A Vaginitis Classification Method Based on Multi-Spectral Image Feature Fusion
Abstract
:1. Introduction
- This paper is the first try to introduce a multi-spectral imaging method for the vaginitis diagnosis;
- For the first time, it is found that each kind of vaginitis has a unique sensitive spectral band;
- A classification approach MIDV is designed, which combines deep learning with multi-spectral image feature fusion in the vaginitis domain.
2. Related Work
2.1. Medical Image Analysis Using Transfer Learning Strategy
2.2. Multi-Spectral Data Fusion
3. Background Knowledge
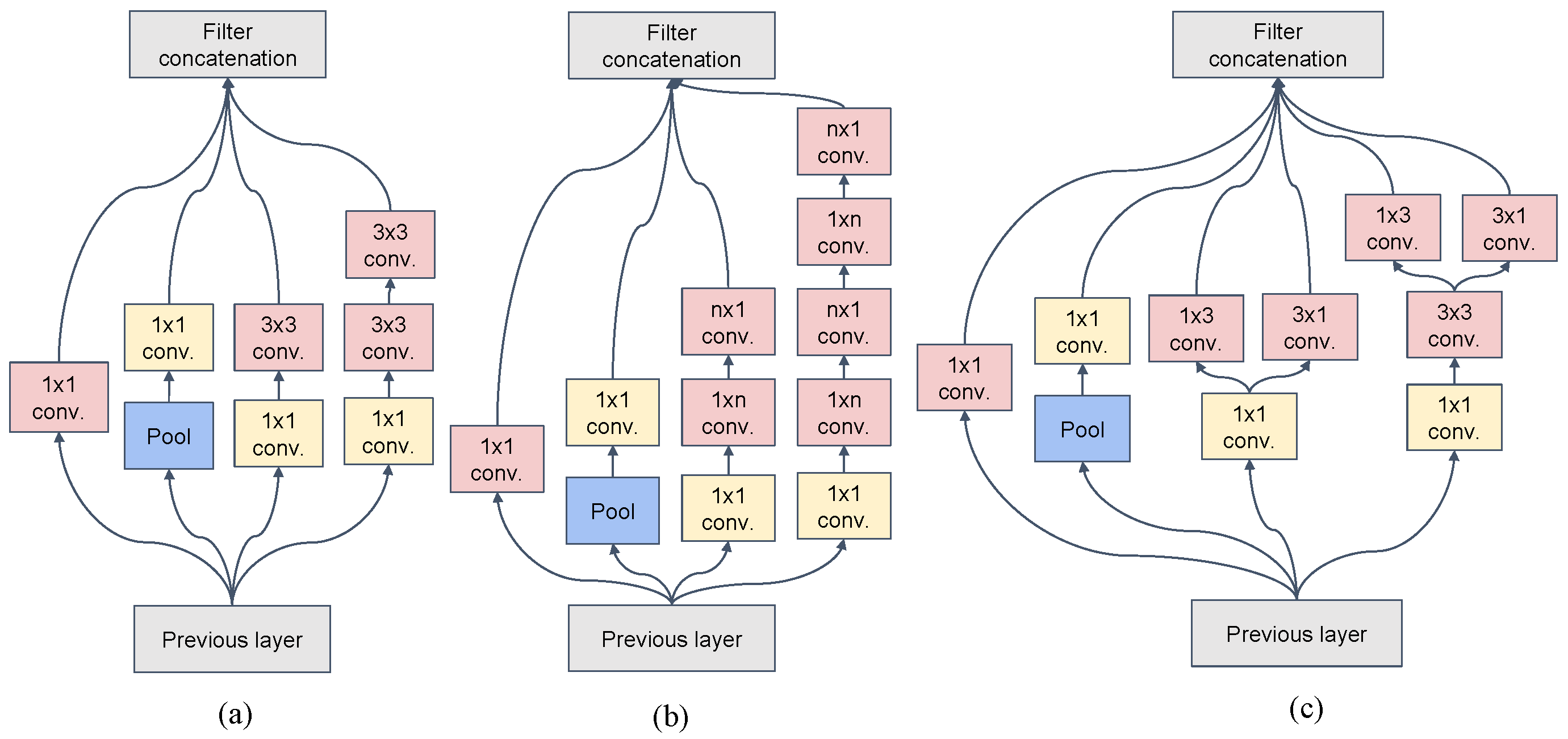
3.1. Inception v3
3.2. Multi-Spectral
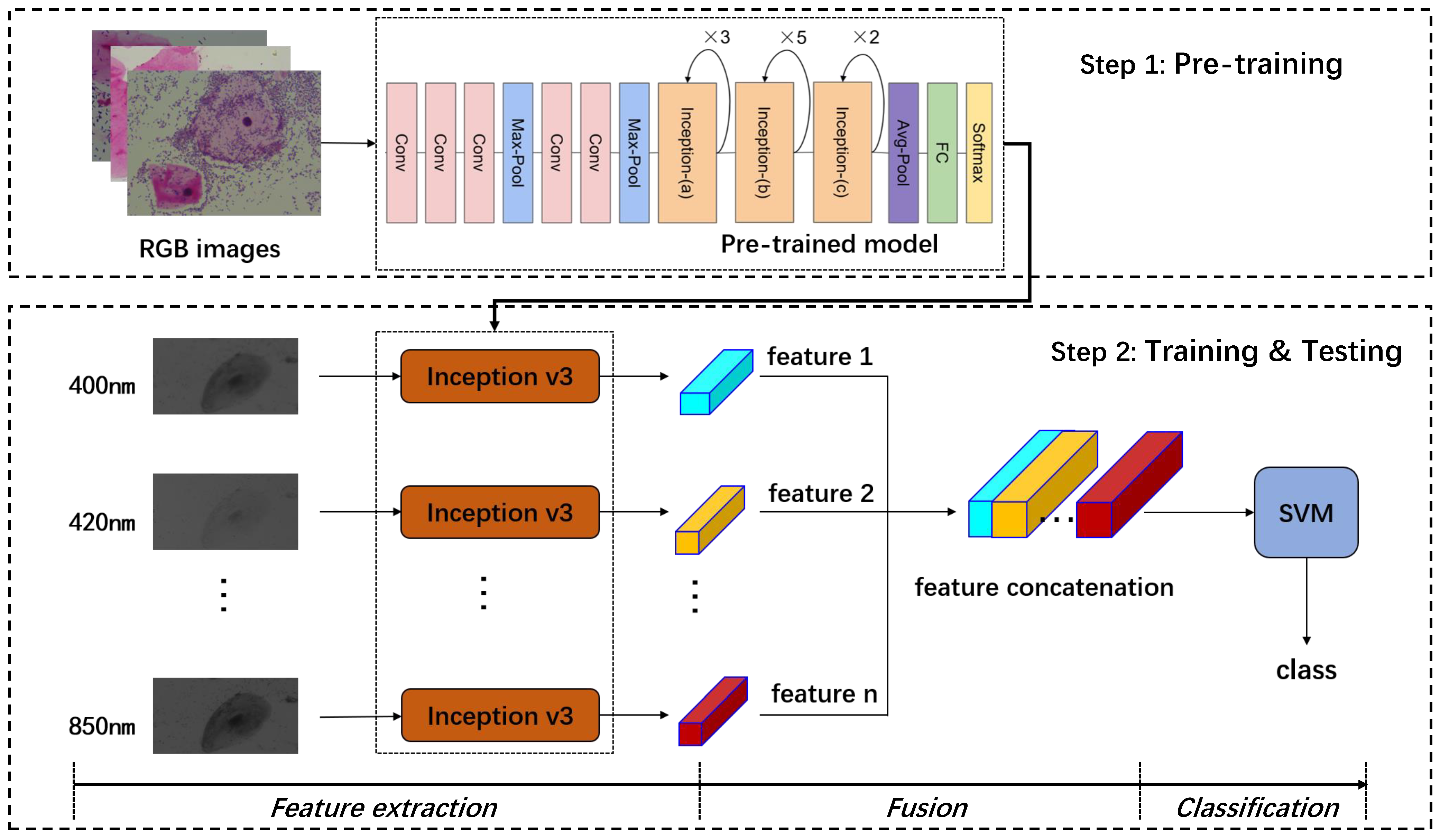
4. Methodology
- Step 1. Train an inception v3 model using RGB images of vaginal microorganisms.
- Step 2. The last layer of the inception v3 model as the classifier is removed, so the left parts are used as a feature extractor for multi-spectral images.
- Step 3. Extract features using the inception v3 extractor in Step 2 for every single spectral image in multi-spectral images.
- Step 4. Arrange the features from small to large according to the wavelength of the corresponding single-spectrum image and connect them together with the concatenation operation.
- Step 5. Input the fused feature vector into the SVM classifier, and get the disease category of vaginitis.
5. Experiment
5.1. Setup
5.1.1. Dataset
5.1.2. Image-Collecting Instrument
5.2. Training Strategy
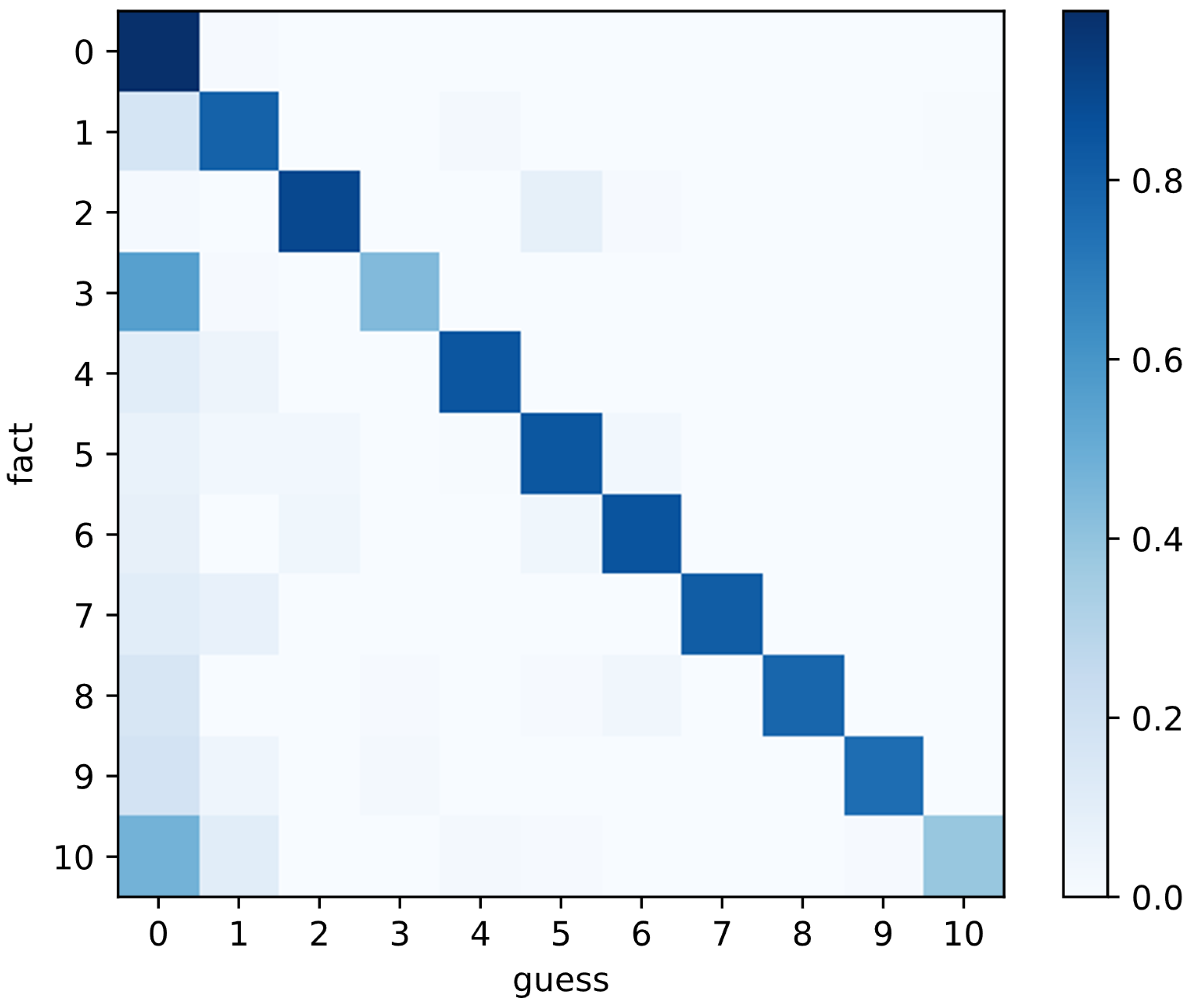
5.3. Results
5.3.1. Comparison with RGB Image
5.3.2. Comparison with Other Fusion Methods
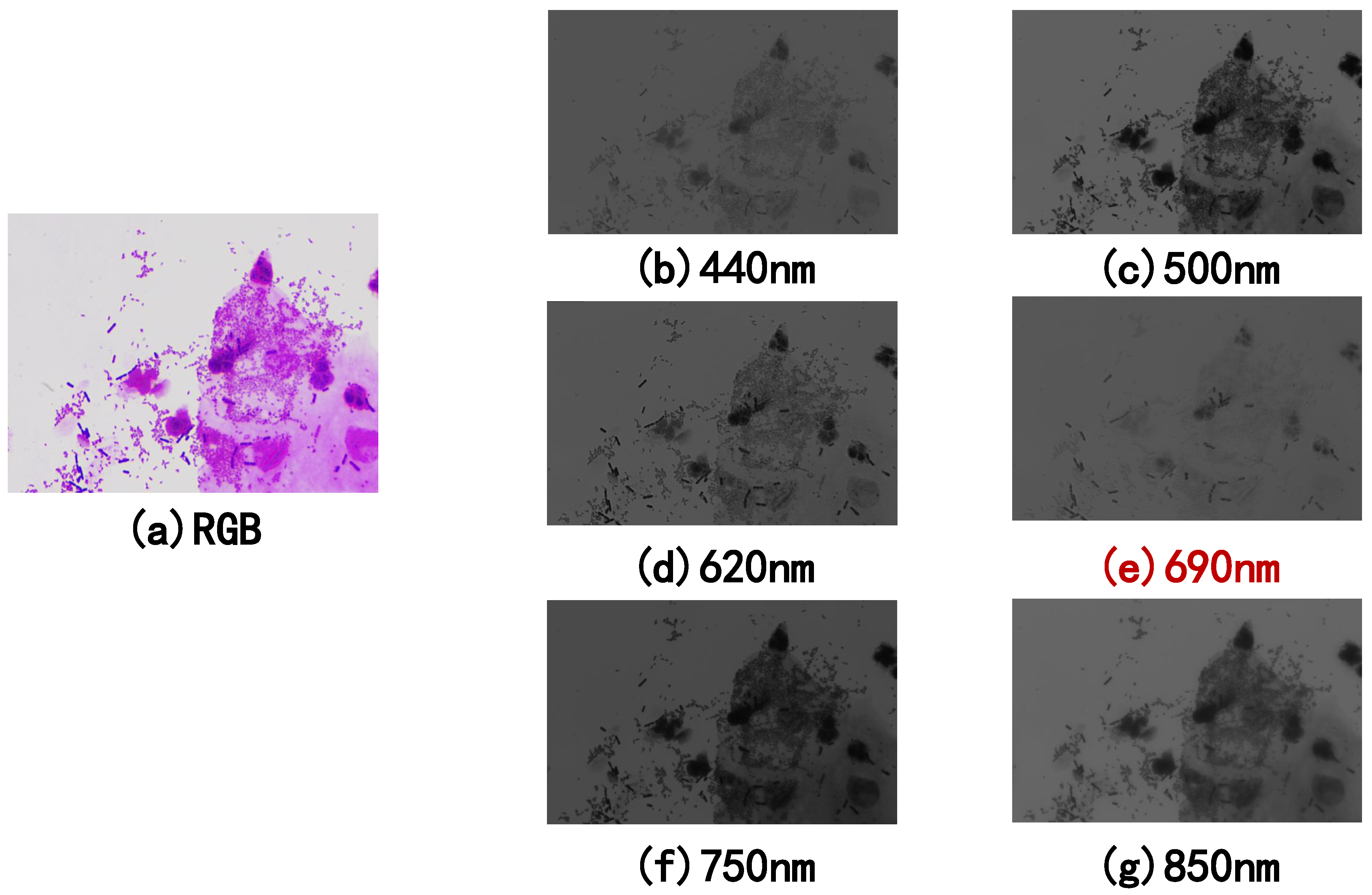
5.3.3. Spectrum Sensitivity
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- China Health Statistical Year Book; Technical Report; Peking Union Medical College Press: Beijing, China, 2020; Available online: https://www.yearbookchina.com/navibooklist-n3020013080-1.html (accessed on 14 December 2021).
- Shen, Y.; Bao, J.; Tang, J.; Liao, Y.; Liu, Z.; Liang, Y. Relationship between vaginitis, HPV infection and cervical cancer. Chin. J. Women Child. Health 2019, 10, 3. [Google Scholar]
- Meng, L.; Xue, Y.; Yue, T.; Yang, L.; Gao, L.; An, R. Relationship of HPV infection and BV, VVC, TV: A clinical study based on 1261 cases of gynecologic outpatients. Chin. J. Obstet. Gynecol. 2016, 51, 730–733. [Google Scholar]
- Yang, X.; Luo, W.; Xing, L.; Liu, D.; He, R. Research progress on the relationship between candidal vaginitis during pregnancy and adverse pregnancy outcome. Chin. J. Mycol. 2019, 14, 3. [Google Scholar]
- Ding, H.; Cao, D. Analysis of related factors of misdiagnosis of vaginitis secretion under microscope. Chin. J. Clin. Lab. Sci. 2007, 25, 89. [Google Scholar]
- Zhu, A. Analysis of related factors affecting the results of vaginal secretion examination. Guide China Med. 2009, 7, 2. [Google Scholar]
- Li, Y.; Feng, W.; Wang, P.; Xu, Z.; Bai, Q.; Liu, C. Analysis of related factors of misdiagnosis of bacterial vaginosis secretions. J. Aerosp. Med. 2014, 25, 2. [Google Scholar]
- Song, Y.; Lei, B.; He, L.; Zeng, Z.; Zhou, Y.; Ni, D.; Chen, S.; Wang, T. Automatic Detection of Vaginal Bacteria Based on Superpixel and Support Vector Machine. Chin. J. Biomed. Eng. 2015, 34, 8. [Google Scholar]
- Guo, R. Pattern Recognition Research of Microscope Wet Leucorrhea Image Based on CNN-SVM. Master’s Thesis, South China University of Technology, Guangzhou, China, 2016. [Google Scholar]
- Zhang, L. The Classification Research on Microscopic Leucocyte Image. Master’s Thesis, Harbin Engineering University, Harbin, China, 2008. [Google Scholar]
- Qin, F.; Gao, N.; Peng, Y.; Wu, Z.; Shen, S.; Grudtsin, A. Fine-grained leukocyte classification with deep residual learning for microscopic images. Comput. Methods Programs Biomed. 2018, 162, 243–252. [Google Scholar] [CrossRef]
- Yan, S. Research on the Effect of Generative Adversarial Network on Improving the Accuracy of Leucorrhea Microscopic Image Detection. Master’s Thesis, East China Normal University, Shanghai, China, 2019. [Google Scholar]
- Wang, Z.; Zhang, L.; Zhao, M.; Wang, Y.; Bai, H.; Wang, Y.; Rui, C.; Fan, C.; Li, J.; Li, N.; et al. Deep neural networks offer morphologic classification and diagnosis of bacterial vaginosis. J. Clin. Microbiol. 2020, 59, e02236-20. [Google Scholar] [CrossRef]
- Guo, Y.; Ma, L.; Li, J. Detection and Statistical Analysis of Lactobacillus in Gynecological Medical Micrographs. J. Data Acquis. Process. 2015, 30, 8. [Google Scholar]
- Ma, L. Components Statistics and Analysis Based on Texture Analysis in Gynecologic Microscopic Image. Master’s Thesis, University of Jinan, Jinan, China, 2016. [Google Scholar]
- Yan, J.; Chen, H.; Liu, L. Overview of hyperspectral image classification. Opt. Precis. Eng. 2019, 27, 3. [Google Scholar]
- Liu, W.; Wang, L.; Liu, J.; Yuan, J.; Chen, J.; Wu, H.; Xiang, Q.; Yang, G.; Li, Y. A comparative performance analysis of multispectral and rgb imaging on her2 status evaluation for the prediction of breast cancer prognosis. Transl. Oncol. 2016, 9, 521–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.L.; Wang, L.W.; Chen, J.M.; Yuan, J.P.; Xiang, Q.M.; Yang, G.F.; Qu, A.P.; Liu, J.; Li, Y. Application of multispectral imaging in quantitative immunohistochemistry study of breast cancer: A comparative study. Tumor Biol. 2016, 37, 5013–5024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Comparative performance analysis of stained histopathology specimens using RGB and multispectral imaging. In Medical Imaging 2011: Computer-Aided Diagnosis; International Society for Optics and Photonics: Lake Buena Vista (Orlando), FL, USA, 2011; Volume 7963, p. 79633B. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Maghdid, H.S.; Asaad, A.T.; Ghafoor, K.Z.; Sadiq, A.S.; Mirjalili, S.; Khan, M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. In Multimodal Image Exploitation and Learning 2021; International Society for Optics and Photonics: Bellingham, WA, USA, 2021; Volume 11734, p. 117340E. [Google Scholar]
- Liu, W.; Cheng, Y.; Liu, Z.; Liu, C.; Cattell, R.; Xie, X.; Wang, Y.; Yang, X.; Ye, W.; Liang, C.; et al. Preoperative prediction of Ki-67 status in breast cancer with multiparametric MRI using transfer learning. Acad. Radiol. 2021, 28, e44–e53. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Wahab, N.; Khan, A.; Lee, Y.S. Transfer learning based deep CNN for segmentation and detection of mitoses in breast cancer histopathological images. Microscopy 2019, 68, 216–233. [Google Scholar] [CrossRef] [Green Version]
- Hall, D.; Llinas, J. Multisensor Data Fusion; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Zhang, H.; Cheng, C.; Xu, Z.; Li, J. Survey of data fusion based on deep learning. Comput. Eng. Appl. 2020, 56, 11. [Google Scholar]
- Meng, T.; Jing, X.; Yan, Z.; Pedrycz, W. A survey on machine learning for data fusion. Inf. Fusion 2020, 57, 115–129. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Cheng, J.; Chen, X.; Wang, Z.J. A multi-scale data fusion framework for bone age assessment with convolutional neural networks. Comput. Biol. Med. 2019, 108, 161–173. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, H.; Wen, J.; Li, S.; Liu, Q. A deep learning-based recognition method for degradation monitoring of ball screw with multi-sensor data fusion. Microelectron. Reliab. 2017, 75, 215–222. [Google Scholar] [CrossRef]
- Fu, H.; Ma, H.; Wang, G.; Zhang, X.; Zhang, Y. MCFF-CNN: Multiscale comprehensive feature fusion convolutional neural network for vehicle color recognition based on residual learning. Neurocomputing 2020, 395, 178–187. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef] [Green Version]

| Method Category | Year | References | Approach | Object | Results |
|---|---|---|---|---|---|
| SVM | 2015 | [8] | superpixel and SVM | vaginal bacteria | Accuracy: 89.27% |
| 2016 | [9] | CNN and SVM | candida | Recall: 72% | |
| deep learning | 2008 | [10] | BP neural network | white blood cells (5 types) | Accuracy: 83.7% |
| 2018 | [11] | deep residual learning theory | white blood cells (40 types) | Accuracy: 76.84% | |
| 2019 | [12] | Faster R-CNN | white blood cells (8 types) | Precision:69.94% Recall: 74.15% mAP: 61.74% | |
| 2020 | [13] | CNN | bacterial vaginosis (3 types) | Accuracy: 75.1% | |
| laws texture energy | 2015 | [14] | laws texture energy and threshold segmentation | lactobacilli | Accuracy: 94.2% |
| 2016 | [15] | CNN and SVM | cue cells, epithelial cells | Accuracy: 90.07% |
| Components | Patch Size/Stride or Remarks | Input Size |
|---|---|---|
| conv | 3 × 3/2 | 299 × 299 × 3 |
| conv | 3 × 3/1 | 149 × 149 × 32 |
| conv padded | 3 × 3/1 | 147 × 147 × 32 |
| pool | 3 × 3/2 | 147 × 147 × 64 |
| conv | 3 × 3/1 | 73 × 73 × 64 |
| conv | 3 × 3/2 | 71 × 71 × 80 |
| conv | 3 × 3/1 | 35 × 35 × 192 |
| 3 × Inception | As in Figure 1a | 35 × 35 × 288 |
| 5 × Inception | As in Figure 1b | 17 × 17 × 768 |
| 2 × Inception | As in Figure 1c | 8 × 8 × 1280 |
| pool | 8 × 8 | 8 × 8 × 2048 |
| linear | logits | 1 × 1 × 2048 |
| softmax | classifier | 1 × 1 × 1000 |
| Label_Index | Label_Name | Image_Count | Percentage |
|---|---|---|---|
| 0 | normal flora | 228,275 | 53.47% |
| 1 | AV | 48,075 | 11.26% |
| 2 | BV | 25,600 | 6.00% |
| 3 | VVC | 30,300 | 7.10% |
| 4 | flora inhibition | 26,950 | 6.31% |
| 5 | BV + AV | 29,950 | 7.02% |
| 6 | BV middle | 11,775 | 2.76% |
| 7 | BV middle + VVC + AV | 5450 | 1.28% |
| 8 | BV middle + VVC | 7275 | 1.70% |
| 9 | AV + TV | 3750 | 0.88% |
| 10 | AFCC | 9500 | 2.23% |
| all | sum | 426,900 | 100.00% |
| Data_Type | Accuracy | Precision | Recall | F1-Score | Kappa |
|---|---|---|---|---|---|
| RGB image | 76.04% | 77.36% | 48.35% | 53.69% | 61.07% |
| multi-spectral image (ours) | 87.43% | 93.18% | 75.60% | 81.66% | 80.42% |
| Model | Data_Type | Accuracy | Precision | Recall | F1-Score | Kappa |
|---|---|---|---|---|---|---|
| VGG16 | RGB image | 61.99% | 42.51% | 32.78% | 30.30% | 38.82% |
| multi-spectral image | 66.56% | 77.49% | 58.14% | 62.89% | 50.97% | |
| ResNet50 | RGB image | 72.68% | 59.98% | 46.77% | 49.12% | 56.14% |
| multi-spectral image | 77.28% | 84.72% | 71.18% | 72.56% | 64.97% | |
| Inception v3 | RGB image | 68.90% | 52.49% | 41.86% | 42.36% | 49.48% |
| multi-spectral image (fewer epochs) | 84.65% | 85.97% | 85.25% | 83.11% | 78.30% |
| Fusion Type | Accuracy | Precision | Recall | F1-Score | Kappa |
|---|---|---|---|---|---|
| data layer fusion | 77.46% | 80.23% | 53.78% | 59.57% | 63.56% |
| decision layer fusion | 79.30% | 89.32% | 55.49% | 62.67% | 65.95% |
| feature layer fusion (ours) | 87.43% | 93.18% | 75.60% | 81.66% | 80.42% |
| Model | Fusion Type | Accuracy | Precision | Recall | F1-Score | Kappa |
|---|---|---|---|---|---|---|
| VGG16 | data layer fusion | 52.41% | 6.49% | 9.61% | 7.36% | 1.22% |
| decision layer fusion | 53.52% | 11.18% | 9.16% | 6.47% | 0.21% | |
| feature layer fusion | 66.56% | 77.49% | 58.14% | 62.89% | 50.97% | |
| ResNet50 | data layer fusion | 53.51% | 8.65% | 9.15% | 6.45% | 0.16% |
| decision layer fusion | 55.16% | 18.50% | 11.46% | 10.29% | 6.03% | |
| feature layer fusion | 77.28% | 84.72% | 71.18% | 72.56% | 64.97% | |
| Inception v3 | data layer fusion | 52.90% | 16.66% | 17.39% | 14.77% | 20.78% |
| decision layer fusion | 57.19% | 41.70% | 17.46% | 19.04% | 11.65% | |
| feature layer fusion(fewer epochs) | 84.65% | 85.97% | 85.25% | 83.11% | 78.30% |
| Label_Name | Most Sensitive Spectrum | Single-Spectrum Precision | RGB Precision | Improve |
|---|---|---|---|---|
| accuracy | 600 nm | 78.57% | 76.04% | 2.53% |
| normal flora | 640 nm | 78.90% | 77.10% | 1.80% |
| AV | 540 nm | 73.60% | 64.90% | 8.70% |
| BV | 850 nm | 87.50% | 82.90% | 4.60% |
| VVC | 580 nm | 95.70% | 85.60% | 10.10% |
| BV-AV | 810 nm | 84.70% | 75.40% | 9.30% |
| BV middle | 850 nm | 79.20% | 62.30% | 16.90% |
| BV middle + VVC + AV | 580 nm | 95.80% | 84.40% | 11.40% |
| BV middle + VVC | 690 nm | 92.30% | 60.40% | 31.90% |
| AV-TV | 480 nm | 100.00% | 84.80% | 15.20% |
| AFCC | 480 nm | 100.00% | 90.00% | 10.00% |
| Label_Name | Most Sensitive Spectrum | Single-Spectrum Precision | RGB Precision | Improve |
|---|---|---|---|---|
| AV-TV | 670 nm | 96.22% | 89.56% | 6.67% |
| VVC | 750 nm | 80.21% | 76.07% | 4.13% |
| BV middle | 640 nm | 98.09% | 95.27% | 2.83% |
| AFCC | 580 nm | 88.16% | 85.44% | 2.72% |
| flora inhibition | 650 nm | 92.41% | 90.09% | 2.32% |
| BV-AV | 810 nm | 97.23% | 95.12% | 2.11% |
| BV middle + VVC | 440 nm | 94.63% | 92.97% | 1.66% |
| BV middle + VVC + AV | 730 nm | 97.86% | 96.26% | 1.60% |
| AV | 540 nm | 89.19% | 87.98% | 1.20% |
| BV | 460 nm | 99.46% | 99.09% | 0.37% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Gao, P.; Liu, S.; Wang, Y.; Li, G.; Wang, Y. A Vaginitis Classification Method Based on Multi-Spectral Image Feature Fusion. Sensors 2022, 22, 1132. https://doi.org/10.3390/s22031132
Zhao K, Gao P, Liu S, Wang Y, Li G, Wang Y. A Vaginitis Classification Method Based on Multi-Spectral Image Feature Fusion. Sensors. 2022; 22(3):1132. https://doi.org/10.3390/s22031132
Chicago/Turabian StyleZhao, Kongya, Peng Gao, Sunxiangyu Liu, Ying Wang, Guitao Li, and Youzheng Wang. 2022. "A Vaginitis Classification Method Based on Multi-Spectral Image Feature Fusion" Sensors 22, no. 3: 1132. https://doi.org/10.3390/s22031132
APA StyleZhao, K., Gao, P., Liu, S., Wang, Y., Li, G., & Wang, Y. (2022). A Vaginitis Classification Method Based on Multi-Spectral Image Feature Fusion. Sensors, 22(3), 1132. https://doi.org/10.3390/s22031132






