A Self-Driven Microfluidic Chip for Ricin and Abrin Detection
Abstract
:1. Introduction
2. Materials and Methods
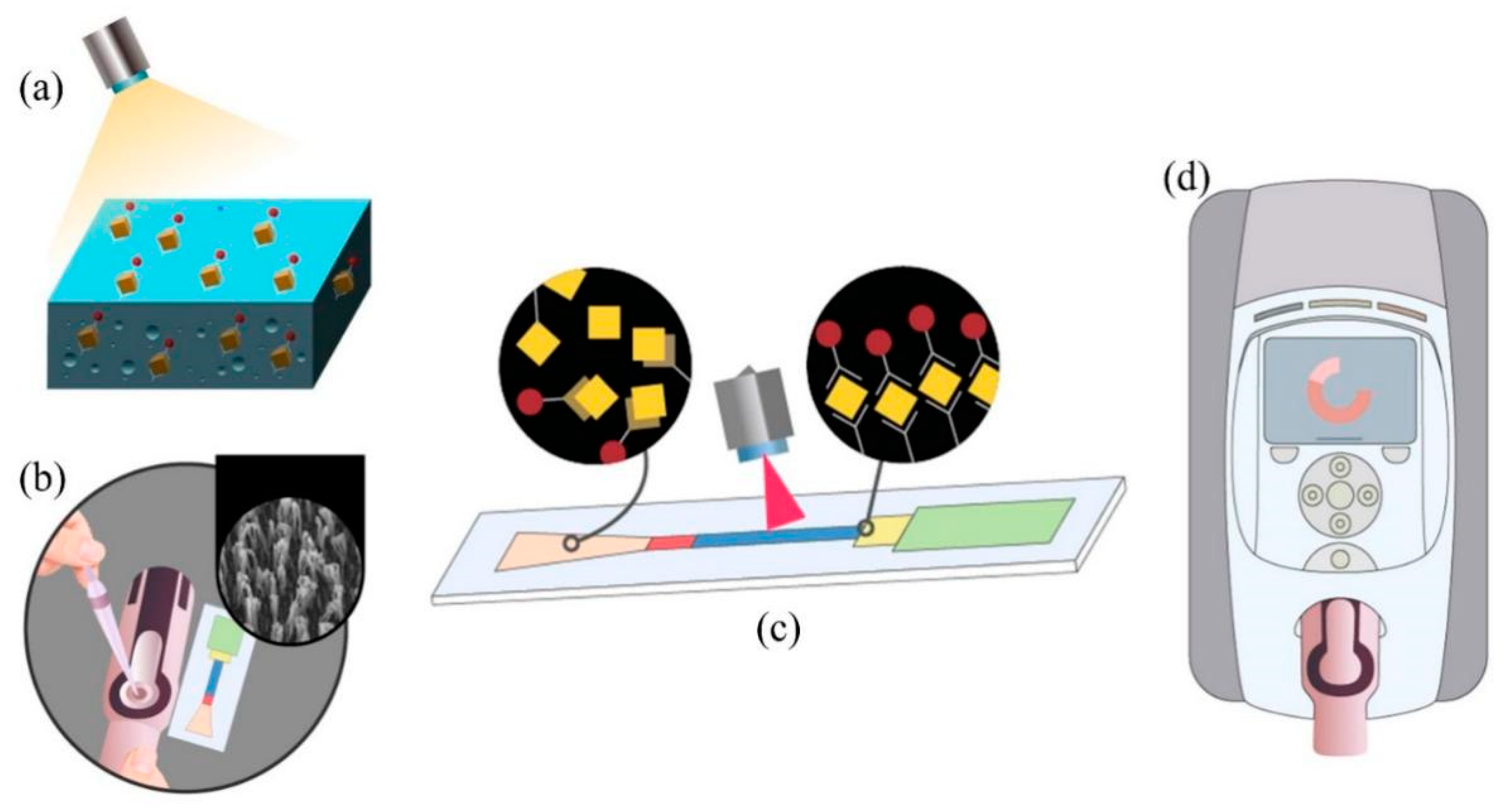
2.1. Preparation and Characterization of Nanoforest Structures
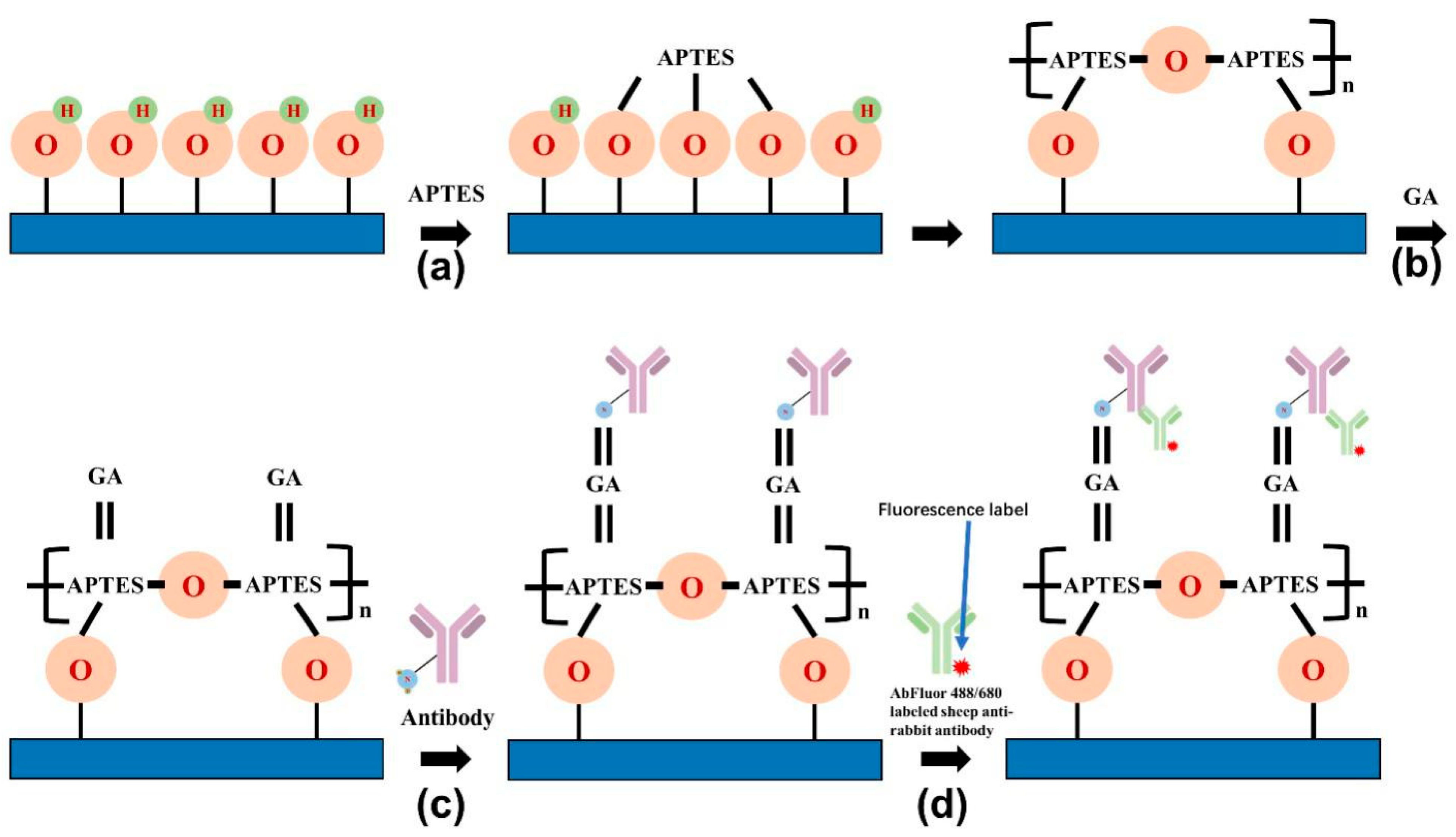
2.2. Surface Modification and Saturation Fluorescence Experiments on Silicon Nanoforest Structured Chips
2.3. Preparation of Microfluidic Chip Sensors
2.4. Microfluidic Chip Sensors Signal Measurement
3. Results and Discussions
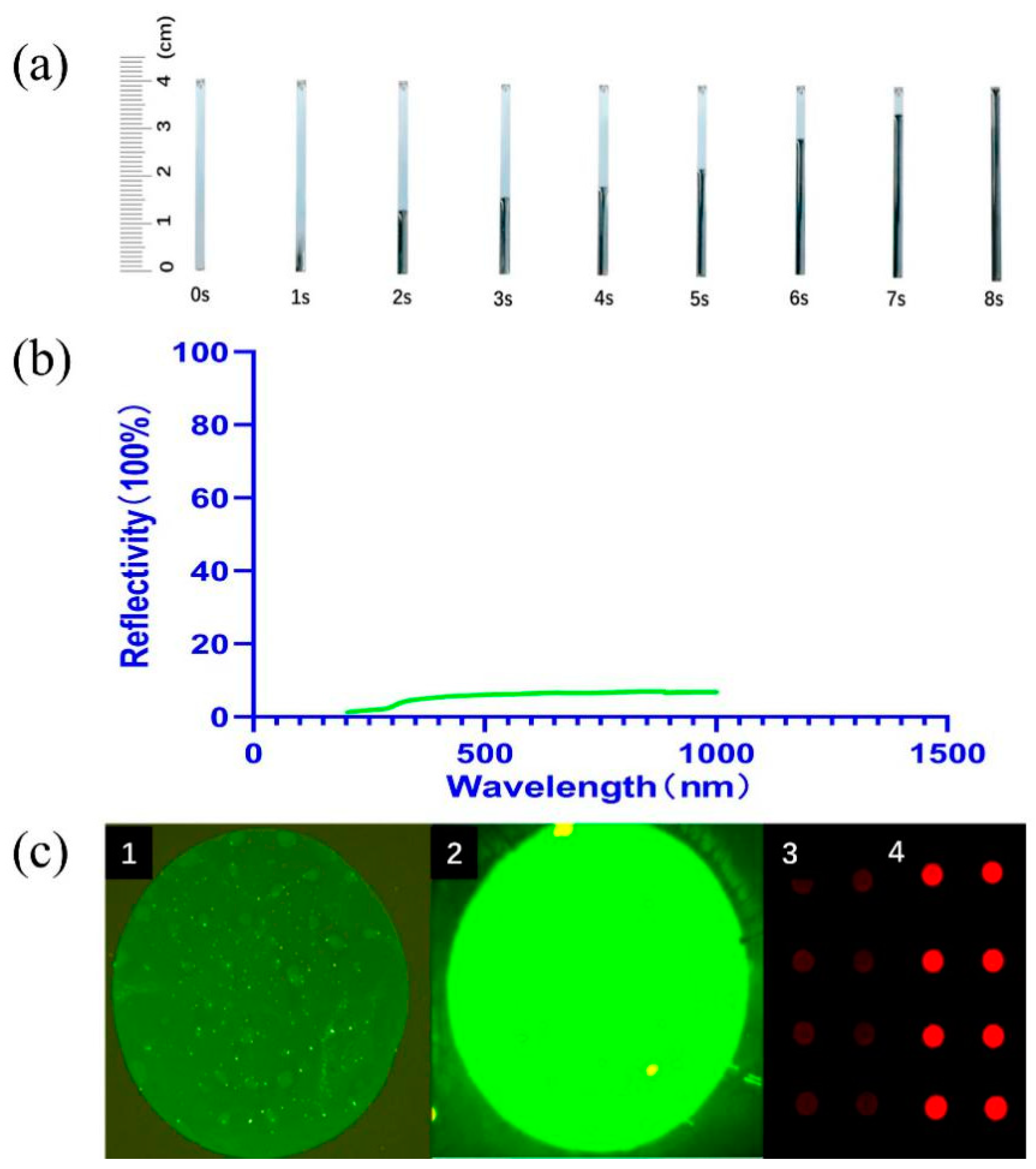
3.1. Characterization of Nanoforest Structure Chip
3.2. Sensor Detection of Two Plant Toxins
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gal, Y.; Sapoznikov, A.; Falach, R.; Mazor, O.; Alcalay, R.; Elhanany, E.; Aftalion, M.; Ehrlich, S.; Kronman, C.; Sabo, T. Equal Neutralization Potency of Antibodies Raised against Abrin Subunits. Antibodies 2020, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isenberg, S.L.; Carter, M.D.; Miller, M.A.; Noras, A.I.; Mojica, M.A.; Carlsen, S.T.; Bulathsinghala, C.P.; Thomas, J.D.; Johnson, R.C. Quantification of Ricinine and Abrine in Human Plasma by HPLC–MS-MS: Biomarkers of Exposure to Ricin and Abrin. J. Anal. Toxicol. 2018, 42, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Worbs, S.; Kampa, B.; Skiba, M.; Hansbauer, E.M.; Stern, D.; Volland, H.; Becher, F.; Simon, S.; Dorner, M.B.; Dorner, B.G. Differentiation, Quantification and Identification of Abrin and Abrus precatorius Agglutinin. Toxins 2021, 13, 284. [Google Scholar] [CrossRef] [PubMed]
- Janik, E.; Ceremuga, M.; Saluk-Bijak, J.; Bijak, M. Biological Toxins as the Potential Tools for Bioterrorism. Int. J. Mol. Sci. 2019, 20, 1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldberg, L.; Elhanany, E.; Laskar, O.; Schuster, O. Rapid, Sensitive and Reliable Ricin Identification in Serum Samples Using LC–MS/MS. Toxins 2021, 13, 79. [Google Scholar] [CrossRef]
- Verougstraete, N.; Helsloot, D.; Deprez, C.; Heylen, O.; Casier, I.; Croes, K. Lethal Injection of a Castor Bean Extract: Ricinine Quantification as a Marker for Ricin Exposure Using a Validated LC–MS/MS Method. J. Anal. Toxicol. 2019, 43, e1–e5. [Google Scholar] [CrossRef]
- Oliveira, G.; Schneedorf, J.M. A Simple, Fast and Portable Method for Electrochemical Detection of Adenine Released by Ricin Enzymatic Activity. Toxins 2021, 13, 238. [Google Scholar] [CrossRef]
- Crompton, R.; Gall, D. Georgi Markov—Death in a pellet. Med. Leg J. 1980, 48, 51–62. [Google Scholar] [CrossRef]
- Benamor, M.; Gharbi, E.; Bouzid, S.; Chakroun-Walha, O.; Rekik, N. Ricin poisoning after oral ingestion of castor beans: A case report and literature review. Afr. J. Emerg. Med. 2020, 10, 274–276. [Google Scholar] [CrossRef]
- Mei, Q.; Fredrickson, C.K.; Lian, W.; Jin, S.; Fan, Z.H. Ricin Detection by Biological Signal Amplification in a Well-in-a-Well Device. Anal. Chem. 2006, 78, 7659–7664. [Google Scholar] [CrossRef]
- Selvaprakash, K.; Chen, Y.-C. Detection of ricin by using gold nanoclusters functionalized with chicken egg white proteins as sensing probes. Biosens. Bioelectron. 2017, 92, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S.; Kozlov, J.V. Ricin. Toxicon 2001, 39, 1723–1728. [Google Scholar] [CrossRef]
- Saxena, N.; Bhutia, Y.D.; Kumar, O.; Phatak, P.; Kaul, R.K. Prophylactic efficacy of some chemoprotectants against abrin induced lethality. Interdiscip. Toxicol. 2018, 11, 169–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunez, O.F.L.; Pizon, A.F.; Tamama, K. Ricin Poisoning after Oral Ingestion of Castor Beans: A Case Report and Review of the Literature and Laboratory Testing. J. Emerg. Med. 2017, 53, e67–e71. [Google Scholar] [CrossRef] [PubMed]
- From, S.; Płusa, T. Today’s threat of ricin toxin. Pol. Merkur Lek. 2015, 39, 162–164. [Google Scholar]
- Rinner, G.R.; Watkins, S.A.; Shirazi, F.M.; Fernández, M.C.; Hess, G.; Mihalic, J.; Runcorn, S.; Waddell, V.; Ritter, J.; Reagan-Steiner, S.; et al. Fatal abrin poisoning by injection. Clin. Toxicol. 2021, 59, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Bagaria, S.; Karande, A.A. A chimeric protein of abrin and Abrus precatorius agglutinin that neutralizes abrin mediated lethality in mice. Toxicon 2017, 127, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Ler, S.G.; Lee, F.K.; Gopalakrishnakone, P. Trends in detection of warfare agents: Detection methods for ricin, staphylococcal enterotoxin B and T-2 toxin. J. Chromatogr. A 2006, 1133, 1–12. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.; Hernlem, B. CCD Based Detector for Detection of Abrin Toxin Activity. Toxins 2020, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Koja, N.; Shibata, T.; Mochida, K. Enzyme-linked immunoassay of ricin. Toxicon 1980, 18, 611–618. [Google Scholar] [CrossRef]
- Ma, X.; Tang, J.; Li, C.; Liu, Q.; Chen, J.; Li, H.; Guo, L.; Xie, J. Identification and quantification of ricin in biomedical samples by magnetic immunocapture enrichment and liquid chromatography electrospray ionization tandem mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 5147–5155. [Google Scholar] [CrossRef] [PubMed]
- He, X.; McMahon, S.; Ii, T.D.H.; Griffey, S.M.; Cheng, L.W. Ricin Toxicokinetics and Its Sensitive Detection in Mouse Sera or Feces Using Immuno-PCR. PLoS ONE 2010, 5, e12858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayan-Kenigsberg, J.; Bertocchi, A.; Garber, E.A. Rapid detection of ricin in cosmetics and elimination of artifacts associated with wheat lectin. J. Immunol. Methods 2008, 336, 251–254. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Deen, B.; Rodda, T.; Ronningen, I.; Blasius, T.; Haynes, C.; Diez-Gonzalez, F.; Labuza, T.P. Rapid Detection of Ricin in Milk Using Immunomagnetic Separation Combined with Surface-Enhanced Raman Spectroscopy. J. Food Sci. 2011, 76, N49–N53. [Google Scholar] [CrossRef]
- Feltis, B.; Sexton, B.; Glenn, F.; Best, M.; Wilkins, M.; Davis, T. A hand-held surface plasmon resonance biosensor for the detection of ricin and other biological agents. Biosens. Bioelectron. 2008, 23, 1131–1136. [Google Scholar] [CrossRef]
- Higashi, Y.; Miyoshi, H.; Takeda, K.; Saruwatari, H.; Kubo, H.; Sakaguchi, I.; Iwata, M.; Uchida, Y.; Tada, K.; Miyamoto, M.; et al. Evaluation of a newly-developed immunochromatography strip test for diagnosing dermatophytosis. Int. J. Dermatol. 2012, 51, 406–409. [Google Scholar] [CrossRef]
- Xia, Q.Y.; Li, M.Y.; Yang, X.L.; Xiao, S.S.; He, P.P.; Guo, A.P. Immunochromatography Test Strip and Its Applications in Detection of Genetically Modified Organisms. China Biotechnol. 2017, 37, 101–110. [Google Scholar]
- Kulinsky, L.; Noroozi, Z.; Madou, M. Present Technology and Future Trends in Point-of-Care Microfluidic Diagnostics. Methods Mol. Biol. 2013, 949, 3–23. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Mao, H.; Wu, D.; Wu, W.; Xu, J.; Hao, Y. The fabrication of diversiform nanostructure forests based on residue nanomasks synthesized by oxygen plasma removal of photoresist. Nanotechnology 2009, 20, 445304. [Google Scholar] [CrossRef]
- Jeon, J.; Mills, C.; Shell, M. Molecular Insights into Diphenylalanine Nanotube Assembly: All-Atom Simulations of Oligomerization. J. Phys. Chem. B 2013, 117, 3935–3943. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mao, H.; Xiong, J.; Jia, Y.; Li, R.; Wang, W. Optical Features of Nanowire Forests Prepared by a Plasma Repolymerization Technique. IEEE Trans. Nanotechnol. 2018, 17, 719–722. [Google Scholar] [CrossRef]
- Wu, W.; Mao, H.; Han, X.; Xu, J.; Wang, W. Fabrication and characterization of SiO2/Si heterogeneous nanopillar arrays. Nanotechnology 2016, 27, 305301. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, T.; Kang, L.; Gao, S.; Xin, W.; Wang, J. Production and Characterization of a Recombinant Chimeric Protein Consisting Mutant Ricin A Chain and Abrin a Chain. J. Tech. Commun. 2011, 22, 453–457. [Google Scholar]
- Zhi, J.; Xin, W.; Huang, J.; Kang, L.; Yuan, Y.; Wang, J. Multiple organ damage induced by Clostridium perfringens ε toxin. Microbiol. China 2020, 47, 1920–1926. [Google Scholar]
- Gao, J.; Wang, J.; Nan, N.; Gong, S.; Tao, L.; Wang, H. Establishment of an intracellular persistence model of botulinum toxin type A light chain. Acta Microbiol. Sin. 2021, 61, 3496–3505. [Google Scholar]
- Pöhlmann, C.; Elßner, T. Multiplex Immunoassay Techniques for On-Site Detection of Security Sensitive Toxins. Toxins 2020, 12, 727. [Google Scholar] [CrossRef]
- Chen, H.Y.; Tran, H.; Foo, L.Y.; Sew, T.W.; Loke, W.K. Development and validation of an ELISA kit for the detection of ricin toxins from biological specimens and environmental samples. Anal. Bioanal. Chem. 2014, 406, 5157–5169. [Google Scholar] [CrossRef]
- He, X.; Patfield, S.; Cheng, L.W.; Stanker, L.H.; Rasooly, R.; McKeon, T.A.; Zhang, Y.; Brandon, D.L. Detection of Abrin Holotoxin Using Novel Monoclonal Antibodies. Toxins 2017, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jin, Z.; Bai, X.; Wan, W.; Li, Y.; Wu, J.; Zhang, J.; Gao, S.; Wang, J. On-site rapid detection method of biotoxins based on high-sensitivity lanthanide fluorescence. Mil. Med. Sci. 2021, 45, 537–542+547. [Google Scholar]
- Gao, S.; Nie, C.; Wang, J.; Wang, J.-L.; Kang, L.; Zhou, Y. Colloidal Gold–Based Immunochromatographic Test Strip for Rapid Detection of Abrin in Food Samples. J. Food Prot. 2012, 75, 112. [Google Scholar] [CrossRef] [PubMed]
- Dapeng, C.; Bryden, W.A.; Catherine, F. Rapid analysis of ricin using hot acid digestion and MALDI-TOF mass spectrometry. J. Mass Spectrom. JMS 2018, 53, 1013–1017. [Google Scholar]
- Livet, S.; Worbs, S.; Volland, H.; Simon, S.; Dorner, M.B.; Fenaille, F.; Dorner, B.G.; Becher, F. Development and Evaluation of an Immuno-MALDI-TOF Mass Spectrometry Approach for Quantification of the Abrin Toxin in Complex Food Matrices. Toxins 2021, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Brandon, D.L. Detection of Ricin Contamination in Ground Beef by Electrochemiluminescence Immunosorbent Assay. Toxins 2011, 3, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, C.; Tong, Z.; Mu, X.; Liu, B.; Xu, J.; Du, B.; Wang, J.; Liu, Z. A highly sensitive electrochemiluminescence method for abrin detection by a portable biosensor based on a screen-printed electrode with a phage display affibody as specific labeled probe. Anal. Bioanal. Chem. 2021, 414, 1095–1104. [Google Scholar] [CrossRef] [PubMed]




| Photoresist Thickness (μm) | Density (Fibers/μm2) | Diameter (nm) | Height (μm) |
|---|---|---|---|
| 2 | 10 | 100 | 0.2 |
| 5 | 15 | 50–100 | 1.8 |
| 10 | 20 | 200–400 | 5 |
| Name | Detectable Toxin Agents | Sensitivity | Time | Cost of Equipment |
|---|---|---|---|---|
| ELISA | RT [38] | 0.093 ng/mL | 4–5 h | Expensive |
| AT [39] | 1 ng/mL | |||
| Immunochromatographic Test Strip | RT [40] | 0.5 ng/mL | 15 min | Cheap |
| AT [41] | 3 ng/mL | |||
| MALDI-TOF MS | RT [42] | 0.2 ng/mL | 3.5–7.5 h | Expensive |
| AT [43] | 40 ng/mL | |||
| Electrochemical Luminescence Method | RT [44] | 0.2 ng/mL | 5.5 h | Expensive |
| AT [45] | 5 pg/mL | |||
| This work | RT, AT | 10 pg/mL | 15 min | Cheap |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, X.; Hu, C.; Chen, L.; Wang, J.; Li, Y.; Wan, W.; Jin, Z.; Li, Y.; Xin, W.; Kang, L.; et al. A Self-Driven Microfluidic Chip for Ricin and Abrin Detection. Sensors 2022, 22, 3461. https://doi.org/10.3390/s22093461
Bai X, Hu C, Chen L, Wang J, Li Y, Wan W, Jin Z, Li Y, Xin W, Kang L, et al. A Self-Driven Microfluidic Chip for Ricin and Abrin Detection. Sensors. 2022; 22(9):3461. https://doi.org/10.3390/s22093461
Chicago/Turabian StyleBai, Xuexin, Chenyi Hu, Liang Chen, Jing Wang, Yanwei Li, Wei Wan, Zhiying Jin, Yue Li, Wenwen Xin, Lin Kang, and et al. 2022. "A Self-Driven Microfluidic Chip for Ricin and Abrin Detection" Sensors 22, no. 9: 3461. https://doi.org/10.3390/s22093461
APA StyleBai, X., Hu, C., Chen, L., Wang, J., Li, Y., Wan, W., Jin, Z., Li, Y., Xin, W., Kang, L., Jin, H., Yang, H., Wang, J., & Gao, S. (2022). A Self-Driven Microfluidic Chip for Ricin and Abrin Detection. Sensors, 22(9), 3461. https://doi.org/10.3390/s22093461





