Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases
Abstract
:1. Introduction
2. Materials and Methods
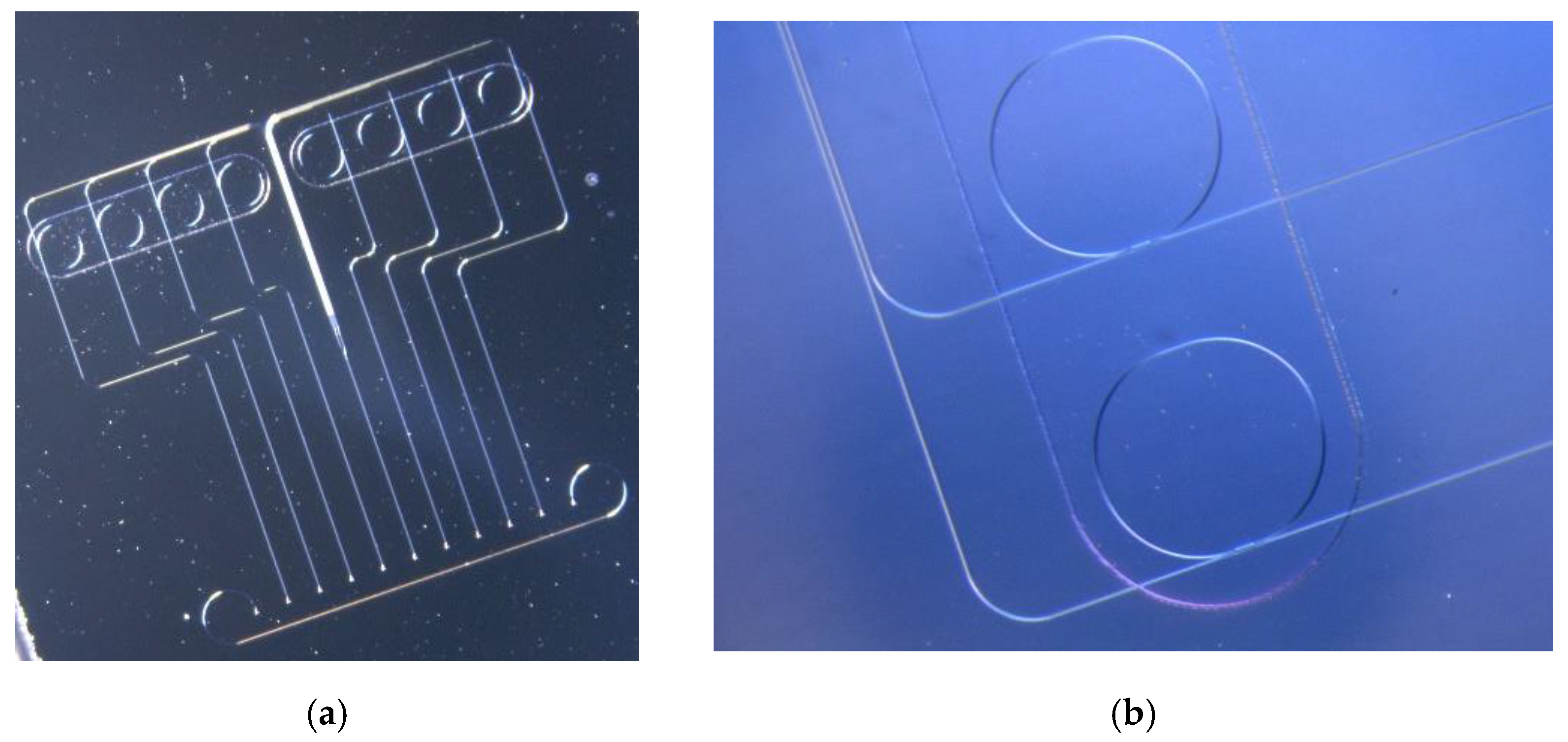
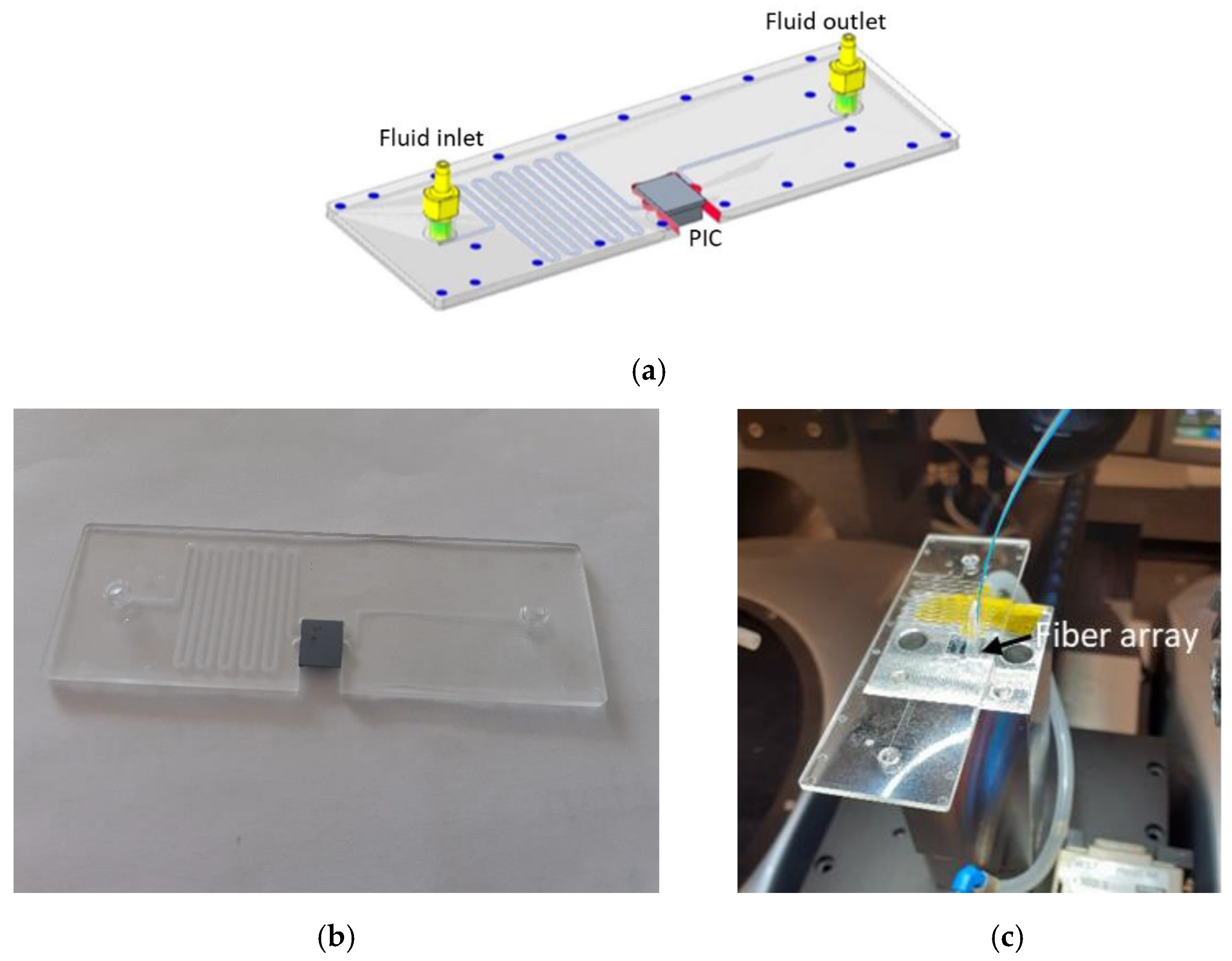
2.1. Biosensors Description
2.2. Identification, Purification and Characterization of the MREs
2.2.1. Purification
2.2.2. Western Blotting Experiments
2.2.3. Indirect and Sandwich ELISA
2.3. Immobilization of the MREs
- −
- Sulfuric acid ≥98% (H2SO4, CAS Number 7664-93-9) from VWR (102765G; es.vwr.com/102765G, accessed on 20 September 2021).
- −
- Hydrogen peroxide 30% (V/V) (H2O2, CAS Number 7722-84-1) from Labbox (HYPE-30A-1K0; ien.labbox.com/hydrogen-peroxide, accessed on 20 September 2021).
- −
- Carboxyethylsilanetriol disodium salt 25% in water (CTES, CAS Number 18191-40-7) from ABCR (AB110934; abcr.com/AB110934, accessed on 20 September 2021).
- −
- 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, CAS Number 25952-53-8) from TCI Chemicals (D1601; www.tcichemicals.com/D1601, accessed on 20 September 2021).
- −
- N-Hydroxysuccinimide (NHS, CAS Number 6066-82-6) from TCI Chemicals (H0623; www.tcichemicals.com/H0623, accessed on 20 September 2021).
- −
- 2-(N-morpholino)ethanesulfonic acid (MES, CAS Number 145224-94-8) from Fisher Scientific (BP300-100; www.fishersci.es/10419123/BP300-100, accessed on 20 September 2021).
- −
- Gelatine from cold water fish skin (fish gelatine, CAS Number 9000-70-8) from Sigma-Aldrich (G7041, www.sigmaaldrich.com/g7041, accessed on 20 September 2021).
- −
- Phosphate Buffered Saline (PBS 10x) from Fisher Scientific (BP399-1; www.fishersci.es/10204733/BP399-1, accessed on 20 September 2021).
2.4. Optofluidics for Biosensing
3. Results
3.1. Identification, Production, and Characterization of the MREs
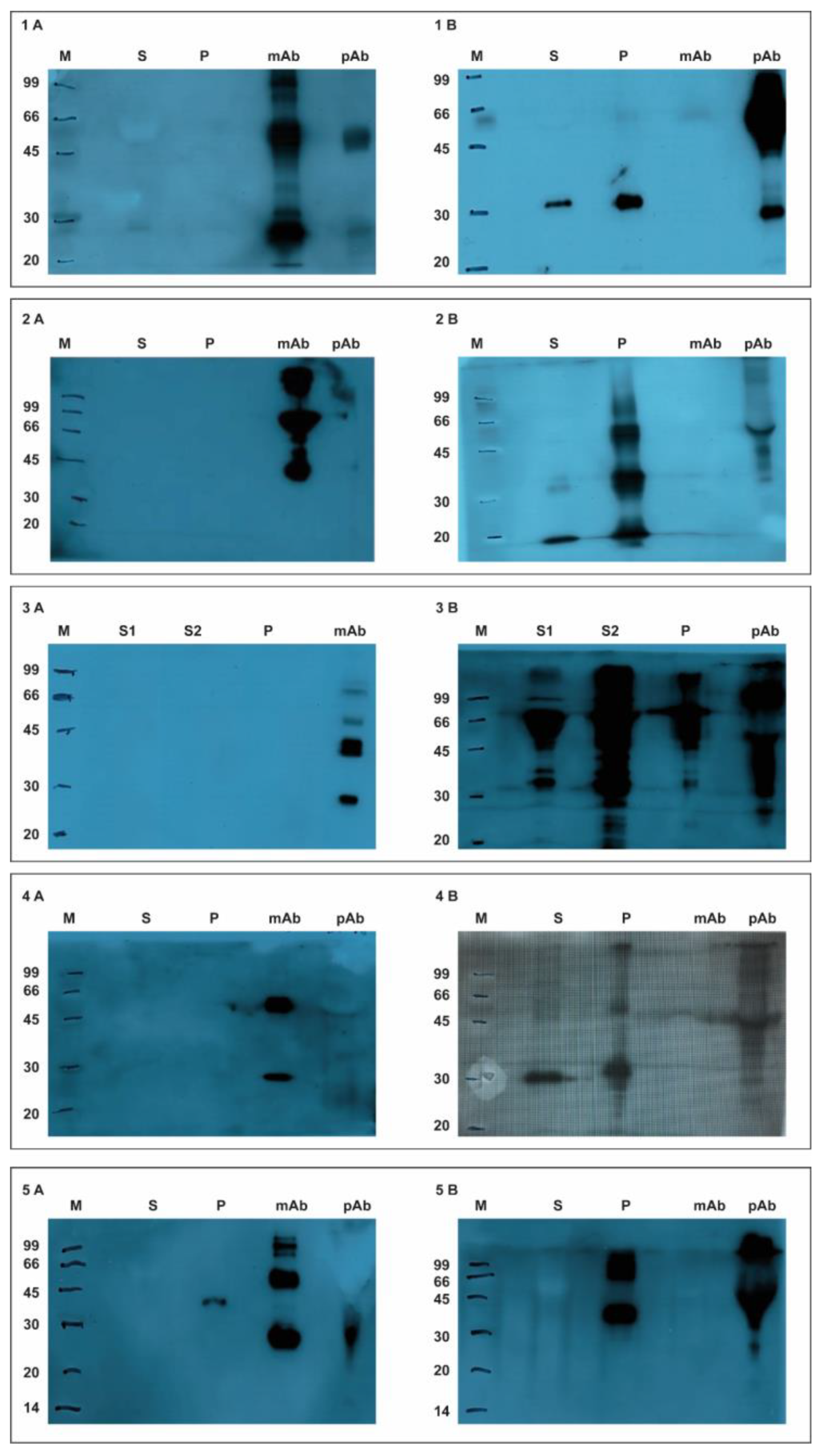
3.1.1. Western Blotting Experiments
3.1.2. ELISA Test Experiments
3.2. Photonic Biosensing
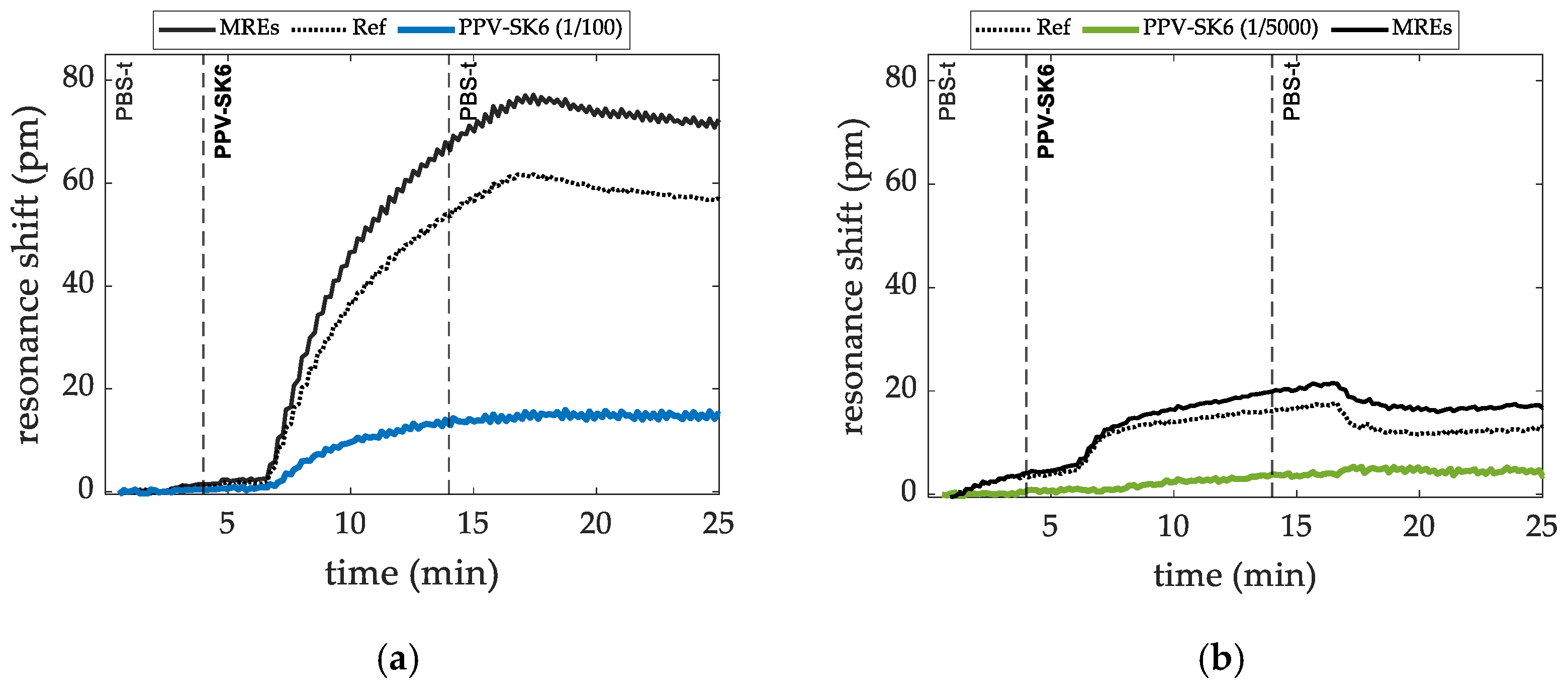
3.2.1. Targeted Virus PPV
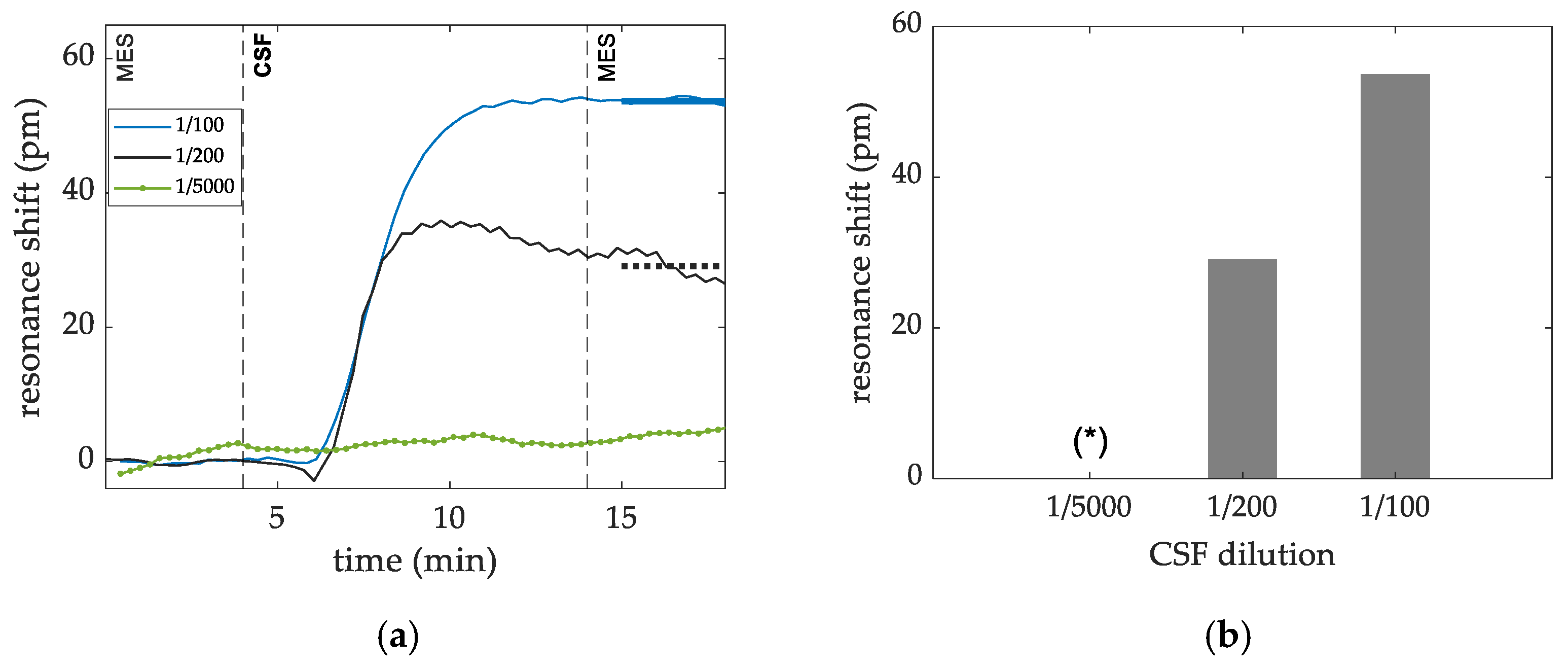
3.2.2. Targeted Virus CSF
3.2.3. Targeted Virus PRRSV
3.2.4. Targeted Virus ASFV and SIV Virus Sensing Measurements
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, K.M.; Machalaba, C.C.; Seifman, R.; Feferholtz, Y.; Karesh, W.B. Infectious disease and economics: The case for considering multi-sectoral impacts. One Health 2019, 7, 100080. [Google Scholar] [CrossRef] [PubMed]
- Jamal, S.M.; Belsham, G.J. Foot-and-mouth disease: Past, present and future. Vet. Res. 2013, 44, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ASF Regionalization in the EU. Available online: https://ec.europa.eu/food/system/files/2021-04/ad_control-measures_asf_pl-lt-regionalisation.pdf. (accessed on 10 July 2021).
- CSF. Available online: https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/CLASSICAL_SWINE_FEVER.pdf (accessed on 10 July 2021).
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bào, Y.; Ng, T.F.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Biernacka, K.; Fan, J.; Gerber, P.F.; Stadejek, T.; Opriessnig, T. Circulation of Porcine Parvovirus Types 1 through 6 in Serum Samples Obtained from Six Commercial Polish Pig Farms. Transbound. Emerg. Dis. 2017, 64, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Oliver-Ferrando, S.; Segalés, J.; López-Soria, S.; Callén, A.; Merdy, O.; Joisel, F.; Sibila, M. Evaluation of natural porcine circovirus type 2 (PCV2) subclinical infection and seroconversion dynamics in piglets vaccinated at different ages. Vet. Res. 2016, 47, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castrucci, M.R.; Donatelli, I.; Sidoli, L.; Barigazzi, G.; Kawaoka, Y.; Webster, R.G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 1993, 193, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Perestam, A.T.; Fujisaki, K.K.; Nava, O.; Hellberg, R.S. Comparison of real-time PCR and ELISA-based methods for the detection of beef and pork in processed meat products. Food Control 2017, 71, 346–352. [Google Scholar] [CrossRef] [Green Version]
- Biagetti, M.; Cuccioloni, M.; Bonfili, L.; Cecarini, V.; Sebastiani, C.; Curcio, L.; Giammarioli, M.; De Mia, G.M.; Eleuteri, A.M.; Angeletti, M. Chimeric DNA/LNA-based biosensor for the rapid detection of African swine fever virus. Talanta 2018, 184, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.S.; Tachikawa, K.; Manz, A. Micro total analysis systems. Latest advancements and trends. Anal. Chem. 2006, 78, 3887–3907. [Google Scholar] [CrossRef] [PubMed]
- Griol, A.; Peransi, S.; Rodrigo, M.; Hurtado, J.; Bellieres, L.; Ivanova, T.; Zurita, D.; Sánchez, C.; Recuero, S.; Hernández, A.; et al. Design and Development of Photonic Biosensors for Swine Viral Diseases Detection. Sensors 2019, 19, 3985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varriale, A.; Pennacchio, A.; Pinto, G.; Oliviero, G.; D’Errico, S.; Majoli, A.; Scala, A.; Capo, A.; Pennacchio, A.; Di Giovanni, S.; et al. A Fluorescence Polarization Assay to Detect Steroid Hormone Traces in Milk. J. Agric. Food Chem. 2015, 63, 9159–9164. [Google Scholar] [CrossRef] [PubMed]
- LAEMMLI, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Montagnese, C.; Barattini, P.; Giusti, A.; Balka, G.; Bruno, U.; Bossis, I.; Gelasakis, A.; Bonasso, M.; Philmis, P.; Dénes, L.; et al. A diagnostic device for in-situ detection of swine viral diseases: The SWINOSTICS project. Sensors 2019, 19, 407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Pérez, P.; Gómez-Gómez, M.; Angelova, T.; Griol, A.; Hurtado, J.; Bellieres, L.; García-Rupérez, J. Continuous detection of increasing concentrations of thrombin employing a label-free photonic crystal aptasensor. Micromachines 2020, 11, 464. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, R.; Lapin, Z.J.; Nichols, A.S.; Braun, R.M.; Fountain, A.W. Photonic integrated circuits for Department of Defense-relevant chemical and biological sensing applications: State-of-the-art and future outlooks. Opt. Eng. 2019, 58, 020901. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Vasiliev, A.; Muneeb, M.; Malik, A.; Sprengel, S.; Boehm, G.; Amann, M.C.; Šimonytė, I.; Vizbaras, A.; Vizbaras, K.; et al. III–V-on-silicon photonic integrated circuits for, spectroscopic sensing in the 2–4 µm wavelength range. Sensors 2017, 17, 1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steglich, P.; Hülsemann, M.; Dietzel, B.; Mai, A. Optical biosensors based on silicon-on-insulator ring resonators: A review. Molecules 2019, 24, 519. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Target | Recombinant Antigen | MREs |
|---|---|---|
| Porcine Circovirus—PCV2(*) | Capsid protein ORF2 (30 kDa) alpha diagnostic PCV2C25-R-10 | pAb—Thermo Fisher PA5-34969 mAb—Ingenasa M.11.PCV.I36A9 |
| Porcine Reproductive and Respiratory Syndrome—PRRSV | Nuclear protein NP (15 kDa) alpha diagnostic PRSNP-15-R-10 | pAb—alpha diagnostic PRSNP11-S mAb—RTI, LLC SDOW17-A |
| Porcine Parvovirus—PPV | VP2 protein (68 kDa) alpha diagnostic PPVVP21-R-10 | pAb—alpha diagnostic PPVVP21-S mAb—VMRD 3C9D11H11 |
| African Swine Fever Virus—ASFV | p30 protein 24.4 kDa alpha diagnostic ASFV15-R-10 | pAb—alpha diagnostic ASFV11-S mAb—Ingenasa M.11.PPA.I18BB11 mAb—Ingenasa M.11.PPA.1BC11 |
| Classical Swine Fever Virus—CSFV | Envelope protein E2 (35 kDa) alpha diagnostic CSFE25-R-10 | pAb—alpha diagnostic CSFE21-S mAb—alpha scientific RAE0826 |
| Swine Influenza A Virus—SIV | mAb—Thermo Fisher MA5-17101 |
| Virus | Indirect ELISA | Sandwich ELISA | Cross-Reactivity of Polyclonal Antibodies | ||||||
|---|---|---|---|---|---|---|---|---|---|
| pAb | mAb | mAb/pAb | Anti-PCV2 | Anti-PPV | Anti-PRRSV | Anti-CSFV | Anti-ASFV | ||
| PCV2 | S | (−) | (−) | (−) | (−) | (−) | (−) | (−) | (−) |
| P | √ | (−) | (−) | √ | (−) | (−) | (−) | (−) | |
| PRRSV | S | √ | (−) | (−) | (−) | (−) | √ | (−) | (−) |
| P | (−) | (−) | (−) | (−) | (−) | (−) | (−) | (−) | |
| PPV | S | √ | (−) | (−) | (−) | √ | √ | (−) | (−) |
| P | √ | (−) | (−) | (−) | √ | √ | (−) | (−) | |
| ASFV | S | √ | √ | (−) | (−) | (−) | (−) | (−) | √ |
| P | √ | √ | (−) | (−) | (−) | (−) | (−) | √ | |
| CSFV | S | (−) | (−) | (−) | (−) | (−) | √ | (−) | (−) |
| P | √ | (−) | (−) | (−) | (−) | (−) | √ | √ | |
| SIV | (−) | √ | (−) | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Gómez, M.; Sánchez, C.; Peransi, S.; Zurita, D.; Bellieres, L.; Recuero, S.; Rodrigo, M.; Simón, S.; Camarca, A.; Capo, A.; et al. Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases. Sensors 2022, 22, 708. https://doi.org/10.3390/s22030708
Gómez-Gómez M, Sánchez C, Peransi S, Zurita D, Bellieres L, Recuero S, Rodrigo M, Simón S, Camarca A, Capo A, et al. Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases. Sensors. 2022; 22(3):708. https://doi.org/10.3390/s22030708
Chicago/Turabian StyleGómez-Gómez, Maribel, Carles Sánchez, Sergio Peransi, David Zurita, Laurent Bellieres, Sara Recuero, Manuel Rodrigo, Santiago Simón, Alessandra Camarca, Alessandro Capo, and et al. 2022. "Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases" Sensors 22, no. 3: 708. https://doi.org/10.3390/s22030708
APA StyleGómez-Gómez, M., Sánchez, C., Peransi, S., Zurita, D., Bellieres, L., Recuero, S., Rodrigo, M., Simón, S., Camarca, A., Capo, A., Staiano, M., Varriale, A., D’Auria, S., Manessis, G., Gelasakis, A. I., Bossis, I., Balka, G., Dénes, L., Frant, M., ... Griol, A. (2022). Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases. Sensors, 22(3), 708. https://doi.org/10.3390/s22030708











