 5, ) |
| Dropout (0.5) | — | — | (5, 5, ) |
| SepConv2D + ReLU + BN | (3, 3) | (1, 1) | (5, 5, 512) |
| SepConv2D + ReLU + BN | (3, 3) | (1, 1) | (5, 5, 320) |
| Dropout (0.5) | — | — | (5, 5, 320) |
| SepConv2D + ReLU + BN | (3, 3) | (1, 1) | (5, 5, 256) |
| Dropout (0.5) | — | — | (5, 5, 256) |
| SepConv2D + ReLU + BN | (3, 3) | (1, 1) | (5, 5, 128) |
| SepConv2D + ReLU + BN | (3, 3) | (1, 1) | (5, 5, 32) |
| if or : | |||
| Conv2D + ReLU | (3, 3) | (N, N, 1) | |
| elif : | |||
| Conv2D + ReLU | (3, 3) | (0, 0) | (3, 3, 1) |
| Reshape | — | — | (9, 1) |
| FC | — | — | N |
| Field | Metric | Hyper3D NetReg N = 1 | Hyper3D NetReg N = 3 | Hyper3D NetReg N = 5 | AdaBoost. App N = 1 | SAE N = 1 | 3D-CNN N = 1 | CNN-LF N = 1 | RF N = 1 | BMLR N = 1 | MLR N = 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | 13.52 | 11.88 | 10.88 | 12.69 | 10.93 | 11.64 | 10.73 | 15.45 | 10.95 | 10.98 | |
| 8.94 | 8.10 | 7.01 | 7.74 | 6.75 | 7.59 | 7.42 | 11.42 | 6.75 | 6.74 | ||
| r | 0.15 | 0.47 | 0.50 | 0.16 | 0.27 | 0.26 | 0.30 | 0.37 | 0.52 | 0.51 | |
| 33.58 | 36.21 | 43.2 | 39.74 | 41.66 | 38.85 | 40.96 | 38.96 | 40.37 | 40.39 | ||
| 51.87 | 56.29 | 62.23 | 58.82 | 60.99 | 57.81 | 60.15 | 57.83 | 59.74 | 59.8 | ||
| G1 | 17.39 | 15.93 | 15.19 | 16.05 | 15.66 | 18.94 | 16.81 | 18.36 | 16.82 | 16.85 | |
| 10.09 | 10.63 | 9.04 | 9.92 | 10.19 | 11.24 | 10.70 | 11.81 | 10.01 | 10.06 | ||
| r | 0.29 | 0.34 | 0.43 | 0.29 | 0.33 | 0.32 | 0.37 | 0.26 | 0.37 | 0.37 | |
| 15.93 | 15.44 | 15.95 | 19.73 | 22.21 | 22.35 | 23.15 | 17.4 | 24.48 | 24.51 | ||
| 34.96 | 36.45 | 37.01 | 40.56 | 44.0 | 44.17 | 46.03 | 37.28 | 46.58 | 46.61 | ||
| G2 | 17.05 | 19.47 | 16.71 | 17.02 | 24.81 | 24.52 | 21.1 | 16.97 | 27.6 | 28.09 | |
| 11.55 | 15.72 | 12.45 | 12.66 | 37.12 | 18.20 | 17.70 | 14.01 | 20.31 | 20.92 | ||
| r | 0.37 | 0.29 | 0.55 | 0.43 | 0.10 | 0.10 | 0.29 | 0.60 | 0.21 | 0.19 | |
| 5.44 | 5.52 | 6.55 | 5.92 | 1.73 | 3.3 | 5.56 | 8.59 | 2.03 | 1.88 | ||
| 13.96 | 12.69 | 15.38 | 14.43 | 5.59 | 8.9 | 13.51 | 18.97 | 3.69 | 3.35 | ||
| G3 | 19.28 | 19.11 | 16.36 | 23.19 | 17.62 | 21.44 | 42.86 | 18.34 | 23.9 | 27.57 | |
| 14.51 | 13.75 | 11.26 | 16.71 | 13.32 | 13.73 | 14.88 | 11.98 | 15.35 | 19.19 | ||
| r | 0.35 | 0.54 | 0.64 | 0.52 | 0.58 | 0.31 | 0.30 | 0.45 | 0.56 | 0.55 | |
| 25.36 | 27.22 | 29.49 | 25.2 | 27.81 | 27.37 | 23.5 | 23.83 | 26.25 | 23.12 | ||
| 48.24 | 51.43 | 54.05 | 48.32 | 52.55 | 47.79 | 37.78 | 47.12 | 45.32 | 39.61 |
| Field | Metric | Hyper3D NetReg N = 1 | Hyper3D NetReg N = 3 | Hyper3D NetReg N = 5 | AdaBoost. App N = 1 | SAE N = 1 | 3D-CNN N = 1 | CNN-LF N = 1 | RF N = 1 | BMLR N = 1 | MLR N = 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | 19.04 | 18.46 | 19.34 | 14.98 | 21.41 | 20.43 | 17.83 | 19.15 | 14.42 | 14.94 | |
| 14.27 | 15.18 | 16.96 | 9.74 | 20.43 | 8.40 | 9.47 | 16.73 | 5.96 | 5.80 | ||
| r | 0.19 | 0.18 | 0.21 | 0.15 | 0.18 | 0.27 | 0.26 | 0.14 | 0.19 | 0.19 | |
| 26.54 | 29.12 | 28.63 | 30.01 | 28 | 32.14 | 29.42 | 30.34 | 30.68 | 30.58 | ||
| 57.24 | 60.47 | 59.98 | 63.35 | 57.67 | 66.5 | 64.68 | 59.62 | 66.36 | 66.28 | ||
| G1 | 23.82 | 26.41 | 27.01 | 24.34 | 19.04 | 33.03 | 23.24 | 19.19 | 19.62 | 20.42 | |
| 14.96 | 17.98 | 20.53 | 17.57 | 12.96 | 26.88 | 13.95 | 12.92 | 13.22 | 14.01 | ||
| r | 0.35 | 0.27 | 0.41 | 0.41 | 0.30 | 0.33 | 0.19 | 0.53 | 0.43 | 0.43 | |
| 16.38 | 18.39 | 14.64 | 22.51 | 25.47 | 24.97 | 23.67 | 28.88 | 26.72 | 26.64 | ||
| 42 | 41.16 | 36.88 | 46.27 | 52.16 | 45.13 | 46.2 | 57.19 | 54.86 | 54.72 | ||
| G2 | 43.42 | 43.36 | 46.49 | 43.6 | 46.64 | 44.92 | 41.22 | 39.6 | 35.68 | 35.4 | |
| 37.74 | 36.47 | 40.04 | 37.09 | 40.48 | 38.18 | 34.35 | 32.88 | 27.71 | 27.34 | ||
| r | 0.17 | 0.21 | 0.23 | 0.03 | 0.04 | 0.28 | 0.18 | 0.24 | 0.03 | 0.03 | |
| 9.69 | 8.96 | 10.01 | 7.46 | 6 | 9.09 | 9.32 | 12.39 | 9.74 | 9.34 | ||
| 21.33 | 20.72 | 20.21 | 19.06 | 15.95 | 19.51 | 21.6 | 25.71 | 24.87 | 24.38 | ||
| G3 | 21.79 | 18.51 | 18.09 | 21.51 | 19.39 | 18.6 | 21.6 | 21.97 | 17.96 | 27.76 | |
| 13.53 | 11.57 | 11.53 | 14.55 | 12.02 | 11.08 | 14.13 | 16.35 | 11.44 | 22.08 | ||
| r | 0.47 | 0.49 | 0.53 | 0.57 | 0.57 | 0.51 | 0.40 | 0.47 | 0.57 | 0.58 | |
| 32.09 | 37.19 | 41.85 | 40.65 | 40.52 | 35.34 | 26.88 | 37.08 | 44.17 | 39.5 | ||
| 54.94 | 60.31 | 64.63 | 62.64 | 62.39 | 60.47 | 50.08 | 60.74 | 66.72 | 59.6 |
| Field | Metric | Hyper3D NetReg N = 1 | Hyper3D NetReg N = 3 | Hyper3D NetReg N = 5 | AdaBoost. App N = 1 | SAE N = 1 | 3D-CNN N = 1 | CNN-LF N = 1 | RF N = 1 | BMLR N = 1 | MLR N = 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | 23.62 | 20.97 | 19.39 | 17.86 | 16.95 | 19.85 | 15.13 | 13.28 | 13.43 | 13.13 | |
| 21.49 | 19.02 | 17.44 | 13.93 | 14.48 | 17.35 | 11.85 | 9.85 | 10.57 | 10.21 | ||
| r | 0.25 | 0.32 | 0.34 | 0.27 | 0.24 | 0.28 | 0.24 | 0.29 | 0.27 | 0.27 | |
| 19.27 | 19.72 | 22.27 | 20.89 | 20.26 | 19.64 | 19.86 | 21.97 | 22.0 | 21.51 | ||
| 42.15 | 46.16 | 49.2 | 45.96 | 47.77 | 45.04 | 46.61 | 50.69 | 50.6 | 50.11 | ||
| G1 | 16.85 | 14.31 | 12.88 | 16.85 | 14.12 | 14.88 | 18.72 | 16.27 | 18.13 | 20.43 | |
| 10.82 | 9.43 | 8.57 | 11.51 | 9.82 | 10.35 | 12.30 | 11.51 | 12.29 | 13.33 | ||
| r | 0.23 | 0.24 | 0.31 | 0.21 | 0.15 | 0.26 | 0.29 | 0.11 | 0.12 | 0.11 | |
| 16.17 | 17.68 | 20.01 | 17.27 | 17.11 | 18.35 | 16.22 | 17.75 | 15.54 | 14.5 | ||
| 37 | 43.91 | 46.91 | 39.62 | 42.47 | 42.25 | 35.64 | 40.86 | 36.2 | 31.21 | ||
| G2 | 18.05 | 17.22 | 16.66 | 16.17 | 18.09 | 16.69 | 22.83 | 30.69 | 16.81 | 16.84 | |
| 11.24 | 11.78 | 10.48 | 11.70 | 12.50 | 11.56 | 18.72 | 23.04 | 11.67 | 11.65 | ||
| r | 0.21 | 0.37 | 0.41 | 0.05 | 0.08 | 0.10 | 0.38 | 0.10 | 0.18 | 0.18 | |
| 4.93 | 8.64 | 7.39 | 5.37 | 2.5 | 6.18 | 7.57 | 4.82 | 5.72 | 5.75 | ||
| 12.86 | 17.91 | 16.98 | 14.05 | 10.11 | 14.18 | 16.95 | 12.93 | 15.16 | 15.2 |
| Split | 1st Year | 2nd Year | 3rd Year |
|---|---|---|---|
| F1 | 86 | 130 | 101 |
| G1 | 82.35 | 199.5 | 66.1 |
| G2 | 78.9 | 92.6 | 60.8 |
| G3 | 66 | 105.5 | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, G.; Sheppard, J.W.; Hegedus, P.B.; Maxwell, B.D. Improved Yield Prediction of Winter Wheat Using a Novel Two-Dimensional Deep Regression Neural Network Trained via Remote Sensing. Sensors 2023, 23, 489. https://doi.org/10.3390/s23010489
Morales G, Sheppard JW, Hegedus PB, Maxwell BD. Improved Yield Prediction of Winter Wheat Using a Novel Two-Dimensional Deep Regression Neural Network Trained via Remote Sensing. Sensors. 2023; 23(1):489. https://doi.org/10.3390/s23010489
Chicago/Turabian StyleMorales, Giorgio, John W. Sheppard, Paul B. Hegedus, and Bruce D. Maxwell. 2023. "Improved Yield Prediction of Winter Wheat Using a Novel Two-Dimensional Deep Regression Neural Network Trained via Remote Sensing" Sensors 23, no. 1: 489. https://doi.org/10.3390/s23010489
APA StyleMorales, G., Sheppard, J. W., Hegedus, P. B., & Maxwell, B. D. (2023). Improved Yield Prediction of Winter Wheat Using a Novel Two-Dimensional Deep Regression Neural Network Trained via Remote Sensing. Sensors, 23(1), 489. https://doi.org/10.3390/s23010489









